Abstract
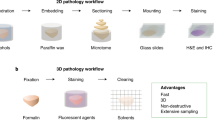
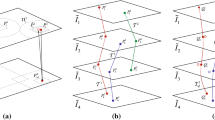
3-D histology has become an attractive technique providing insights into morphology of histologic specimens. However, existing techniques in generating 3-D views from a stack of whole slide images are scarce or suffer from poor co-registration performance when displaying diagnostically important areas at sub-cellular resolution. Our team developed a new scale-invariant feature transform (SIFT)-based workflow to co-register histology images and facilitate 3-D visualization of micro-structures important in histopathology of lung adenocarcinoma. The co-registration accuracy and visualization capacity of the workflow were tested by digitally perturbing the staining coloration seven times. The perturbation slightly affected the co-registration but overall the co-registration errors remained very small when compared to those published to date. The workflow yielded accurate visualizations of expert-selected regions permitting confident 3-D evaluation of the clusters. Our workflow could support the evaluation of histologically complex tumors such as lung adenocarcinomas that are currently routinely viewed by pathologists in 2-D on slides, but could benefit from 3-D visualization.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Arganda-Carreras, I., Sorzano, C.O.S., Marabini, R., Carazo, J.M., Ortiz-de Solorzano, C., Kybic, J.: Consistent and elastic registration of histological sections using vector-spline regularization. In: Beichel, R.R., Sonka, M. (eds.) Computer Vision Approaches to Medical Image Analysis, pp. 85–95. Springer, Berlin (2006). https://doi.org/10.1007/11889762_8
Falk, M., Ynnerman, A., Treanor, D., Lundström, C.: Interactive visualization of 3D histopathology in native resolution. IEEE Trans. Vis. Comput. Graph. 25(1), 1008–1017 (2019). https://doi.org/10.1109/TVCG.2018.2864816
Farahani, N., Braun, A., Jutt, D., Huffman, T., Reder, N., Liu, Z., Yagi, Y., Pantanowitz, L.: Three-dimensional imaging and scanning: current and future applications for pathology. J. Pathol. Inf. 8(1), 36 (2017). https://doi.org/10.4103/jpi.jpi_32_17
Gertych, A., Swiderska-Chadaj, Z., Ma, Z., Ing, N., Markiewicz, T., Cierniak, S., Salemi, H., Guzman, S., Walts, A.E., Knudsen, B.S.: Convolutional neural networks can accurately distinguish four histologic growth patterns of lung adenocarcinoma in digital slides. Sci. Rep. 9(1) (2019). https://doi.org/10.1038/s41598-018-37638-9
Gibson, E., Gaed, M., Gómez, J., Moussa, M., Pautler, S., Chin, J., Crukley, C., Bauman, G., Fenster, A., Ward, A.: 3D prostate histology image reconstruction: quantifying the impact of tissue deformation and histology section location. J. Pathol. Inf. 4(1), 31 (2013)
Kadota, K., Nitadori, J., Sima, C.S., Ujiie, H., Rizk, N.P., Jones, D.R., Adusumilli, P.S., Travis, W.D.: Tumor spread through air spaces is an important pattern of invasion and impacts the frequency and location of recurrences after limited resection for small stage 1 lung adenocarcinomas. J. Thoracic Oncol. 10(5), 806–814 (2015). https://doi.org/10.1097/JTO.0000000000000486
Kartasalo, K., Latonen, L., Vihinen, J., Visakorpi, T., Nykter, M., Ruusuvuori, P.: Comparative analysis of tissue reconstruction algorithms for 3D histology. Bioinformatics 34(17), 3013–3021 (2018). https://doi.org/10.1093/bioinformatics/bty210
Lowe, D.G.: Object recognition from local scale-invariant features. In: Proceedings of the International Conference on Computer Vision, ICCV ’99, vol. 2, p. 1150. IEEE Computer Society, USA (1999)
Ma, Z., Shiao, S.L., Yoshida, E.J., Swartwood, S., Huang, F., Doche, M.E., Chung, A.P., Knudsen, B.S., Gertych, A.: Data integration from pathology slides for quantitative imaging of multiple cell types within the tumor immune cell infiltrate. Diagn. Pathol. 12(1) (2017). https://doi.org/10.1186/s13000-017-0658-8
Magee, D., Song, Y., Gilbert, S., Roberts, N., Wijayathunga, N., Wilcox, R., Bulpitt, A., Treanor, D.: Histopathology in 3D: from three-dimensional reconstruction to multi-stain and multi-modal analysis. J. Pathol. Inf. 6(1), 6 (2015). https://doi.org/10.4103/2153-3539.151890
Morales-Oyarvide, V., Mino-Kenudson, M.: Taking the measure of lung adenocarcinoma: towards a quantitative approach to tumor spread through air spaces (STAS). J. Thorac. Dis. 9(9) (2017). https://doi.org/10.21037/jtd.2017.07.97
Onozato, M., Klepeis, V., Yagi, Y., Mino-Kenudson, M.: A role of three-dimensional (3D)-reconstruction in the classification of lung adenocarcinoma. Anal. Cell. Pathol. 35(2), 79–84 (2012). https://doi.org/10.3233/ACP-2011-0030
Onozato, M.L., Hammond, S., Merren, M., Yagi, Y.: Evaluation of a completely automated tissue-sectioning machine for paraffin blocks. J. Clin. Pathol. 66(2), 151–154 (2013). https://doi.org/10.1136/jclinpath-2011-200205
Pantanowitz, L., Valenstein, P., Evans, A., Kaplan, K., Pfeifer, J., Wilbur, D., Collins, L., Colgan, T.: Review of the current state of whole slide imaging in pathology. J. Pathol. Inf. 2(1), 36 (2011). https://doi.org/10.4103/2153-3539.83746
Pichat, J., Iglesias, J.E., Yousry, T., Ourselin, S., Modat, M.: A survey of methods for 3D histology reconstruction. Med. Image Anal. 46, 73–105 (2018). https://doi.org/10.1016/j.media.2018.02.004
Reinhard, E., Adhikhmin, M., Gooch, B., Shirley, P.: Color transfer between images. IEEE Comput. Grap. Appl. 21(5), 34–41 (2001). https://doi.org/10.1109/38.946629
Roberts, N., Magee, D., Song, Y., Brabazon, K., Shires, M., Crellin, D., Orsi, N.M., Quirke, R., Quirke, P., Treanor, D.: Toward routine use of 3D histopathology as a research tool. Am. J. Pathol. 180(5), 1835–1842 (2012). https://doi.org/10.1016/j.ajpath.2012.01.033
Ruifrok, A.C., Johnston, D.A.: Quantification of histochemical staining by color deconvolution. Anal. Quant. Cytol. Histol. 23(4), 291–299 (2001)
Song, Y., Treanor, D., Bulpitt, A., Magee, D.: 3D reconstruction of multiple stained histology images. J. Pathol. Inf. 4(2), 7 (2013). https://doi.org/10.4103/2153-3539.109864
Song, Y., Treanor, D., Bulpitt, A.J., Wijayathunga, N., Roberts, N., Wilcox, R., Magee, D.R.: Unsupervised content classification based nonrigid registration of differently stained histology images. IEEE Trans. Biomed. Eng. 61(1), 96–108 (2014). https://doi.org/10.1109/TBME.2013.2277777
Travis, W.D., Brambilla, E., Nicholson, A.G., Yatabe, Y., Austin, J.H., Beasley, M.B., Chirieac, L.R., Dacic, S., Duhig, E., Flieder, D.B., Geisinger, K., Hirsch, F.R., Ishikawa, Y., Kerr, K.M., Noguchi, M., Pelosi, G., Powell, C.A., Tsao, M.S., Wistuba, I.: The 2015 world health organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J. Thor. Oncol. 10(9), 1243–1260 (2015). https://doi.org/10.1097/JTO.0000000000000630
Walts, A.E., Marchevsky, A.M.: Current evidence does not warrant frozen section evaluation for the presence of tumor spread through alveolar spaces. Arch. Pathol. Lab. Med. 142(1), 59–63 (2018). https://doi.org/10.5858/arpa.2016-0635-OA
Warth, A.: Spread through air spaces (STAS): a comprehensive update. Transl. Lung Cancer Res. 6(5), 501 (2017)
Xu, Y., Pickering, J.G., Nong, Z., Ward, A.D.: 3D morphological measurement of whole slide histological vasculature reconstructions. In: Gurcan, M.N., Madabhushi, A. (eds.) Medical Imaging 2016: Digital Pathology, vol. 9791, pp. 181–187. International Society for Optics and Photonics, SPIE (2016). https://doi.org/10.1117/12.2214871
Yagi, Y., Aly, R.G., Tabata, K., Barlas, A., Rekhtman, N., Eguchi, T., Montecalvo, J., Hameed, M., Manova-Todorova, K., Adusumilli, P.S., Travis, W.D.: Three-dimensional histologic, immunohistochemical and multiplex immunofluorescence analysis of dynamic vessel co-option of spread through air spaces (STAS) in lung adenocarcinoma. J. Thor. Oncol. (2019). https://doi.org/10.1016/j.jtho.2019.12.112
Yagi, Y., Tabata, K., Rekhtman, N., Eguchi, T., Fu, X., Montecalvo, J., Adusumilli, P., Hameed, M., Travis, W.: Three-dimensional assessment of spread through air spaces in lung adenocarcinoma: insights and implications. J. Thor. Oncol. 12(11), S1797 (2017)
Acknowledgements
This work has been supported by in part by the Precision Health Grant at C-S and seed grants from the Department of Surgery at Cedars-Sinai Medical Center. The authors would like to thank Dr. Mari Mino-Kenudson from the Massachusetts General Hospital for her help in data preparation and input on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 The Editor(s) (if applicable) and The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Pyciński, B., Yagi, Y., Walts, A.E., Gertych, A. (2021). 3-D Tissue Image Reconstruction from Digitized Serial Histologic Sections to Visualize Small Tumor Nests in Lung Adenocarcinomas. In: Pietka, E., Badura, P., Kawa, J., Wieclawek, W. (eds) Information Technology in Biomedicine. Advances in Intelligent Systems and Computing, vol 1186. Springer, Cham. https://doi.org/10.1007/978-3-030-49666-1_5
Download citation
DOI: https://doi.org/10.1007/978-3-030-49666-1_5
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-49665-4
Online ISBN: 978-3-030-49666-1
eBook Packages: Intelligent Technologies and RoboticsIntelligent Technologies and Robotics (R0)