Abstract
Breast cancer is one of the main causes of cancer death worldwide. Early diagnostics significantly increases the chances of correct treatment and survival, but this process is tedious and often leads to disagreement between pathologists. Computer-aided diagnosis systems show potential for improving the diagnostic accuracy. In this work, we develop the computational approach based on deep convolution neural networks for breast cancer histology image classification. Hematoxylin and eosin stained breast histology microscopy image dataset is provided as a part of the ICIAR 2018 Grand Challenge on Breast Cancer Histology Images. Our approach utilizes several deep neural network architectures and gradient boosted trees classifier. For 4-class classification task, we report 87.2% accuracy. For 2-class classification task to detect carcinomas we report 93.8% accuracy, AUC 97.3%, and sensitivity/specificity 96.5/88.0% at the high-sensitivity operating point. To our knowledge, this approach outperforms other common methods in automated histopathological image classification. The source code for our approach is made publicly available at https://github.com/alexander-rakhlin/ICIAR2018.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
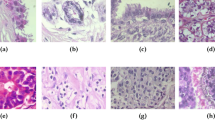
Breast cancer is the most common cancer diagnosed among women in the United States (excluding skin cancers) [23]. Breast tissue biopsies allow the pathologists to histologically assess the microscopic structure the tissue. Histopathology aims to distinguish between normal tissue, non-malignant (benign) and malignant lesions (carcinomas), and to perform a prognostic evaluation [7]. A combination of hematoxylin and eosin (H&E) is the principal stain of tissue specimens for histopathological diagnostics. There are multiple types of breast carcinomas that embody characteristic tissue morphology, see Fig. 1. Breast carcinomas arise from the mammary epithelium and cause a pre-malignant epithelial proliferation within the ducts, called ductal carcinoma in situ. Invasive carcinoma is characterized by the cancer cells gaining the capacity to break through the basal membrane of the duct walls and infiltrate into surrounding tissues [20].
Morphology of tissue and cells is regulated by complex biological mechanisms related to cell development and pathology [13]. Traditionally, morphological assessment were visually performed by a pathologist. This process is tedious and subjective, causing inter-observer variations even among senior pathologists [6, 16]. The subjectivity of morphological criteria in visual classification motivates the use of computer-aided diagnosis (CAD) to improve the diagnosis accuracy, reduce human error, increase inter-observer agreement and reproducibility [20].
There are many methods developed for the digital pathology image analysis, from rule-based to applications of machine learning [20]. Recently, deep learning based approaches were shown to outperform conventional machine learning methods in many image analysis tasks, automating end-to-end processing [4, 10, 12]. In the domain of medical imaging, convolutional neural networks (CNN) have been successfully used for diabetic retinopathy screening [19], bone disease prediction [26] and age assessment [11], and other problems [4, 22]. Previous deep learning based applications in histological microscopic image analysis have demonstrated their potential to provide utility in diagnosing breast cancer [1, 2, 20, 24].
In this paper, we present an approach for histology microscopy image analysis for breast cancer type classification. Our approach utilizes deep CNNs for feature extraction and gradient boosted trees for classification and, to our knowledge, outperforms other similar solutions.
2 Methods
2.1 Dataset
The dataset is an extension of the dataset from [1] and consists of 400 H&E stain images (\(2048\,\times \,1536\) pixels) of 4 classes. All the images are digitized with the same acquisition conditions: a magnification of \(200{\times }\) and a \(0.42\,\upmu \mathrm{m}\times 0.42\,\upmu \mathrm{m}\) pixel size. Each image is labeled with one of the four balanced classes: normal, benign, in situ carcinoma, and invasive carcinoma, where class is defined as a predominant cancer type in the image, see Fig. 1. The image-wise annotation was performed by two medical experts [9]. The goal of the challenge is to provide an automatic classification of each input image.
2.2 Approach Overview
The limited size of the dataset poses a significant challenge for the training of a deep CNN [4]. Very deep architectures (ResNet, Inception) that contain millions of parameters have achieved state-of-the-art results in many computer vision tasks [25]. However, training these models from scratch requires a large number of images, as training on a small dataset leads to overfitting. A typical remedy in these circumstances is fine-tuning, when only a part of the pre-trained neural network is being fitted to a new dataset. Since fine-tuning did not demonstrate good performance on this task, we employed a different approach known as deep convolutional feature representation [8]. It uses deep CNNs, trained on large datasets like ImageNet (10M images, 20K classes) [5] for unsupervised feature representation extraction. In this study, images are encoded with state-of-the-art general-purpose networks to obtain sparse descriptors of low dimensionality (1408 or 2048). This unsupervised dimensionality reduction step significantly reduces the risk of overfitting on the next stage of supervised learning.
We use LightGBM, the fast, distributed, high performance implementation of gradient boosted trees, for supervised classification [14]. Gradient boosting models are widely used in machine learning due to their speed, accuracy, and robustness against overfitting [17].
2.3 Data Pre-processing and Augmentation
To bring the microscopy images into a common space to enable improved quantitative analysis, we normalize the amount of H&E stained on the tissue as described in [15]. For each image, we perform 50 random color augmentations. Following [21] the amount of H&E is adjusted by decomposing the RGB color of the tissue into H&E color space, followed by multiplying the magnitude of H&E of every pixel by two random uniform variables from the range [0.7, 1.3]. Furthermore, we downscale images in half to \(1024\times 768\) pixels. From the downscaled images we extract crops of \(400\times 400\) pixels and \(650\times 650\) pixels. Thereby, each image was represented by 20 crops that are encoded into 20 descriptors. Then, the set of 20 descriptors is combined through 3-norm pooling [3] into a single descriptor:
where the hyperparameter \(p=3\) as suggested in [3, 27], N is the number of crops, \(\mathbf {d}_i\) is descriptor of a crop and \(\mathbf {d}_{pool}\) is pooled descriptor of the image. The p-norm of a vector gives the average for \(p = 1\) and the max for \(p\rightarrow \infty \). As a result, for each original image, we obtain 50 (number of color augmentations) \(\times 2\) (crop sizes) \(\times 3\) (CNN encoders) \(=300\) descriptors.
2.4 Feature Extraction
Overall pre-processing pipeline is depicted in Fig. 2. For features extraction, we use pre-trained ResNet-50, InceptionV3 and VGG-16 networks. We remove fully connected layers from each model to allow the networks to consume images of an arbitrary size. In ResNet-50 and InceptionV3, we convert the last convolutional layer consisting of 2048 channels via GlobalAveragePooling into a one-dimensional feature vector with a length of 2048. With VGG-16 we apply the GlobalAveragePooling operation to the four internal convolutional layers: block2, block3, block4, block5 with 128, 256, 512, 512 channels respectively. We concatenate them into one vector with a length of 1408, see Fig. 3.
2.5 Training
We split the data into 10 stratified folds to preserve class distribution, while all descriptors of the same image are contained in the same fold to prevent information leakage. Augmentations increase the size of the dataset \(\times 300\) (2 patch sizes x 3 encoders x 50 color/affine augmentations). For each combination of the encoder, crop size and scale we train 10 gradient boosting models per fold. This allows us to increase the diversity of the models with limited data (bagging). Furthermore, we recycle each dataset 5 times with different random seeds in LightGBM adding augmentation on the model level. As a result, we train 10 (number of folds) \({\times }5\) (seeds) \({\times }4\) (scale and crop) \({\times }3\) (CNN encoders) \(= 600\) gradient boosting models. At the cross-validation stage, we predict every fold only with the models not trained on this fold. For the test data, we similarly extract 300 descriptors for each image and use them with all models trained for particular patch size and encoder. The predictions are averaged over all augmentations and models. Finally, the predicted class is defined by the maximum probability score.
(a) Non-carcinoma vs. carcinoma classification, ROC. High sensitivity setpoint = 0.33 (green): 96.5% sensitivity and 88.0% specificity to detect carcinomas. Setpoint = 0.50 (blue): 93.0% sensitivity and 94.5% specificity (b) Confusion matrix, without normalization. Vertical axis - ground truth, horizontal - predictions. (Color figure online)
3 Results
To validate the approach we use 10-fold stratified cross-validation.
For 2-class non-carcinomas (normal and benign) vs. carcinomas (in situ and invasive) classification accuracy was 93.8 ± 2.3\(\%\), the area under the ROC curve was 0.973, see Fig. 4a. At high sensitivity setpoint 0.33 the sensitivity of the model to detect carcinomas was 96.5\(\%\) and specificity 88.0\(\%\). At the setpoint 0.50 the sensitivity of the model was 93.0\(\%\) and specificity 94.5\(\%\), Fig. 4a. Out of 200 carcinomas cases only 9 in situ and 5 invasive were missed, Fig. 4b.
Table 1 shows classification accuracy for 4-class classification. Accuracy averaged across all folds is 87.2 ± 2.6\(\%\). Data augmentation and model fusion are particularly evident. The fused model accuracy is by 4–5% higher than any of its individual constituents. The standard deviation of the ensemble across 10 folds is as twice as low than the average standard deviation of individual models. Moreover, all results are slightly improved by averaging across 5 seeded models.
4 Conclusions
In this paper, we propose a simple and effective method for the classification of H&E stained histological breast cancer images in the situation of very small training data (few hundred samples). To increase the robustness of the classifier we use strong data augmentation and deep convolutional features extracted at different scales with publicly available CNNs pretrained on ImageNet. On top of it, we apply accurate and prone to overfitting implementation of the gradient boosting algorithm. Unlike some previous works, we purposely avoid training neural networks on this amount of data to prevent suboptimal generalization.
To our knowledge, the reported results are superior to the automated analysis of breast cancer images reported in literature.
References
Araújo, T., Aresta, G., Castro, E., Rouco, J., Aguiar, P., Eloy, C., Polónia, A., Campilho, A.: Classification of breast cancer histology images using convolutional neural networks. PloS One 12(6), e0177544 (2017)
Bejnordi, B.E., Veta, M., van Diest, P.J., van Ginneken, B., Karssemeijer, N., Litjens, G., van der Laak, J.A., Hermsen, M., Manson, Q.F., Balkenhol, M., et al.: Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 318(22), 2199–2210 (2017)
Boureau, Y.L., Ponce, J., LeCun, Y.: A theoretical analysis of feature pooling in visual recognition. In: Proceedings of the 27th International Conference on Machine Learning (ICML-10), pp. 111–118 (2010)
Ching, T., Himmelstein, D.S., Beaulieu-Jones, B.K., Kalinin, A.A., Do, B.T., Way, G.P., Ferrero, E., Agapow, P.M., Zietz, M., Hoffman, M.M., Xie, W., Rosen, G.L., Lengerich, B.J., Israeli, J., Lanchantin, J., Woloszynek, S., Carpenter, A.E., Shrikumar, A., Xu, J., Cofer, E.M., Lavender, C.A., Turaga, S.C., Alexandari, A.M., Lu, Z., Harris, D.J., DeCaprio, D., Qi, Y., Kundaje, A., Peng, Y., Wiley, L.K., Segler, M.H.S., Boca, S.M., Swamidass, S.J., Huang, A., Gitter, A., Greene, C.S.: Opportunities and obstacles for deep learning in biology and medicine. J. R. Soc. Interface 15(141), 20170387 (2018)
Deng, J., Dong, W., Socher, R., Li, L.J., Li, K., Fei-Fei, L.: Imagenet: a large-scale hierarchical image database. In: IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2009, pp. 248–255. IEEE (2009)
Elmore, J.G., Longton, G.M., Carney, P.A., Geller, B.M., Onega, T., Tosteson, A.N., Nelson, H.D., Pepe, M.S., Allison, K.H., Schnitt, S.J., et al.: Diagnostic concordance among pathologists interpreting breast biopsy specimens. JAMA 313(11), 1122–1132 (2015)
Elston, C.W., Ellis, I.O.: Pathological prognostic factors in breast cancer. i. the value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology 19(5), 403–410 (1991)
Guo, Y., Liu, Y., Oerlemans, A., Lao, S., Wu, S., Lew, M.S.: Deep learning for visual understanding: a review. Neurocomputing 187, 27–48 (2016)
ICIAR 2018 Grand Challenge on Breast Cancer Histology Images. https://iciar2018-challenge.grand-challenge.org/. Accessed 31 Jan 2018
Iglovikov, V., Mushinskiy, S., Osin, V.: Satellite imagery feature detection using deep convolutional neural network: a kaggle competition. arXiv preprint arXiv:1706.06169 (2017)
Iglovikov, V., Rakhlin, A., Kalinin, A., Shvets, A.: Pediatric bone age assessment using deep convolutional neural networks (2017). arXiv preprint arXiv:1712.05053
Iglovikov, V., Shvets, A.: Ternausnet: U-net with vgg11 encoder pre-trained on imagenet for image segmentation (2018). arXiv preprint arXiv:1801.05746
Kalinin, A.A., Allyn-Feuer, A., Ade, A., Fon, G.V., Meixner, W., Dilworth, D., Jeffrey, R., Higgins, G.A., Zheng, G., Creekmore, A., et al.: 3d cell nuclear morphology: microscopy imaging dataset and voxel-based morphometry classification results. bioRxiv, 208207 (2017)
Ke, G., Meng, Q., Finley, T., Wang, T., Chen, W., Ma, W., Ye, Q., Liu, T.Y.: Lightgbm: a highly efficient gradient boosting decision tree. In: Advances in Neural Information Processing Systems, pp. 3149–3157 (2017)
Macenko, M., Niethammer, M., Marron, J., Borland, D., Woosley, J.T., Guan, X., Schmitt, C., Thomas, N.E.: A method for normalizing histology slides for quantitative analysis. In: IEEE International Symposium on Biomedical Imaging: From Nano to Macro, ISBI 2009, pp. 1107–1110. IEEE (2009)
Meyer, J.S., Alvarez, C., Milikowski, C., Olson, N., Russo, I., Russo, J., Glass, A., Zehnbauer, B.A., Lister, K., Parwaresch, R.: Breast carcinoma malignancy grading by bloom-richardson system vs proliferation index: reproducibility of grade and advantages of proliferation index. Mod. Pathol. 18(8), 1067 (2005)
Natekin, A., Knoll, A.: Gradient boosting machines, a tutorial. Front. Neurorobot. 7, 21 (2013)
Open Data Science (ODS). https://ods.ai. Accessed 31 Jan 2018
Rakhlin, A.: Diabetic retinopathy detection through integration of deep learning classification framework. bioRxiv, 225508 (2017)
Robertson, S., Azizpour, H., Smith, K., Hartman, J.: Digital image analysis in breast pathology-from image processing techniques to artificial intelligence. Transl. Res. 194, 19–35 (2017)
Ruifrok, A.C., Johnston, D.A., et al.: Quantification of histochemical staining by color deconvolution. Anal. Quant. Cytol. Histol. 23(4), 291–299 (2001)
Shvets, A., Rakhlin, A., Kalinin, A.A., Iglovikov, V.: Automatic instrument segmentation in robot-assisted surgery using deep learning. arXiv preprint arXiv:1803.01207 (2018)
Siegel, R.L., Miller, K.D., Jemal, A.: Cancer statistics, 2018. CA Cancer J. Clin. 68(1), 7–30 (2018). https://doi.org/10.3322/caac.21442
Spanhol, F.A., Oliveira, L.S., Petitjean, C., Heutte, L.: Breast cancer histopathological image classification using convolutional neural networks. In: 2016 International Joint Conference on Neural Networks (IJCNN), pp. 2560–2567. IEEE (2016)
Szegedy, C., Ioffe, S., Vanhoucke, V., Alemi, A.A.: Inception-v4, inception-resnet and the impact of residual connections on learning. In: AAAI, vol. 4, p. 12 (2017)
Tiulpin, A., Thevenot, J., Rahtu, E., Lehenkari, P., Saarakkala, S.: Automatic knee osteoarthritis diagnosis from plain radiographs: a deep learning-based approach. Sci. Rep. 8, 1727 (2018)
Xu, Y., Jia, Z., Ai, Y., Zhang, F., Lai, M., Eric, I., Chang, C.: Deep convolutional activation features for large scale brain tumor histopathology image classification and segmentation. In: 2015 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), pp. 947–951. IEEE (2015)
Acknowledgments
The authors thank the Open Data Science community [18] for useful suggestions and other help aiding the development of this work.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer International Publishing AG, part of Springer Nature
About this paper
Cite this paper
Rakhlin, A., Shvets, A., Iglovikov, V., Kalinin, A.A. (2018). Deep Convolutional Neural Networks for Breast Cancer Histology Image Analysis. In: Campilho, A., Karray, F., ter Haar Romeny, B. (eds) Image Analysis and Recognition. ICIAR 2018. Lecture Notes in Computer Science(), vol 10882. Springer, Cham. https://doi.org/10.1007/978-3-319-93000-8_83
Download citation
DOI: https://doi.org/10.1007/978-3-319-93000-8_83
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-92999-6
Online ISBN: 978-3-319-93000-8
eBook Packages: Computer ScienceComputer Science (R0)