Abstract
A Breast cancer diagnosis provides prevention and treatment to save lives or improve the life quality of patients, and a recent tool with good performance for this diagnosis is deep learning methods to process breast histology images. However, these methods are based on Convolutional Neural Networks (CNN) with a high computational cost that reduces usability. Therefore, this paper proposes an optimized CNN for breast cancer diagnosis named Lightweight CNN for Histology Image Processing (LCIP). LCIP is based on the MobileNet V2 architecture adapted with four inverted residual convolutions to find cell features. LCIP was validated with the BreakHis database, reporting an accuracy of 99.73%, the best result in the literature. Additionally, LCIP is the Histology Image Processing Deep learning method with fewer parameters than recent state-of-the-art methods. These results demonstrate that LCIP is a method that can be used as a feasible, portable, and accessible method to develop novel tools for breast cancer diagnosis.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
According to the World Health Organization (WHO), there exist 2.3 million people with breast cancer and 685,000 deaths related to this disease in 2020. Therefore, the early diagnosis is essential for patients, correct treatment, and care. The first stage of diagnosis is breast self-examination, and the second stage is the analysis with ultrasound, mammography, or magnetic resonance. The final stage is the biopsy, which is a histologic tissue sample analyzed with an expert [1].
Many deep learning methods for histology image processing have been proposed to develop novel breast cancer analysis methods. According to the literature, these methods achieve good results, and they are novel methodologies to prevent breast tumor growth [2]. Regard to histologic image processing with deep learning, different CNN architectures achieve accuracies higher than 90% like Inception-ResNet [3, 4] and Xception [5, 6]. However, these CNN have high computational costs, and they can be implemented into expensive computational platforms [7]. Therefore, this paper proposes a novel CNN with low computational cost named Lightweight CNN for Histology Image Processing (LCIP). LCIP classifies breast tissue on benign or malignant cells and is based on the MobileNet V2 architecture presented in [8] and inverted residual convolutions layers to analyze histological images with different magnifications and cell features. The architecture of LCIP brings a tool to analyze histological breast tissue with embedded machine learning systems. This tool is useful to reduce clinical costs and supports telemedicine for fast breast cancer diagnosis. According to [9, 12], developing tools for telemedicine and breast cancer diagnosis is a paramount topic for health in the next years.
The rest of the paper is organized as follows: Sects. 2 and 3 present the BreakHis dataset and the LCIP proposed method. Section 4 reports the results, and finally, Sect. 5 presents the conclusions.
2 Dataset
There are many breast histologic image datasets to propose tissue analysis algorithms for the literature. Some of them are Grand Challenge on Breast Cancer Histology Images (BACH) [13], Breast Histopathology Images [11], Breast Cancer Histopathological Annotation and Diagnosis (BreCaHAD) [10], and Breast Cancer Histopathological Database (BreakHis) [14]. We select BreakHis because it is the most popular in literature. Also, this database has histologic samples with different magnification levels, which is helpful to train networks with different feature sizes. This aspect is important because the histologic analysis is developed with different magnification observations to diagnose the tissue characteristics.
BreakHis was designed to evaluate the different histologic processing methods. This database is composed of 7,909 microscopic images of breast tumor tissue collected from 82 patients using various magnification factors (40X, 100X, 200X, and 400X). It contains 2,480 benign and 5,429 malignant samples of color images with \(700\times 460\) pixels, 8-bit resolution, and PNG format. Table 1 shows the sample distribution according to magnification and the classes of benign and malignant cells.
3 Lightweight CNN for Histology Image Processing
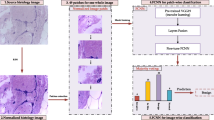
Figure 1 shows a general scheme of the proposed method, where the input is an RGB histological image, \(I(x,y)^{RGB}\). The first stage is preprocessing, which consists of color normalization. The next stage is the deep CNN, which analyzes the properties of the image to classify the tissue as Benign or Malignant cells. The deep CNN is based on a MobileNet V2 network, but we add four inverted residual convolutions to generate features with different magnification levels.
Then, the feature extraction of LCIP has a convolution layer and inverted residual block composed of parallel dilation convolutions to find features in different magnification levels. The following average pooling and convolution layers are placed to reduce the feature dimension. The classification stage of LCIP is based on two fully connected and a convolution layer of 1\(\times \)1. The next subsections explain each layer.
3.1 Preprocessing
The input of LCIP is \(I(x,y)^{RGB}\), which is an image variant to color respect other histological images due to the staining and the acquisition protocol. Then, it is necessary to normalize the images \(I(x,y)^{RGB}\) with the method of Macenko [15], which is the most popular in literature for staining normalization. The output of the Macenko method is an image \(I_M(x,y)^{RGB}\). The following step is to normalize \(I_M(x,y)^{RGB}\) regarding color level intensity with:
where \(m_{max}\) is the maximum value of the image and \(m_{min}\) is the minimum.
3.2 First Convolutional Layer
This layer finds the abstract properties of the cells with the convolution given by:
where \(\rho \) is the layer of the network (\(\rho =1\) means the first layer), \(\tau \) is the depth of the kernels, \(F_{\rho -1}(x,y)\) is the feature map of the last layer. The input \(F_0(x,y)\) is \(M(x,y)^{RGB}\). The activation function f(.) is ReLU 6 [16] because this function generate best generalization results than other activation functions. This layer has a batch normalization to accelerate the deep training by reducing internal covariate shift [17].
3.3 Residual Block
This layer has seven Inverted Residual blocks that consist of a set of convolutions with kernel sizes of 1 \(\times \) 1, 3 \(\times \) 3, 5 \(\times \) 5, and 7 \(\times \) 7. These kernels find features of the cells from different magnification images.
Figure 2 shows the scheme of this block, where the first layer of this block is a convolution given by (2), where \(\rho =2\), \(\tau =3\), \(l=1 \times 1\). This layer reduces the computational cost by combining the color image in one channel but preserving the information. The next layer is a set of parallel convolutions given by:
where \(\otimes _l\) is a depth separable convolution with dilation l. Figure 2 shows that this block has three convolution given by (3) with a dilation factor of \(l=1 \times 1\), \(l=3\times 3\), \(l=5\times 5\), and \(l=7\times 7\) to find properties and features of tissue cells from different magnification levels. In parallel to the dilation convolutions, there are an average pooling [18] and a convolution given by (2), \(\rho =3\), \(\tau =1\), \(l=1\times 1\) to find global features. The convolutions of (2) and the next parallel line of the average pooling with the convolution of 1\(\times \)1 are concatenated to generate a tensor feature map \(F_\rho (x,y,k)\), \(k=1,...,4\) where from \(k=1\) to \(k=3\) are the dilation convolution outputs \(\otimes _l\), \(\rho =4\) \(l=\{1,3,5,7\}\), and \(k=4\) is the average pooling [18] with the 1\(\times \)1 convolution.
The next step is a convolution 3 of \(F_\rho (x,y,k)\) where \(\rho =5\), \(l=1\times 1\), and the input is the concatenated map \(F_4(x,y,k)\). Finally, the feature maps are added to fuse the features and find the patterns of the tissue cells. The addition is defined as follows:
The result in this layer is a set of abstract properties that map tissue composition of different magnification levels. This composition is based on texture, cell corpuscles, and cell nucleus features.
3.4 Convolutional Layer for Feature Compression
The next layer is a Convolutional layer defined by (2), where \(\rho =6\), \(l=1\times 1\), and the activation function is ReLU 6. This layer has Batch normalization to normalize the data of all the layers within the same dynamic range. The abstract tissue features are normalized in a single map with this layer.
3.5 Global Average Pooling
This layer compresses the information of the features as possible but keeps the tissue properties. The average pooling is defined as follows:
where N is the number of windows, \((\nu ,\mu )\) is the size of each windows that compress the features, (P, Q) is the number of windows, \(p=1,...,P\) and \(q=1,...,Q\). This layer is the output of the feature extraction stage of LCIP.
3.6 First Fully Connected Layer
This is the first layer of the classification stage of LCIP, and it is defined as follows:
where \(\tau =1\), \(l=\nu \times \mu \), and \(W_{\tau ,8,l}(n,m)\) is a set of weights that learns the benign properties of the compress tissue features. Equation 6 is the dot product between the weights and the features \(F_7(n,m)\). If \(I(x,y)^{RGB}\) has information of benign cells, \(F_8(n,m)\) generates a vector with values close to zero, but if \(I(x,y)^{RGB}\) has information of malignant cells, \(F_8(n,m)\) generate values also close to one, and they surround the feature vectors generated by benign cells. Then, \(F_8(n,m)\) generates a nonlinear classification subspace.
3.7 Convolutional Layer for Classification
The next layer is a Convolutional layer that separates the vector values of both classes and works as a new feature map with linear separation. This layer is defined by the Eq. 2, where \(\rho =9\), \(\tau =1\), \(l=3\times 3\).
3.8 Second Fully Connected Layer
This layer classifies \(I(x,y)^{RGB}\) on benign or malignant cells with the following expression:
where \(\tau =1\), \(l =\nu \times \mu \), and \(W_{\tau ,10,l}(n,m)\) is a the prototype that represent the pattern of benign cells. Equation 7 represents the dot product between this prototype and the features \(F_9(n,m)\). Then, if the result is positive, \(I(x,y)^{RGB}\) has information of benign cells, but if the result is negative, \(I(x,y)^{RGB}\) has information of malignant cells. In this case, f(.) is a softmax activation function defined in [19]. This activation function generates two magnitudes that represent the classes of benign or malignant tissue.
4 Results
This section presents information about the implementation of LCIP, a comparison of LCIP with the most popular methods in the literature, and a brief Cross-Validation explanation to understand the learning of LCIP.
4.1 Training and Computer Platform
LCIP was trained with backpropagation by considering 1000 epochs with early stopping (the training was stopped in 70 epochs). The BreakHis dataset was divided into 70% of images for training, 15% for the test, and 15% for validation. LCIP was implemented in Python 3.7.0, and the computer has an i7-8750H Intel processor and an NVIDIA GPU GeForce GTX 1060 with a Max-Q design.
4.2 Comparison of LCIP with Other State of the Art Methods
The metrics used to compare LCIP with the state-of-the-art methods were accuracy (Acc), F measure (F1) [20], and Number of parameters (Np). Np is the number of variables that the network processes during the inference. The networks selected for the comparisons have the best results in literature in Acc and Np. These methods are the ResNet-50 [5] network published in 2020, a Capsule Neural Network (CapsNet)[21], and two Inception ResNet published in 2019 [3, 4]. Also, we added the MobileNet V2, which is the foundation of our proposed model. Other CNNs were not considered in this comparison because they have low accuracy or the number of parameters is complicated to calculate due to their architecture. Next, we describe the networks used in the comparisons.
The MobileNet V2 [8] is a CNN for mobile devices or embedded systems. This network has an inverted residual structure with shortcut connections between the bottleneck layers. The intermediate layers use lightweight depthwise convolutions. According to Table 2, MobileNet V2 has the lowest performance because the histologic images have patterns that are not processed adequately with linear operations. However, MobileNet V2 has significantly fewer parameters than ResNet or Inception-ResNet.
CapsNet [21] presents an Acc of 86% but does not report F1. CapsNet has capsules, which are vector structures generated from the outputs of the neuron group. The capsules generate invariant features to spatial and orientation, which help find the nucleus and other cell properties. However, the performance is lower than ResNet or Inception ResNet.
Inception-ResNet [3, 4] is an architecture widely used for histologic image processing. The architecture of [3] extracts features constructed with a new autoencoder network that transforms the features to a low dimensional. The model of [4] is an ensemble of VGG19, MobileNet, and DenseNet. This ensemble generates a model similar to the Inception-ResNet network. However, the result is 92.4% with BreakHis, and the Np is the highest.
ResNet-50 presents an Acc of 99% in [5]. This network has pre-trained kernels with ImageNet and was trained with BreakHis, but the Np is high compared to other networks.
LCIP achieves the best results with the highest Acc and the lowest Np. These results are because LCIP combines the architecture of MobileNet with a block that extracts abstract features according to the magnification level. LCIP finds the necessary features describing the cells with the first convolutional layer and the inverted residual block. The following convolutional layer and the average pooling reduce the dimension of the features. Finally, the classification stage generates the hyperplanes to find the subspace where the images can be separated into benign or malignant cells.
4.3 Cross Validation
The methods of ResNet, Inception-ResNet, and LCIP report accuracies higher than 90%, but it is essential to know if a result higher than 90% is due to the network learns. However, none of the articles reported in the literature present an analysis to validate the obtained accuracy, like Cross-Validation (CV). For this reason, this subsection presents the average results of a CV analysis of LCIP, ResNet, and MobileNet V2. The CV was developed with 70 epochs and five k-folds because these parameters were enough to know the generalization capability of the networks. Table 3 shows the average of the five k-folds of the networks. LCIP achieves the best Acc and F1 metrics. ResNet has low F1, which is very different than the result shown in Table 2. On the other hand, MobileNet V2 achieved better results in the CV than the results reported in Table 2. Inception-ResNet 1 and 2 do not generate conclusive results because the CV reports lower performance than MobileNet V2.
5 Conclusion
This paper presents a novel method named Lightweight CNN for Histology Image Processing (LCIP), a network for benign and malignant cell detection in histological breast tissue samples obtained from digital images. LCIP is based on the architecture of MobileNet V2 and a block with dilated convolution in parallel to extract cell features of different magnification levels. The second convolutional layer and the average pooling reduce the dimension of the features. Finally, the classification stage generates the subspaces where the images can be separated into benign or malignant cells. According to the results, LCIP achieves the best accuracy and F1 measure, with fewer parameters in the BreakHis dataset compared to network models reported in the literature. LCIP has a low computational cost architecture that includes a set of layers that find cell features in the different magnification levels. The accuracy of LCIP was 99.73% with 70 epochs, and the average of the five k-folds in CV was 86.66% with 70 epochs. On the other hand, the average accuracy of Xception falls from 99% to 84.75% in the CV, and MobileNet V2 increases its performance from 54.18% to 63.55%. These results mean that the performances obtained with the backpropagation generate overfitting in all the networks due to \(M(x,y)^{RGB}\) do not distinguish features at different magnification levels. However, LCIP achieves better results in the CV than any other method reported in the literature. Furthermore, the number of parameters of LCIP is significantly fewer than MobileNet V2, CapsNet, Inception-ResNet, and Xception-50. These LCIP results are because in the case of images with different magnifications levels, the increase in the number of parallel operations, the network extracts descriptive features of the histological tissue with fewer parameters. Then, based on the accuracy results of LCIP, the CV validation, and the number of parameters, we conclude that LCIP is a feasible network for histologic image processing. Future work will test LCIP in embedded GPU devices to generate embedded machine learning technology for telemedicine.
References
Breast Cancer (2021). https://www.who.int/news-room/fact-sheets/detail/breast-cancer
Shen, L., Margolies, L.R., Rothstein, J.H., Fluder, E., McBride, R., Sieh, W.: Deep learning to improve breast cancer detection on screening mammography. Nature 9(12495), 1–12 (2019)
Xie, J., Liu, R., Luttrell, J., Zhang, C.: Deep learning based analysis of histopathological images of breast cancer. Front. Genet. 10(80), 1–19 (2019)
Kassani, S.H. Wesolowski, M.J., Schneider, K.A.: Classification of Histopathological biopsy images using ensemble of deep learning networks. In: 29th Annual International Conference on Computer Science and Software Engineering, pp. 92–99. ACM, Toronto (2019)
Al-Haija, Q.A., Adebanjo, A.: Breast cancer diagnosis in histopathological images using ResNet-50 convolutional neural network. IEEE International IOT, Electronics and Mechatronics Conference, pp. 1–8. IEEE, Vancouver (2020)
Bhowalm, P., Sen, S., Velasquez, J.D., Sarkar, R.: Fuzzy ensemble of deep learning models using choquet fuzzy integral, coalition game and information theory for breast cancer histology classification. Expert Syst. App. 190(1), 116167 (2022)
Bianco, S., Cadene, R., Celona, L., Napoletano, P.: Benchmark analysis of representative deep neural network architectures. IEEE Access 6(1), 64270–64277 (2018)
Akay, M., et al.: Deep learning classification of systemic sclerosis skin using the mobilenetv2 model. IEEE Open J. Eng. Med. Biol. 2(1), 104–110 (2021)
Wang, Y., et al.: Improved breast cancer histological grading using deep learning. Ann. Oncol. 190(1), 89–98 (2022)
Aksac, A., Demetrick, D.J., Ozyer, T., et al.: BreCaHAD: a dataset for breast cancer histopathological annotation and diagnosis. BMC Res Notes 12, 82 (2019)
Janowczyk, A., Madabhushi, A.: Deep learning for digital pathology image analysis. A comprehensive tutorial with selected use cases. J. Pathol. Inform. 7(29), 1–10 (2016)
The Lancet Rheumatology: Telemedicine: is the new normal fit for purpose? Lancet Rheumatolol. 4(1), e1 (2022)
Aresta, G., Araujo, T.: BACH Grand challenge on breast cancer histology images. Med. Image Anal. 56(1), 122–139 (2019)
Spanhol, F., Cavalin, P., Oliveira, L., Petitjean, C., Heutte, L.: Deep features for breast cancer histopathological image classification. In: International Conference on Systems, Man, and Cybernetics, pp. 1868–1873. Banff, IEEE (2017)
Macenko, M., et al.: A method for normalizing histology slides for quantitative analysis. In: International Symposium Biomedical Imaging From Nano to Macro, pp. 1107–1110. IEEE, Boston (2009)
Duan, C., Zhang, T.: Two-stream convolutional neural network based on gradient image for aluminum profile surface defects classification and recognition. IEEE Access 81, 172152–172165 (2020)
Kalayeh, M.M., Shah, M.: Training faster by separating modes of variation in batch-normalized models. IEEE Trans. Pattern Anal. Mach. Intell. 42(6), 1483–1500 (2020)
Theodoridis, T., Loumponias, K., Vretos, N., Daras, P.: Zernike pooling generalizing average pooling using Zernike moments. IEEE Access 9(1), 121128–121136 (2021)
Gao, F., Li, B., Chen, L., Shang, Z., Wei, X., He, C.: A softmax classifier for high-precision classification of ultrasonic similar signals. Ultrasonics 112(1), 1–8 (2021)
Liu, M., Xu, C., Luo, Y., Xu, C., Wen, Y., Tao, D.: Cost-sensitive feature selection by optimizing F-measures. IEEE Trans. Image Process. 27(3), 1323–1335 (2018)
Anupama, M., Sowmya, V., Soman, K.P.: Breast cancer classification using capsule network with preprocessed histology images. In: International Conference on Communication and Signal Processing (ICCSP), pp. 1–8. IEEE, Chennai (2019)
Acknowledgements
This work was supported by the Tecnologico Nacional de Mexico under grants TecNM. The number of funding support is TecNM 14044.22-P.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Ramirez-Quintana, J., Acosta-Lara, I., Ramirez-Alonso, G., Chacon-Murguia, M., Corral-Saenz, A. (2022). A Lightweight Convolutional Neural Network for Breast Cancer Diagnosis with Histology Images. In: Vergara-Villegas, O.O., Cruz-Sánchez, V.G., Sossa-Azuela, J.H., Carrasco-Ochoa, J.A., Martínez-Trinidad, J.F., Olvera-López, J.A. (eds) Pattern Recognition. MCPR 2022. Lecture Notes in Computer Science, vol 13264. Springer, Cham. https://doi.org/10.1007/978-3-031-07750-0_30
Download citation
DOI: https://doi.org/10.1007/978-3-031-07750-0_30
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-07749-4
Online ISBN: 978-3-031-07750-0
eBook Packages: Computer ScienceComputer Science (R0)