Abstract
Automated Diagnosis in healthcare is becoming an interesting study in recent time among the data scientists to predict and diagnose the conditions in patients. In this manner, analysis of plays a major part in detection and classification of disease and accurately diagnose the medical condition in patients. Most of the data mining task is held up with poor classification accuracy due to the presence of redundant or irrelevant data items. In this research, the issue of poor classification accuracy is addressed and is solved by developing a framework that involves a series of stages. This includes pre-processing, feature extraction and classification of data items. The study uses Optimal Feature Selection Method (OFSM) as its feature selection tool and Multi-Support Vector Machine as its classification tool. The experimental validation is carried out to study the efficacy of the proposed method over various datasets and the outputs are evaluated in terms of accuracy, specificity, sensitivity and f-measure.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Biomedical computing offers a significant role in the field of healthcare and this includes several theoretical models for the diagnosis of automatic diagnostic frameworks. In healthcare systems, there exist various problems on automated diagnostic framework that includes its cost, multiple alternative therapies, inadequate diagnostic information etc. [1]. Hence the researchers used various techniques on classification to assists the physicians to facilitate and check medical diagnosis and to reduce the vulnerability of diseases [2]. The majority of the classification model operates by optimal selection of features and such selects boost the classification process by securing the primitive information [3, 4].
In order to determine the type of disease and to assist the surgical notions, it is necessary to optimally classify the type and nature of disease [5]. Conventional machine learning classifiers are employed to classify the attributes on the basis of the characteristic weight to diagnose the problems [6]. The decision tree is the first classifier used in medical history for the diagnosis of diseases [7]. Machine Learning (ML) is also applied as a supervised tool for the classification of type of disease [9]. Among the classification tools, the SVM is the best-known supervised classification model which maps the sample vector into a space onto high dimensions and searches for an ideal isolation hyperplane to classify the type. The support vector machine acts as a pattern identification technique that is used commonly as machine learning technique for various disease classification [8] [10]. Network-based SVM (NSVM) is an enhancement model that overcomes the limitations of the classification of SVM and provides effective class of the classified samples [11–13].
It is impossible to analyze the data with redundant and incomplete items because of the existence of huge volume of data in real time [23]. Feature Selection (FS) is a most appropriate technique that removes the replicated and undesired features and provides the most insightful final data variables, leading to successful prediction or classification. The objective is to minimize the amount of features in order to minimize the area size directly and allow classification models to use only the necessary features. This is regarded as an optimization problem and it is fixed using support vector machine [23, 24]. SVM acts as a generalized algorithm in making decisions to diagnose type of disease. It avoids the problems associated with the high-dimensional features with overfitting issue and it further reduces the cost of computation.
The major purpose of this research is discussed in the subsequent sections: In this paper, a framework that consists of a pre-processing, feature extraction using optimal feature selection model (OFSM) and classification of data items using multi-support vector machine is developed.
2 Related Works
The MRI classification of coded brain structures has primarily been tested for patient monitoring and examination of different brain conditions. In an audit of the best programmed methods accessible from the brain structure to the complete brain percolation techniques, the research by Villa et al. [16] showed specific segmentation strategies. No single programmed segmentation model has been developed that will enhance normal clinical work on a precise characterization of the brain structures. The next step should concentrate primarily on the integration of multi- strategies with methodologies that are focused on learning. In text mining, the ML system is used to investigate doctor studies focused on cancer, which have been partly studied.
In enhancing data extraction out of cancer data, Napolitano et al. [17] suggested some rules for pre-trained breast cancer reports. The general objective of Molina et al. [18], in view of the revelation of repeated trends is to distinguish time schemes. First, the numerical time arrangements have been transformed into standard time groups in which the images are supposed to characterize the respective ideas in space. These images may be characterized using either the open or expert domain information.
Tosas et al. [19] presented another worldview of the creation of an activity of clinical classification from different data sources to anticipate a recovery of patients. Various classification strategies embrace a metric of each unit which must be usable during prediction.
In the present scenario, chronic data was graded as ordinary, fringe or obsessed in terms of clinical suggestions for disease according to De Bruin et al. [20]. The classification results often test the built-in fuzzy sets where instructions are used as rules for classification of data from time to time.
The ML models for the detection of CKD are considered by Salekins and Stankovic [21] to have 24 prediction parameters. The study focuses on the cost-accuracy trade-off in order to identify the most accurate and least expensive reliable CKD prediction model.
Yildirim et al. [22] investigate the effect of data imbalance while developing the CKD model. The tests showed that the sampling models would increase the classification performance, and a significant variable affects the multi-computer perception.
Elhoseny, M., et al. [26] proposed the Density based Feature Selection using ant colony optimization algorithm for CKD as an intelligent classification method. Before the ACO-based classifier construction, the given intelligent system uses DFS to remove invalid or redundant features.
Compared to other algorithms while classifying the different stages for patients with CKD, Rady, E. H. A., and Anwar, A. S. [27] found that the Probabilistic Neural Networks (PNN) algorithm provides the overall maximum classifier accuracy of 96.7%. But the Multilayer Perceptron requires a short execution period, while the PNN takes 12 s to complete the study. The above algorithms were correlated to classifier accuracy depending on the number of correctly identified phases for patients with CKD, the time it took to construct the model as well the time it took for testing it.
Subasi et al. [28] used a variety of ML classifiers that were experimentally tested on the data set from UCI Repository and the results were compared to those published in latest literature. The results are presented quantitatively and qualitatively and the observations show that the random forest classifier performs near-optimally in identifying CKD patients.
Random forest also was used by Qin, J., et al. [29] to obtain the highest results, with 99.75% diagnostic precision. It was suggested an optimized model which incorporates random forest and logistic regression with perceptron, that can averagely attain a 99.83% accuracy after simulating ten times, after evaluating the misclassified instances produced by the existing models.
The SVM algorithm was used to detect CKD by Polat, H., et al. [30]. To minimize the dimension of the CKD dataset, two basic forms of feature selection strategies namely wrapper and filter approaches were chosen to diagnose the disease. Classifier subset evaluator with greedy stepwise search engine and wrapper subset evaluator with Best First search engine were used in the wrapper method.
3 Proposed Method
Data Classification is another method which involves different information sorting techniques and conditions within a storage capacity or a database. This is done essentially using a database or a business knowledge program which enables information to be examined, recognized and isolated.
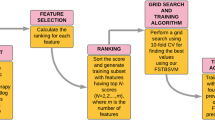
With the aid of an optimal method of extraction, the existing medical classification technique used hybrid classifier. The efficiency is improved and time complexity issues are minimized significantly by the modern classification model. The presented model applies OFSM (Optimal Feature Selection Method) with M-SVM to address these daunting challenges. In this case, the input dataset is preprocessed. Feature selection is then carried out with OFSM from the pre-processed results. Finally, the classification task is conducted using the M-SVM system for clinical data. Figure 1 shows the overall classification process.
Preprocessing
The unprocessed medical input data is provided as input for preprocessing. These unprocessed results tend to be very noisy, with no values and inconsistencies. Classification accuracy is influenced by the dominance of these unprocessed results. Unprocessed data can be preprocessed to improve the standard of patient data.
Preprocessing in this article is more important in order to obtain data from non-numerical information in the context of a mathematical structure. For additional information, non-numerical data is collected and the arithmetic data collection is retrieved.
The pre-processing removes missing values from the input dataset. The discrete traits are built by synchronizing the models if continuous features tends to exist. The pre-processing further aims to reduce the presence of missing and noisy values in each instances using data cleaning operation. It further avoids the inconsistencies in the input dataset. The missing values are replaced with the value 0 and it gets transformed to make the data suitable for the process of mining. The values are further normalized via coding in data transformation process and hence the values take the binary form 0 or 1.
Dataset Description
Three datasets from medical domain are taken for experimentation. Dataset for Heart disease, CKD dataset and Hepatitis dataset are used in this proposed work. All these datasets are available in the UCI machine learning repository. The Cleveland Heart disease dataset has 76 attributes and 303 instances. But all the published experiments concentrate only on 14 attributes. The CKD dataset consists of 24 attributes plus a class attribute that finds out the presence or not of CKD. 400 samples of data are given in CKD dataset. The hepatitis dataset contains 19 attributes and 155 instances.
Feature Selection Using Differential Evolution Optimization
One type of evolutionary algorithmic ways is Differential evolution (DE) in which feature exploration is based on ant colony optimization. DE features various actions, including 1) the capacity to manage multimodal, nonlinear and non-differentiable values, 2) parallelization that deals with high computational costs, 3) ease of use, and 4) good convergence properties. DE, like Genetic Algorithm, makes use of elements such as mutation, crossover, and selection. Performance of DE is subject to how the target vector and difference are handled during the searching operation in order to obtain a task vector. DE is a population-based metaheuristic technique for solving optimization problems using numerical vectors.
Feature Selection Using Optimal Feature Selection Method
Instead of mutation and crossover, Optimal Feature Selection Method (OFSM), encodes solutions to reduce the computational time in order of optimal classification on medical datasets. The process of OFSM is as follows:
Stage 1: Encoding of Solution
Each specific solution from the population is represented as binary string. The solution length is equal to various features in the datasets. The binary code 1 indicates that the feature is selected and vice versa. Thus S = [F1,F2,…,Fm] with m being the features of different datasets.
Stage 2: Initial Population
Set total population size of the OFSM as 50, where it produces random solution varying between 0 and 1 with real values. The real valued solution is then converted to binary value using the following digitization step:
where.
Rand - uniformly distributed random number between [0,1].
Stage 3: Fitness Function
This process measures a single positive integer output. The fitness of the obtained solution is formulated as below that assists M-SVM to correctly perform instance classification that with lesser classifier error.
The solution obtained is the error rate of a classifier, which is otherwise defined as the testing error rate:
Stage 4: Finding New Solutions
Based on the fitness values, the best and worst solution is identified to produce new solution. The solution with the lowest value of the fitness stage is regarded as the best solution because it is obtained with a lesser error rate at a generation i. Here, \({s}_{wt}^{(i)}\) represents the worst solution and \({s}_{bt}^{(i)}\) represents the best solution at an iteration i. With these constraints, the qth position of an old solution \({s}_{p,q}^{(i)}\) is hence given as below:
When random numbers A and B lie in the range of 0 to 1, then digitalization process transforms real into binary values for every position of the subsequent generation i + 1 based on following equation:
Stage 5: Termination Criteria
The termination criteria is satisfied when any one of the given condition is achieved:
1) fitness rate. 2) Threshold value of the iteration process and 3) overall count of iterations.
Algorithm of OFSM.
-
1:
Start
-
2:
Encode the solution
-
3:
Generate initial populations
-
4:
Evaluate the fitness function in terms of error rate
\(fitness({S}_{p}^{\left(i\right)})\)=Error_rate \({(S}_{p}^{\left(i\right)})\).
-
5:
Find the new solutions using the fitness function
$${s}_{p,q}^{(i)}= {s}_{p,q}^{(i)}+A\left|{s}_{bt,q}^{\left(i\right)}- {s}_{p,q}^{\left(i\right)}\right|+B|{s}_{wt,q}^{\left(i\right)}-{s}_{p,q}^{(i)}$$ -
6:
Convert the solutions to binary form
$${S}_{p,q }^{(i)}\left\{\begin{array}{l}1\;\; {S}_{p,q}^{(i+1)}>rand\\ 0\;\; otherwise\end{array}\right.$$ -
7:
End
M-SVM Classification
M-SVM is a useful data classification technique. While Neural Networks are considered more user-friendly than this, they often produce unsatisfactory outcomes. In general, a classification task consists of training and evaluating results, consisting of such data instances [21]. There is one objective value and multiple characteristics in every instance of the training package. The aim of M-SVM is to generate a model which predicts the objective function of data instances provided in a test set only by the attributes [8].
Classification in M-SVM is a working example for monitoring learning. Known labels help to determine whether or not the device works properly. This knowledge leads to the right answer, validates the system's accuracy or is used to assist the system in proper action. One phase in the M-SVM classification includes identifying the known groups closely. This is called collection of functions or extraction of functionality. Even if unknown samples are not required, feature collection and M-SVM classification together will be beneficial. It can be used to classify key sets involved in any classification process [8].
A hyperplane may be used to split the data while the data is linear. But usually the data is non-linear and there are inseparable datasets. To move this kernel, the input data is mapped to a high-dimensional area in a non-linear way.
The data points are converted to a high dimensional space by the nonlinear mapping φ(x), which solves the nonlinear problem between classes xi. This helps to separate points using the mark yi in the solution space. M-SVM solves the resulting problem with the function derived from data xi of N-point and is shown as follows:
where,
ϵi - slack variables and.
c ≥ 0 – tradeoff factor.
A dual Lagrangian optimizes M-SVM and it is expressed as below:
where
K - kernel vector.
K(xi,xj) = < φ(xi), φ(xj) > with < φ(xi), φ(xj) >
Hence for a data point x, the predicted class from the text document is given as below:
At the end of the classification, undesirable data samples are extracted using M-SVM, which eliminates support vectors that are not important. The prediction limits are then graded according to consistent and most important characteristics.
For M-SVM classification with twice parameterized trigonometric kernel function [25] is expressed with dual formulation:
where 0 ≤ αi ≤ C, for all i;
When the amount of training points is great, training SVM becomes very demanding. The twice parameterized trigonometric kernel function helps in locating the solution within the best known bound that eases the process of classification within the limits and the solution tends to remain within bound. Therefore no extra-bound solutions are obtained and it reduces the iteration bound.
Transforming the data to a function space enables a similarity measure to be defined on the basis of the dot product. Pattern recognition can be easy [1] if the function space is selected appropriately.
The definition of the kernel function is not the high-dimensional feature space, but allows for the application of the input region. Therefore, it is not necessary to determine the inner product in the function space. The algorithm should map the input field attributes to the function space. In SVM and its output, the kernel is important. The Kernel Hilbert Spaces is reproduced.
The kernel then represents a valid internal product. With an entry space, the training set cannot be separated linearly. In the functional space, the training set is linearly separable. The kernel trick is referred to as [8, 12].
4 Results and Discussions
In this section, a simulation of the proposed model on different datasets collected from UCI repository that includes heart disease, CKD and hepatitis is provided. The proposed algorithm is simulated in a high end computing system running on i7 processor with 8 GB RAM. The performance of the recommended model is tested under classification accuracy, specificity, sensitivity and f-measure.
Table 1 presents the comparative performance results of heart disease dataset using different classifiers and two feature selection methods.
Figure 2 presents the comparison of classifiers and feature selection strategies for the heart disease data set with respect to accuracy metric. Other classifiers like LR, RF, NB and SVM have accuracy values of 90.71%, 92.76%, 93.93%, and 95.47%, respectively, while the proposed OFSM + M-SVM based feature selection technique gives the best accuracy value of 96.47% (Refer Table 1).
Table 2 presents the comparative performance results of CKD dataset using different classifiers and two feature selection methods.
Figure 3 presents the comparison of classifiers and feature selection strategies for the CKD data set with respect to accuracy metric. Other classifiers like RF, GBT, ANN, and SVM have accuracy values of 91.64%, 93.27%, 93.72%, and 94.51%, respectively, while the proposed OFSM + M-SVM based feature selection technique gives the best accuracy value of 97.14% (Refer Table 2).
Table 3 presents the comparative performance results of Hepatitis disease dataset using different classifiers and two feature selection methods.
Figure 4 presents the comparison of classifiers and feature selection strategies for the Hepatitis disease data set with respect to accuracy metric. Other classifiers like LR, RF, NB and SVM have accuracy values of 93.57%, 92.63%, 95.15%, and 90.43%, respectively, while the proposed OFSM + M-SVM based feature selection technique gives the best accuracy value of 96.61% (Refer Table 3).
5 Conclusions
In this paper, the OFSM based M-SVM is utilized to improve the classification of disease in humans that includes heart disease, kidney disease and liver disease. The dataset following the series of stages in the proposed model including pre-processing, feature selection and classification enables improved classification of instances than existing methods. The OFSM obtains optimal features from the input dataset that improves the accuracy of classifier than existing methods. The M-SVM on other hand classifies with higher precision than other methods. The simulated outputs present that the proposed OFSM-M-SVM gives enhanced classification accuracy, specificity, sensitivity and f-measure. In future, the use of deep learning on large datasets could be applied to enhance the efficiency of OFSM and the classifier.
References
Hassan, C.A.U., Khan, M.S., Shah, M.A.: Comparison of machine learning algorithms in data classification. In: 2018 24th International Conference on Automation and Computing (ICAC), pp. 1–6. IEEE, September 2018
Kling, C.E., Perkins, J.D., Biggins, S.W., Johnson, C.K., Limaye, A.P., Sibulesky, L.: Listing practices and graft utilization of hepatitis C–positive deceased donors in liver and kidney transplant. Surgery 166(1), 102–108 (2019)
Khadidos, A., Khadidos, A.O., Kannan, S., Natarajan, Y., Mohanty, S.N., Tsaramirsis, G.: Analysis of COVID-19 Infections on a CT Image Using DeepSense Model. Frontiers in Public Health, 8 (2020)
Owada, Y., et al.: A nationwide survey of Hepatitis E virus infection and chronic hepatitis in heart and kidney transplant recipients in Japan. Transplantation 104(2), 437 (2020)
Mariappan, L.T.: Analysis on cardiovascular disease classification using machine learning framework. Solid State Technol. 63(6), 10374–10383 (2020)
Reyentovich, A., et al.: Outcomes of the Treatment with Glecaprevir/Pibrentasvir following heart transplantation utilizing hepatitis C viremic donors. Clin. Transplant. 34(9), e13989 (2020)
Raja, R.A., Kousik, N.V.: Privacy Preservation Between Privacy and Utility Using ECC-based PSO Algorithm. In Intelligent Computing and Applications, pp. 567–573. Springer, Singapore (2021). https://doi.org/10.1007/978-981-15-5566-4_51
Gidea, C.G., et al.: Increased early acute cellular rejection events in hepatitis C-positive heart transplantation. J. Heart Lung Transplant. 39(11), 1199–1207 (2020)
Ramana, B.V., Boddu, R.S.K.: Performance comparison of classification algorithms on medical datasets. In: 2019 IEEE 9th Annual Computing and Communication Workshop and Conference (CCWC), pp. 0140–0145. IEEE, January 2019
Lazo, M., et al.: Confluence of epidemics of hepatitis C, diabetes, obesity, and chronic kidney disease in the United States population. Clin. Gastroenterol. Hepatol. 15(12), 1957–1964 (2017)
Gowrishankar, J., Narmadha, T., Ramkumar, M., Yuvaraj, N.: Convolutional neural network classification on 2d craniofacial images. Int. J. Grid Distributed Comput. 13(1), 1026–1032 (2020)
Ariyamuthu, V.K., et al.: Trends in utilization of deceased donor kidneys based on hepatitis C virus status and impact of public health service labeling on discard. Transpl. Infect. Dis. 22(1), e13204 (2020)
Yuvaraj, N., Vivekanandan, P.: An efficient SVM based tumor classification with symmetry non-negative matrix factorization using gene expression data. In 2013 International Conference on Information Communication and Embedded Systems (Icices), pp. 761–768. IEEE, February 2013
Bowring, M.G., et al.: Center-level trends in utilization of HCV-exposed donors for HCV-uninfected kidney and liver transplant recipients in the United States. Am. J. Transplant. 19(8), 2329–2341 (2019)
Wasuwanich, P., et al.: Hepatitis E-Associated Hospitalizations in the United States: 2010–2015 and 2015–2017. J. Viral Hepatitis 28(4), 672–681 (2021)
González-Villà, S., Oliver, A., Valverde, S., Wang, L., Zwiggelaar, R., Lladó, X.: A review on brain structures segmentation in magnetic resonance imaging. Artif. Intell. Med. 73, 45–69 (2016)
Napolitano, G., Marshall, A., Hamilton, P., Gavin, A.T.: Machine learning classification of surgical pathology reports and chunk recognition for information extraction noise reduction. Artif. Intell. Med. 70, 77–83 (2016)
Molina, M.E., Perez, A., Valente, J.P.: Classification of auditory brainstem responses through symbolic pattern discovery. Artif. Intell. Med. 70, 12–30 (2016)
Last, M., Tosas, O., Cassarino, T.G., Kozlakidis, Z., Edgeworth, J.: Evolving classification of intensive care patients from event data. Artif. Intell. Med. 69, 22–32 (2016)
de Bruin, J.S., Adlassnig, K.P., Blacky, A., Koller, W.: Detecting borderline infection in an automated monitoring system for healthcare-associated infection using fuzzy logic. Artif. Intell. Med. 69, 33–41 (2016)
Salekin, A., Stankovic, J.: Detection of chronic kidney disease and selecting important predictive attributes. In: 2016 IEEE International Conference on Healthcare Informatics (ICHI), pp. 262–270. IEEE, October 2016
Yildirim, P.: Chronic kidney disease prediction on imbalanced data by multilayer perceptron: Chronic kidney disease prediction. In: 2017 IEEE 41st Annual Computer Software and Applications Conference (COMPSAC), vol. 2, pp. 193–198. IEEE, July 2017
Wong, G.L.H., et al.: Chronic kidney disease progression in patients with chronic hepatitis B on tenofovir, entecavir, or no treatment. Aliment. Pharmacol. Ther. 48(9), 984–992 (2018)
Kaul, D.R., Tlusty, S.M., Michaels, M.G., Limaye, A.P., Wolfe, C.R.: Donor-derived hepatitis C in the era of increasing intravenous drug use: a report of the Disease Transmission Advisory Committee. Clin. Transplant. 32(10), e13370 (2018)
Bouafia, M., Yassine, A.: An efficient twice parameterized trigonometric kernel function for linear optimization. Optim. Eng. 21(2), 651–672 (2019). https://doi.org/10.1007/s11081-019-09467-w
Elhoseny, M., Shankar, K., Uthayakumar, J.: Intelligent diagnostic prediction and classification system for chronic kidney disease. Sci. Rep. 9(1), 1–14 (2019)
Rady, E.H.A., Anwar, A.S.: Prediction of kidney disease stages using data mining algorithms. Inform. Med. Unlocked 15, 100178 (2019)
Subasi, A., Alickovic, E., Kevric, J.: Diagnosis of chronic kidney disease by using random forest. In: CMBEBIH 2017, pp. 589–594. Springer, Singapore (2017). https://doi.org/10.1007/978-981-10-4166-2_89
Qin, J., Chen, L., Liu, Y., Liu, C., Feng, C., Chen, B.: A machine learning methodology for diagnosing chronic kidney disease. IEEE Access 8, 20991–21002 (2019)
Polat, H., Mehr, H.D., Cetin, A.: Diagnosis of chronic kidney disease based on support vector machine by feature selection methods. J. Med. Syst. 41(4), 55 (2017)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Priscila, S.S., Kumar, C.S. (2022). Classification of Medical Datasets Using Optimal Feature Selection Method with Multi-support Vector Machine. In: Rajagopal, S., Faruki, P., Popat, K. (eds) Advancements in Smart Computing and Information Security. ASCIS 2022. Communications in Computer and Information Science, vol 1759. Springer, Cham. https://doi.org/10.1007/978-3-031-23092-9_18
Download citation
DOI: https://doi.org/10.1007/978-3-031-23092-9_18
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-23091-2
Online ISBN: 978-3-031-23092-9
eBook Packages: Computer ScienceComputer Science (R0)