Abstract
Jawed vertebrate species (Gnathostomata) are all characterized by an adaptive immune system based on B and T cells along with the huge diversity and specificity of their antigen receptors, the immunoglobulins (IG) or antibodies and the T-cell receptors (TCRs), respectively. The availability of genome assemblies of many species has recently provided valuable information on the complexity and diversity of teleost germline IG loci. The development of deep sequencing technologies has also favored a growing interest for immunoglobulin repertoires, to address basic questions about immune mechanisms in teleost or applied concerns such as the identification of molecular markers of protection after vaccination. This work provides an overview on the germline configuration of teleost IG loci, IG repertoire studies, and recent findings on IG functional roles in this group of vertebrates.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
7.1 Introduction
The emergence of vertebrates was accompanied by major morphological and functional innovations, such as the development of an adaptive immune system (Flajnik and Du Pasquier 2004). In jawed vertebrates (Gnathostomata), including bony fish (superclass Osteichthyes), the adaptive immune system relies on B and T cells and on the huge diversity and specificity of their antigen receptors, the immunoglobulins (IG) or antibodies, and the T-cell receptors (TCR), respectively (Cooper and Alder 2006; Flajnik 2018).
IGs are produced by B lymphocytes, either as a secretory form (antibody) or as a membrane form (B-cell receptor). As described in humans or mice, they constitute a key factor for fish-specific immunity and for the protection afforded by vaccines. The mechanisms underlying the humoral immunity mediated by IGs include opsonization of pathogens to be eliminated by phagocytes, neutralization of toxins and viruses, and activation of the complement cascade (Magadan et al. 2015; Mashoof and Criscitiello 2016). The canonical IG structure is a tetramer that consists of two identical heavy (H) chains and two identical light (L) chains. Both H and L chains contain one N-terminal variable domains (VH or VL) and one or more C-terminal constant domains that form the constant region (CH or CL) and define the isotypes. The variable region that engages the antigen is formed by one VH and one VL domain. So, one IG molecule has two antigen-binding sites, which can recognize the cognate antigen in its native form. Constant regions consist of constant domains of both IGH and IGL as well. While IGL have one CL domain, the number of constant domains of IGH chain vary among different immunoglobulin classes and species. Interestingly, the domains that comprise the constant regions mediate effector functions of the antibody molecule (Sun et al. 2020). As shown in this chapter, the availability of genome assemblies of many species has provided much information on the IGH and IGL chain gene loci in teleost fish.
Compared to mammals, teleosts have slower antibody-mediated responses, only three classes have been described (IgM, IgD, and IgT), and they do not present immunoglobulin class switching. Moreover, there is a considerable debate on whether the IG affinity maturation and clonal selection during B-cell responses in fish (extended to ectothermic vertebrates) are less efficient compared to mammals (Magor 2015), likely because microenvironments do not allow the fast and powerful selection of cells expressing high affinity antibodies as in mammals (Fillatreau et al. 2013; Magadan et al. 2015). However, further studies are required to confirm this hypothesis. The teleost immune system is adapted to particular anatomical and physiological constraints. Therefore, features conserved in common ancestors of fish and mammals are likely to be essential, while other characteristics may represent teleost-specific original solutions. The aim of this chapter is to present an overview of the different IGs expressed by teleost, considering the germline configuration of teleost IG loci and the application of new approaches such as new generation sequencing to decipher the dynamics of the humoral adaptive immune response in teleost. Recent studies that shed light on the functional role of teleost IG classes are also discussed.
7.2 IG Loci
Bony fishes are classified into lobe-finned fishes (Sarcopterygii) and ray-finned fishes (Actinopterygii). Lobe-finned fishes include the coelacanth and lungfish, which are considered to be the closest living species of the land vertebrates (Shan and Gras 2011). Ray-finned fishes include the infraclass Teleostei, which comprises 95% of extant fish species (approximately 35,000 species) (Fricke et al. 2020), and they account for more than half of all extant vertebrate species. The success of this infraclass, along with their ability to thrive in a wide range of ecological niches, suggests that teleost species developed an immune arsenal to counter pathogen and environmental challenges.
Research on the immune system of fish has generally been limited by the lack of reagents suitable for classical cellular immunology research. However, complete genome sequences are now available for many species, allowing thorough analysis of immune mechanisms. In particular, a cycle of tetraploidization and rediploidization occurred during the early evolution of fish. The genome diversity was further increased by lineage-specific events of genome duplication and/or contraction. All these genome remodeling processes affected genes involved in the immune system, such as the major histocompatibility complex or immunoglobulin genes (Bradshaw and Valenzano 2020; Malmstrøm et al. 2016). A deep knowledge about the structure of IG loci (and T-cell receptor gene loci as well) is essential to comprehend the adaptive immune response developed against antigens in a given species. Furthermore, teleost fish can be considered a good model for comparative studies between distant living groups. They can provide clues about the primordial immune system of extinct vertebrates, and new insights into the functional development of adaptive immune systems throughout vertebrate evolution.
The structure of bony fish IGH loci follows the general pattern found in most vertebrates, i.e., the translocon organization, with a large region containing all IGHV genes in 5′, followed by several D, J, and then C region genes at the 3′ end (Fig. 7.1). However, the genomic organization of teleost IGL loci is quite different. They are arranged in multiclusters of VL, JL, and CL region segments: (VL–JL–CL)n or (VL–VL–JL–CL)n (Bengtén and Wilson 2015; Ghaffari and Lobb 1993, 1999; Magadán-Mompó et al. 2013; Rego et al. 2020a). Thus, bony fishes possess a chimeric gene organization for the H- and L-chain genes (Flajnik 2002; Sun et al. 2020).
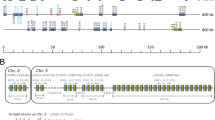
Schematic structure of the genomic organization of IGH loci in representative vertebrate species. The schemes are not drawn to scale and depict only the genomic configuration of V (black boxes), D and J (light gray boxes), and CH gene segments. The number of IGHV, D, and J gene segments are not indicated, and except for chicken, functional V and pseudo V (ψV) segments are not distinguished. Different CH genes are represented as different colored boxes: IGHM (light blue), IGHD (purple), IGHT (orange), IGHX (light green), IGHY (dark green), IGHA (yellow), IGHG (dark blue), and IGHE (brown). The pseudo CH genes are indicated by a ψ. The CH genes with the opposite transcriptional orientation to the whole gene locus are indicated as an arrow. In Atlantic salmon, both iso loci (IGHA and IGHB) are represented. (Asterisk) IGHF has been only identified in Xenopus laevis
Comparative analysis of IG loci structure in closely related species, and different lines or stock, provides information about IG loci variation that may have significant implications for practical issues in aquaculture. In addition, these analyses may also elucidate the mechanisms of gene conversion, somatic hypermutation, and memory in these species and during vertebrate evolution. Furthermore, the development of high-throughput DNA sequencing technologies has enabled large-scale characterization of functional antibody repertoires in human and mice. This way of understanding protective and pathogenic immune responses has started to develop in teleost species, reflecting a growing interest for an accurate and comprehensive description of the antibody-mediated response.
7.2.1 IGH Locus
The immunoglobulin heavy chain determines the antibody effector function, and also contributes to the antibody diversity. Thus, the structure of IGH locus has a relevant effect on the adaptive immune response for any species. Previous works have characterized the IGH locus structure in teleost species, which are used as animal models in biomedicine, such as zebrafish (Danio rerio) (Danilova et al. 2005), or in ecological and evolutionary research, like medaka (Oryzias latipes) (Magadán-Mompó et al. 2011), three-spined stickleback (Gasterosteus acuelatus) (Bao et al. 2010; Gambón-Deza et al. 2010), turquoise killifish (Nothobranchius furzeri), and southern platyfish (Xiphophorus maculatus) (Bradshaw and Valenzano 2020). The IGH loci have been also elucidated in a number of fish species relevant for the aquaculture industry, i.e., rainbow trout (Oncorhynchus mykiss) (Hansen et al. 2005; Magadan et al. 2019b), fugu (Takifugu rubripes) (Fu et al. 2015), and Atlantic salmon (Salmo salar) (Yasuike et al. 2010). These characterizations have revealed remarkable diversity in the size and structure of teleost IGH loci (Bengtén and Wilson 2015; Bradshaw and Valenzano 2020). However, the number of studied IGH loci are very low compared to the total evolutionary diversity of teleost, and are mainly focused on major aquaculture species and established research models.
In all studied bony fish, the IGH locus is organized in translocon configuration. This IGH configuration is characterized by multiple tandemly organized IGHV, D, and J gene segments, followed by a series of IGHC genes, encoding diverse H chain isotypes or subclasses associated with different effector functions (Fig. 7.1). It was initially thought that the IGH locus in teleosts was a simplified version of the translocon IGH locus found in mammals. However, research in teleost fishes has revealed a remarkable degree of diversity in the length and organization (Bengtén and Wilson 2015; Fillatreau et al. 2013). After the discovery of the IgZ/T in zebrafish (Danilova et al. 2005) and rainbow trout (Hansen et al. 2005), the overall organizations of the IGH locus in teleosts have been better understood. Three sets of CH exons define the three Ig classes encoded by isotypes μ, δ, and τ/ζ. As in mammals, IgM and IgD are coproduced through alternative splicing of a long pre-mRNA containing the VDJ region, the Cμ exons and the Cδ exons. IgT/Z is encoded by an additional Dτ-Jτ-Cτ cluster located in the 5′ regions of the IGHD and IGHJ gene segments of Igμ and Igδ in zebrafish (Danilova et al. 2005) and stickleback (Bao et al. 2010; Gambón-Deza et al. 2010) or inserted within the IGHV segments close to IGHM-IGHD genes, as in rainbow trout (Hansen et al. 2005; Magadan et al. 2019a, b) and Atlantic salmon (Yasuike et al. 2010). This IGH configuration has led to the evolution of a distinct B-cell lineage in addition to IgM, which expresses IgT/Z (Danilova et al. 2005; Hansen et al. 2005; Salinas et al. 2011). However, all three classes (IgM, IgD, and IgT/Z) when expressed from a unique IGH locus can use the same set of IGHV genes, which can rearrange either to DHτ or Dμ/δ. (Fig. 7.1).
The number of IGH loci are quite variable among fish species. Among salmonids, Atlantic salmon and rainbow trout possess two IGH isoloci (IGHA and IGHB), due to the tetraploidization of Salmonidae (Hansen et al. 2005; Yasuike et al. 2010). Two IgM isotypes were found in Atlantic salmon and brown trout (Salmo trutta), while it has been suggested through gel filtration analysis that rainbow trout possesses a single IgM (Hordvik 2002), suggesting that one of the two isoloci may be nonfunctional. However, the annotation of IGH loci using Atlantic salmon and rainbow trout assembly genomes resulted in the presence of two functional IGHM genes in both species (Magadan et al. 2019b). Authors found eight IGHT genes in Atlantic salmon, which were previously described by Yasuike et al. (2010), and three IGHT genes in rainbow trout, but only two of them are functional. These results are not in line with the three IgT subclasses previously reported in rainbow trout (Zhang et al. 2017). Nevertheless, it is important to note that different sequencing technologies (Illumina, PacBio long reads, etc.) can be used for genome sequencing, which likely affects the quality of the genome assembly and may generate sequencing gaps and artifacts that have to be clarified by further studies.
Only one IGH locus has been identified in zebrafish (Danilova et al. 2005), in torafugu (Fu et al. 2015), in turquoise killifish, and southern platyfish (Bradshaw and Valenzano 2020). In other species like channel catfish (Ictalurus punctatus), medaka (Magadán-Mompó et al. 2011), and three-spined stickleback (Bao et al. 2010; Gambón-Deza et al. 2010), tandem duplications of the IGH locus have been found (Fig. 7.1). Interestingly, no IGHT gene (or transcript) has been found in channel catfish, medaka (Bengtén et al. 2006; Magadán-Mompó et al. 2011). A recent comparative analysis performed in 12 genomes from species belonging to Cyprinidontiformes suggests that the IGHT gene has undergone duplication and convergent loss in the course of cyprinodontiform evolution (Bradshaw and Valenzano 2020). This work, in addition to confirm the IGHT absence in medaka, has identified other teleost that appears to lack IGHT constant regions, such as the N. furzeri and, two fresh water species used in aquariums, Aphyosemion australe and Nothobranchius orthonotus. All these results demonstrate the remarkable plasticity of the IGH locus across evolutionary time.
In addition to such diverse numbers of IGH loci, the structure of different IGH constant genes also shows a remarkable plasticity among teleosts. While the six-exon genomic structure of the IGHM constant region (four CH, and two transmembrane TM exons) is highly conserved across the jawed vertebrates, with similar configurations observed in mammals, teleosts, and elasmobranchs (Flajnik 2018; Sun et al. 2020), IGHD and IGHT genes have undergone repeated duplications and deletions in the course of teleost evolution. For instances, Cδ2-Cδ3-Cδ4 domains are repeated four times in zebrafish, three in catfish, and twice in Atlantic salmon and Atlantic halibut (Hippoglossus hippoglossus) (Fillatreau et al. 2013; Yasuike et al. 2010). A different structure has been reported in Atlantic cod (Gadus morhua) with two duplicated Cδ1-Cδ2 separated by a short exon, and followed by Cδ7 (Fillatreau et al. 2013). The rainbow trout IgD gene is also particular: the configuration, Cδ1–Cδ2a–Cδ3a–Cδ4a–Cδ2b–Cδ7, seems to be the result of a first duplication of Cδ2–Cδ4 followed by deletion of the Cδ3–Cδ6 domains (Hansen et al. 2005). In other species, including Japanese flounder (Olive flounder) and three-spined stickleback, there is no Cδ duplication (Bengtén and Wilson 2015; Hirono et al. 2003). Interestingly, in catfish and takifugu, there are two different genes that encode the membrane IgD and the secreted IgD (Aparicio et al. 2002; Bengtén et al. 2002; Bengtén and Wilson 2015; Edholm et al. 2010). Based on these studies, we can speculate that the different IgD structures observed in a variety of species may have specific effector functions by adapting to the specific immune environments.
Different number of CH exons have been also identified in IGHT genes: four CH exons are found in most species (Danilova et al. 2005; Hansen et al. 2005; Yasuike et al. 2010), whereas the three-spined stickleback has three (Bao et al. 2010; Gambón-Deza et al. 2010) and fugu two (Aparicio et al. 2002). In carp (Cyprinus carpio), the secreted IgT/Z appears to be a chimera with a variable region and two constant domains, a Cμ1 domain and a Cτ/ζ domain (Savan et al. 2005). While the number of C domains differ among species, it is worthy to note that Cτ/ζ1 (Cμ1 in the carp IgT/chimera) and Cτ/ζ4 seem to be conserved (Bradshaw and Valenzano 2020; Gambón-Deza et al. 2010). If additional C domains may provide different functional proprieties, is a question to be resolved.
Alternative splicing of pre-mRNAs may contribute to an additional level of IGH isotypic variation in fish. In mice and humans, membranes versus secreted IgM H chains are typically produced from the same pre-mRNA through alternative splicing. A secreted Igμ transcript is produced when the mRNA is cleaved and polyadenylated between the constant region domain Cμ4 and the TM exons, and a membrane Igμ transcript is made if a cryptic splice site located within Cμ4 is spliced to the acceptor site of the TM1 exon. In fish, membrane Ig μ transcripts have the TM exons spliced to the donor site located at the 3′ end of the Cμ3 exon, hence lack the last Cμ domain that is present in the secreted IgM forms (Hikima et al. 2011). Exceptions to this rule have been found in different species, including Antarctic Notothenioids fish (Coscia et al. 2010). Transcripts which encode transmembrane IgM with only one or two CH domains have been reported in zebrafish (Hu et al. 2011) and medaka (Magadán-Mompó et al. 2011).
7.2.2 IGL Loci
To date, in all studied vertebrate species, with the exception of chickens, ducks, and snakes, more than one immunoglobulin light chain isotype can be found (Das et al. 2010; Gambón-Deza et al. 2012; Lundqvist et al. 2006). The conventional classification of IGL into kappa and lambda isotypes was initially designated as a means to classify mammalian IgL (Edelman and Poulik 1961). Over the years, additional isotypes have been described, and the classification system groups all vertebrate IGLs into four main ancestral branches: kappa (mammalian κ, elasmobranch type III/NS4, teleost L1/L3/F/G, Xenopus ρ), lambda (mammalian λ, elasmobranch type II/NS3), sigma (Xenopus σ, teleost L2, elasmobranch type IV), and sigma-2 (elasmobranch type I/NS5, variant sigma-type in coelacanth). However, this classification can be modified when new vertebrate genomes are analyzed. A new clustering was proposed by Guselnikov et al. (2018), concluding that there were five ancient IGL isotypes (kappa, lambda, lambda-2, sigma, and sigma-2) which evolved differentially in various lineages of jawed vertebrates.
At first, it was very unclear to establish a relationship between teleost IgL isotypes and the mammalian κ and λ chains so, the nomenclature found in the literature is diverse and in some bony fish species they were named in order of discovery. Finally, a comprehensive phylogenetic analysis of different vertebrate VL and CL regions suggests that previously named L1 and L3 chains are κ orthologous (Criscitiello and Flajnik 2007) and the teleost L2 immunoglobulin light chains are σ orthologous (Partula et al. 1996). According to this, the teleost IGLs have been subdivided into σ-like (previous L2) and two groups of κ-like chains (L1/κG and L3/κF). The lambda chain isotype was thought to be lost in the teleosts until its characterization in channel catfish, Atlantic cod, and rainbow trout (Edholm et al. 2009). Furthermore, a recent phylogenetic analysis of extended IGL datasets indicates that teleostean lambda orthologs actually represent a distinct isotype designated as lambda-2 (Guselnikov et al. 2018). This isotype is also present in holostean and polypterid fish, which suggests its emergence before the radiation of ray-finned fish (see Table 7.1).
To date, the κ, λ (or λ-2), and σ isotypes identified in teleosts have been found to exist on different chromosomes in a cluster assemblage, it means multiple VL-JL-CL units (Bao et al. 2010; Daggfeldt et al. 1993; Edholm et al. 2011; Zimmerman et al. 2011, 2008). In a genome wide study, such clusters have been found in five different chromosomes in zebrafish (Zimmerman et al. 2011). Interestingly, recombination between clusters was reported in this species, which might offer a greater potential combinatorial diversity. A characterization of the fugu IGL genomic loci and southern blot analysis has shown a minimum of 12 clusters (classified as kappa and sigma) spread on three different chromosomes (Fu et al. 2017). Interestingly, a recent genomic analysis using nine rainbow trout lines revealed that kappa, lambda, and sigma light chain isotypes are present in all trout lines studied, with highly conserved constant region nucleotide sequences (Rego et al. 2020b). However, the authors detected differences between the trout lines in the number and size of clusters (mainly sigma-like) that could affect humoral immune responses. In addition, the presence of multiple clusters on one or more chromosomes also indicates that cluster duplication and expansions likely played a major role in the generation of antibody diversity in teleost fishes.
Regardless of the diversity in number or length of teleost IGL loci, the genomic organization is almost conserved. In the kappa locus (L1 and L3), the V segments are in opposite transcriptional orientation as for the J and C segments, which implies that the VJ arrangement happens rather by inversion than deletion. While the sigma (L2) and lambda gen clusters present V segments in same orientation. One exception is the stickleback (Bao et al. 2010), with σ clusters in which the 3′VL segment is in opposite polarity. This difference in genomic configuration would imply diverse mechanisms for generating antibody diversity.
7.3 Immunoglobulin Repertoires in Teleost
An organism is exposed to a huge diversity of antigens, and the generation of different IG receptors required to specifically recognize them is the result of multiple processes that happen before and after antigen recognition. The germline structure of IGH and IGL loci provides the basic pieces from which the IG is assembled, and the diversity is generated. The theoretical diversity of the IG repertoire is a result of a number of different stages in the development of the B cell, and of the gene segments selection and assembly mechanisms therein. Briefly, the IG loci are subject to genomic rearrangements of variable (V), joining (J), diversity (D) gene segments, which along with the allelic exclusion, lead to the expression of a unique antigen receptor by each lymphocyte (Hozumi and Tonegawa 1976; Tonegawa 1983). The V(D)J junction is not exact, and the deletion and insertion of nucleotides at the joint region are commonly observed. This process, along with the pairing of heavy and light chains forming the IG, results in a vast repertoire of B cells bearing structurally diverse receptors (potentially >108 sequences in zebrafish, and >1013 in humans) for specific antigen recognition. During the immune response, each lymphocyte produces a clone of cells specific to one antigenic motif and expressed IG receptors may diversify further by the process of gene hypermutation. Thus, the collection of IG sequences within an organism, also known as the immunoglobulin repertoire, reflect the current state of the humoral adaptive immune system of an animal. Moreover, the changes generated in response to external (i.e., vaccines or pathogens) or internal (i.e., aging) immune challenges, are also depicted.
The development of high-throughput sequencing protocols to analyze the immunoglobulin rearrangements expressed in a lymphocyte population overcomes the previous technical limitations that only allowed the analysis of small fractions of B-lineage lymphocytes (Chaudhary and Wesemann 2018; Pabst et al. 2015; Turchaninova et al. 2016). Sequencing of adaptive immune receptor repertoires (IGs and TCRs) may elucidate the highly complex adaptive immune response in teleost. The analysis of immune repertoire sequences and their quantitative composition on a nucleotide-level within and across individuals shed light on the dynamics of adaptive immune cells in healthy conditions or during an immune response.
The analysis of IG repertoire by Weinstein et al. (2009) reported that in adult naive zebrafish, where the total number of antibody-producing B cells are less than half million (a small number compared to an estimated 1012 cells in humans), a large portion of potential VDJ rearrangements is indeed present in the available IgM repertoire. The number of unique VDJ sequences for IgM were estimated between 1200 and 3500 per fish, most of them were presented at low frequency, and a few of them were found to be highly abundant. Using the same animal model and sequencing technology Jiang et al. (2011), described the structure of zebrafish IG repertoire across development. At early stage (2 weeks old fish), authors found a highly stereotyped state with preferential use of a small number of V, D, and J gene segment combinations, and frequent VDJ combinations shared between different fish. This stereotypy decreases dramatically as the zebrafish mature. Authors suggest that the main process that causes differentiation between adult repertoires is apparently random clonal expansion. In both studies, RNA was obtained from full organisms, but results should be treated with caution as it cannot be stated that the complete IG repertoire was characterized. Thus, RNA capture efficiency, sequencing depth (the platform used was 454 sequencing), the accuracy of IGH locus annotation, and the fact that there are no data about IGL chain or IGHD chain data, and further efforts are required to achieve this challenge.
Deeper sequencing platforms (mainly Illumina technology) with outputs ranging from 15Gb and 25 million reads (Illumina MiSeq) to 6000Gb and 20 billion reads (NovaSeq) are currently available. This technology is applied to decipher the humoral adaptive immune response in important species for aquaculture, such as Atlantic salmon, rainbow trout, and takifugu (Castro et al. 2013; Fu et al. 2018; Krasnov et al. 2017; Magadan et al. 2018). A common feature is the presence of identical heavy chain sequences among several fishes, as described in mouse or human (Galson et al. 2015). This observation indicates that repertoires are not simply determined by equally likely random rearrangements of IG V(D)J gene segments. Thus, certain receptors might be shared between unchallenged controls simply due to their high generation probability.
The analysis of IG repertoires from virus-immunized rainbow trout showed modifications that persist at least for 5 months, with expanded public and private clonotypes (Castro et al. 2013; Magadan et al. 2018). Analysis of trout public responses suggested that repeated immunizations with a heterologous antigen do not lead to a significant attrition of pre-existing responding B cells specific for the primary vaccination (Navelsaker et al. 2019). Same technology was recently applied to characterize the nasal B-cell repertoire in rainbow trout, in which well-organized mucosal-associated lymphoid structures such as tonsils and adenoids are absent (Sepahi and Salinas 2016; Tacchi et al. 2014). In rainbow trout, nasal IgM and IgT repertoires comprise both low frequency and highly expanded clonotypes, being the nasal IgT repertoire dominated by expanded clonotypes Magadan et al. 2019a, b). Interestingly, these characteristics are reminiscent of those described for IgM and IgA mucosal repertoires in humans and mice (Holtmeier 2000; Lindner et al. 2012, 2015).
New teleost genome assemblies are coming, and they will provide a rich source of knowledge for the comparative immunology community, to address the mechanisms of gene conversion, somatic hypermutation, and memory in these species and during vertebrate evolution. In this sense, data available for Atlantic salmon have been recently used to establish a unique and consistent standardized nomenclature of salmonid IGH genes. These efforts led to the elaboration of a sequence reference directory at the international ImMunoGeneTics information system (IMGT, www.imgt.org) that allows the accurate annotation of rainbow trout and Atlantic salmon IG deep sequencing data (Magadan et al. 2019b). However, the full assembly and annotation of IG remain challenging, and more efforts are required to improve accuracy of annotation of adaptive immune receptor repertoire dataset generated by high-throughput sequencing, and to facilitate comparisons between studies and species. Furthermore, most bioinformatics tools for decoding immune receptor repertoires data have been developed for human and mice, and they have to be adapted to other models of interest, such as teleost.
7.4 Teleost Ig Classes: An Overview on Functional Roles
Compared to mammals, a smaller number of antibody classes have been reported in lower vertebrates. In cartilaginous fish, three heavy chain isotypes have been detected, which lead three IG classes: IgM, IgD/W, and IgNAR (Bengtén and Wilson 2015). In teleost fish, three different Ig classes have been identified: IgM, IgD, and IgT/Z. However, not every class is expressed in each fish species studied (Bengtén and Wilson 2015; Fillatreau et al. 2013). Furthermore, the antibody classes expressed in the few extant species of lobe-finned bony fish seem to be different from those found in ray-finned bony fish. No IgT/Z has been identified in the African lungfish (Protopterus annectens and Protopterus aethiopicus), and both IgM and IgT/Z seem to be absent in the African coelacanth (Latimeria chalumnae) (Amemiya et al. 2013; Zhang et al. 2014).
7.4.1 IgM
The IgM class has long been considered the most ancient and was the first Ig class identified in fish. It can be expressed at the surface of B cells or secreted. Secreted tetrameric IgM is the most prevalent serum Ig in fish (Flajnik 2018, 2002), with a concentration between 800 and 9000 μg/mL (Uchida et al. 2000). While in mammals the IgM is mainly secreted as a pentameric form, teleost IgM is a tetramer apparently formed in the absence of the Ig joining (J) chains. These IgM tetramers are secreted at systemic and mucosal level (Elcombe et al. 1985). They are found in different oxidation states that depend on the degree of interdisulfide bond formation among the IgM monomers (Kaattari et al. 1998; Ye et al. 2013). Variability in the degree of inter-heavy chain disulfide polymerization has been observed in more than 15 species (Bromage et al. 2004; Dacanay et al. 2006; Kaattari et al. 1998; Morrison and Nowak 2001; Uchida et al. 2000).
In rainbow trout, the simultaneous structural analyses of mucosal and systemic antibodies (Bromage et al. 2006), suggested that mucosal IgM has considerably less disulfide polymerization than serum IgM. Additional studies also reported that antigen-binding affinity and the Ig half-life are associated with levels of polymerization (Kaattari et al. 2002; Ye et al. 2010). Via an in vitro antigen-driven selection experiment, Ye et al. (2011) shown that the BCR affinity for the inducing antigen affects to post-translational modification that results in both increased polymerization and glycosylation. These authors proposed an integrated model where the antigen-sensitive B lymphocyte operates as an affinity-based transducer, enabling each B cell to modulate the structure of its antibody in order to optimally suit its function with its affinity for the inducing antigen (Ye et al. 2011). The effector functions activated by fish IgM include complement activation, antibody-dependent cellular cytotoxicity (ADCC), and phagocytosis through effector cells (Lobb and Hayman 1989; Mashoof and Criscitiello 2016; Shen et al. 2003). Several studies also reported serum and mucosal noncovalently associated IgM monomers in different fish species, such as the grouper, Epinephelus itaira, the sheepshead, Archosargus probatocephalus, and rainbow trout, O. mykiss (Clem 1971; Clem and McLean 1975; Lobb and Clem 1981; Lobb and William Clem 1981). However, further research is required to shed light on monomeric IgM functionality.
7.4.2 IgT
In 2005, a new teleost fish immunoglobulin isotype was identified. In rainbow trout, this Ig was named IgT (Hansen et al. 2005), whereas in zebrafish it was called IgZ (Danilova et al. 2005). This Ig has been identified in most of studied teleost fish, except in medaka (Magadán-Mompó et al. 2011), in channel catfish (Bengtén et al. 2006), and in turquoise killifish (Bradshaw and Valenzano 2020). The recent analysis of genomes from 10 species belonging to Cyprinodontiformes order also suggests the absence of IGHZ/T exons in A. austral or N. orthonotus (Bradshaw and Valenzano 2020). These results need further confirmation when higher-quality genome sequences from these two species are available.
Most research on IgT has been developed in rainbow trout. In this host, at least three subclasses are expressed (Zhang et al. 2017). At transcriptional level, IgT1 subclass is expressed both in mucosal and systemic lymphoid tissues; IgT2 seems to be mainly expressed in systemic lymphoid organs. Zhang et al. (2017) reported IgT3 protein in rainbow trout serum level. Previous studies performed by Oriol Sunyer and collaborators (Zhang et al. 2010), provided a detailed characterization of rainbow trout IgT and revealed its function in mucosal immunity. It was shown that while plasma IgT is a monomeric Ig (~180 kDa), gut mucus IgT is polymeric (4–5 monomers) formed in absence of J chain. In the same study, a previously unknown IgT+ IgM- IgD- B-cell lineage was identified. This B-cell subset constitutes the first vertebrate B-cell lineage devoid of surface IgD expression. In addition, authors identified a polymeric Ig receptor in rainbow trout (tpIgR) whose putative secretory component was found associated with gut mucus but not serum IgT and IgM (Zhang et al. 2010). This result strongly suggested that like in mammals, pIgR in fish is involved in the transport of polymeric IgT and IgM from the mucosal epithelium into the gut lumen.
Although IgA is absent in lower-jawed vertebrates, a series of studies showed that amphibians and bony fish express specialized mucosal antibody isotypes (IgX in amphibian, and IgT/Z in bony fish), independently, by convergent evolution (Du et al. 2012; Mussmann et al. 1996). In this sense, IgT represents the most ancient vertebrate mucosal Ig identified to date. Its concentration in serum is much lower, approximately 1000 times less, than that of IgM, but in mucus the IgT/IgM ratio is almost 100 times higher than in serum. Results reported in different studies show that secreted IgT is highly induced at mucosal surfaces by pathogens and vaccines (Buchmann 2020; Magadan et al. 2019a, b; Tacchi et al. 2014; Xu et al. 2013, 2016; Yu et al. 2018; Zhang et al. 2010). In addition, secreted IgT seems to be the predominant Ig isotype coating a large portion of the fish microbiota (Xu et al. 2016, 2020). A recent study, Xu et al. (2020) have shed more light on the specialization of secreted Igs in protection of mucosal tissues from pathogens and, in the establishment of healthy microbiota. To address this question, an elegant teleost fish (rainbow trout) model was developed, in which the secreted IgT in adult fish was selectively depleted with the injection of IgT monoclonal antibodies and rainbow trout antiserum against mice IgG. Then, authors performed the analysis of microbiome composition and parasite infections, to evaluate the specific contribution of IgT on mucosal tissue homeostasis, and in the protection of fish mucosal tissues against pathogens, respectively. In rainbow trout, the IgT depletion induced the loss of IgT-coated beneficial taxa, expansion of pathobionts, tissue damage, and inflammation. In addition, an increased susceptibility of fish to the mucosal parasite Ichthyophthirius multifiliis, without developing compensatory IgM responses, was observed. These results support further the notion that IgT and IgA are phylogenetically distant immunoglobulins that specialized in mucosal immune responses and reveal the existence of primordially conserved principles by which mucosal immunoglobulins control both pathogens and microbiota. Moreover, in mammals secreted IgA seems to be less essential in control of mucosal pathogens than IgT in rainbow trout. In patients with selective-secreted IgA deficiency, there is a modest increased susceptibility to respiratory and intestinal infections (Yel 2010). In mammals, the class switch recombination process, which is not present in fish, may allow different class antibodies (i.e., IgM, IgD, and IgA) to share their antigen-binding sites. So, in IgA-deficient individuals, there may be compensatory mechanisms mediated by IgM or IgD that would prevent disease manifestations in some IgA-deficient individuals (Choi et al. 2017; Yel 2010).
7.4.3 IgD
IgD was first discovered in human serum in 1965, and it was considered a recently evolved Ig class expressed in mammals. However, IgD was found in other vertebrates, including teleost, suggesting it may be as old as IgM (Mashoof and Criscitiello 2016; Ohta and Flajnik 2006; Wilson et al. 1997). In 1997, bony fish IgD was identified in the channel catfish, as Ig with a chimeric heavy chain containing a rearranged variable domain, the first constant domain of mu (Cμ1), and seven constant domains encoded by a delta gene (Cδ) (Wilson et al. 1997). A similar splicing pathway was retrieved in Atlantic salmon (Hordvik 2002; Hordvik et al. 1999), and in other fish species (Gambón-Deza et al. 2010; Susana Magadán-Mompó et al. 2011; Stenvik and Jørgensen 2000; Wang et al. 2016). The chimeric Cμ1-Cδ structure has been also described in pigs and other Artiodactyls (Zhao et al. 2002, 2003) and may facilitate the formation of a covalent bond with the light chain. Fish IgD differs from eutherian IgD by a large number (7–17) of Cδ domains and by the absence of a hinge. In contrast to IgM, IgD displays marked structural plasticity, which can be explained by different underlying mechanisms, such as diverse copy number of IgD-codifying gene, different number of germline Cδ exons, and the expression of various splicing forms.
The specific functions of IgD have only recently begun to be elucidated (Gutzeit et al. 2018). Interestingly, secreted form of IgD has been identified in mammals, catfish, and trout, and its structure is different (Bengtén et al. 2006; Mountz et al. 1990; Ramirez-Gomez et al. 2012). In humans, secreted IgD can be detected in the circulation, nasopharyngeal, oral and lachrymal secretions (Chen et al. 2009; Gutzeit et al. 2018; Sun et al. 2020). Moreover, in the upper respiratory tract, the secreted IgD binds to basophils and mast cells, and activates these cells to produce antimicrobial, opsonizing, proinflammatory factors. The cross-linking of basophil-bound IgD by an isotype-specific antibody triggers the IL4 cytokine and thereafter the stimulation of B cells (Chen et al. 2009). These results suggest that IgD orchestrates an immune surveillance system at the interface between innate and adaptive immune systems. In 2010, Edholm et al. (2010) found that all catfish-secreted IgD transcripts from IgM+/IgD+ and IgM−/IgD+ B cells lacked the variable domain, and began with a leader spliced to Cδ1. The authors concluded that IgM−/IgD+ B cells most likely expand in response to certain pathogens, and the secreted V-less IgD Fc region may function as an innate pattern recognition molecule. These results also support the dual nature of the IgD molecule, which may represent an early transition of the innate pattern recognition receptors to adaptive Ig molecules.
In contrast to what was found in catfish, rainbow trout-secreted IgD is expressed with a V domain (Ramirez-Gomez et al. 2012). The rainbow trout-secreted IgD transcripts were found at the highest levels in the spleen, followed by the blood and gills (Ramirez-Gomez et al. 2012). Moreover, rainbow trout-secreted IgD has been shown to coat gill microbiota, albeit at a significantly lower level than secreted IgT, and seems to have the ability to interact with the pIgR, which is required for its mucosal transport to the gill. This is the first description of a pIgR being implicated in IgD secretion in a vertebrate respiratory tract (Xu et al. 2016).
7.5 Conclusion
With very different lymphoid anatomical structures and microenvironments, teleost and mammals provide a very good subject for comparative approaches to distinguish fundamental conserved properties of adaptive immunity and convergent adaptations driven by the necessity to fight pathogens in critical tissues or at entry points. With the development of new reagents and deep sequencing technologies, there has been substantial advancement in our knowledge on the structural characteristics and effector functions of immunoglobulins present in teleost. We get more insight into the plasticity of teleost IG gene loci and repertoires. Comparative studies will provide important insights about the interplay between environment and the evolution of the adaptive immune system in vertebrates.
Abbreviations
- IG:
-
Immunoglobulin
- TCR:
-
T-cell receptor
- BCR:
-
B-cell receptor
- IGH:
-
Immunoglobulin Heavy chain
- IGL:
-
Immunoglobulin Light chain
- C:
-
Constant
- V:
-
Variable
References
Amemiya CT, Alfoldi J, Lee AP, Fan S, Philippe H, MacCallum I, Braasch I, Manousaki T, Schneider I, Rohner N, Organ C, Chalopin D, Smith JJ, Robinson M, Dorrington RA, Gerdol M, Aken B, Biscotti MA, Barucca M, Baurain D, Berlin AM, Blatch GL, Buonocore F, Burmester T, Campbell MS, Canapa A, Cannon JP, Christoffels A, De Moro G, Edkins AL, Fan L, Fausto AM, Feiner N, Forconi M, Gamieldien J, Gnerre S, Gnirke A, Goldstone JV, Haerty W, Hahn ME, Hesse U, Hoffmann S, Johnson J, Karchner SI, Kuraku S, Lara M, Levin JZ, Litman GW, Mauceli E, Miyake T, Mueller MG, Nelson DR, Nitsche A, Olmo E, Ota T, Pallavicini A, Panji S, Picone B, Ponting CP, Prohaska SJ, Przybylski D, Saha NR, Ravi V, Ribeiro FJ, Sauka-Spengler T, Scapigliati G, Searle MJ, Sharpe T, Simakov O, Stadler PF, Stegeman JJ, Sumiyama K, Tabbaa D, Tafer H, Turner-Maier J, Van Heusden P, White S, Williams L, Yandell M, Brinkmann H, Volff JN, Tabin CJ, Shubin N, Schartl M, Jaffe DB, Postlethwait JH, Venkatesh B, Di Palma F, Lander ES, Meyer A, Lindblad-Toh K (2013) The African coelacanth genome provides insights into tetrapod evolution. Nature 496:311–316. https://doi.org/10.1038/nature12027
Aparicio S, Chapman J, Stupka E, Putnam N, Ming CJ, Dehal P, Christoffels A, Rash S, Hoon S, Smit A, Sollewijn Gelpke MD, Roach J, Oh T, Ho IY, Wong M, Detter C, Verhoef F, Predki P, Tay A, Lucas S, Richardson P, Smith SF, Clark MS, Edwards JK, Doggett N, Zharkikh A, Tavtigian SV, Pruss D, Barnstead M, Evans C, Baden H, Powell J, Glusman G, Rowen L, Hood L, Tan YH, Elgar G, Hawkins T, Venkatesh B, Rokhsar D, Brenner S (2002) Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297:1301–1310. https://doi.org/10.1126/science.1072104
Bao Y, Wang T, Guo Y, Zhao Z, Li N, Zhao Y (2010) The immunoglobulin gene loci in the teleost Gasterosteus aculeatus. Fish Shellfish Immunol 28:40–48. https://doi.org/10.1016/j.fsi.2009.09.014
Bengtén E, Wilson M (2015) Antibody repertoires in fish: results and problems in cell differentiation. Springer, pp 193–234. https://doi.org/10.1007/978-3-319-20819-09
Bengtén E, Quiniou MA, Stuge TB, Katagiri T, Miller NW, Clem LW, Warr GW, Wilson M (2002) The IgH locus of the channel catfish Ictalurus punctatus contains multiple constant region gene sequences: different genes encode heavy chains of membrane and secreted IgD. J Immunol 169:2488–2497. https://doi.org/10.4049/jimmunol.169.5.2488
Bengtén E, Clem LW, Miller NW, Warr GW, Wilson M (2006) Channel catfish immunoglobulins: repertoire and expression. Dev Comp Immunol 30:77–92. https://doi.org/10.1016/j.dci.2005.06.016
Bradshaw WJ, Valenzano DR (2020) Extreme genomic volatility characterizes the evolution of the immunoglobulin heavy chain locus in cyprinodontiform fishes. Proc R Soc B Biol Sci 287:20200489. https://doi.org/10.1098/rspb.2020.0489
Bromage ES, Ye J, Owens L, Kaattari IM, Kaattari SL (2004) Use of staphylococcal protein A in the analysis of teleost immunoglobulin structural diversity. Dev Comp Immunol 28:803–814. https://doi.org/10.1016/j.dci.2003.12.001
Bromage ES, Ye J, Kaattari SL (2006) Antibody structural variation in rainbow trout fluids. Comp. Biochem. Physiol. Part B Biochem. Mol Biol 143:61–69. https://doi.org/10.1016/j.cbpb.2005.10.003
Buchmann K (2020) Immune response to Ichthyophthirius multifiliis and role of IgT. Parasite Immunol 42:e12675. https://doi.org/10.1111/pim.12675
Castro R, Jouneau L, Pham HP, Bouchez O, Giudicelli V, Lefranc MP, Quillet E, Benmansour A, Cazals F, Six A, Fillatreau S, Sunyer O, Boudinot P (2013) Teleost fish mount complex clonal IgM and IgT responses in spleen upon systemic viral infection. PLoS Pathog 9(1):e1003098. https://doi.org/10.1371/journal.ppat.1003098
Chaudhary N, Wesemann DR (2018) Analyzing immunoglobulin repertoires. Front Immunol 9:1–18. https://doi.org/10.3389/fimmu.2018.00462
Chen K, Xu W, Wilson M, He B, Miller NW, Bengtén E, Edholm ES, Santini PA, Rath P, Chiu A, Cattalini M, Litzman J, Bussel JB, Huang B, Meini A, Riesbeck K, Cunningham-Rundles C, Plebani A, Cerutti A (2009) Immunoglobulin D enhances immune surveillance by activating antimicrobial proinflammatory and B cell-stimulating programs in basophils. Nat Immunol 10:889–898. https://doi.org/10.1038/ni.1748
Choi JH, Wang K, Zhang D, Zhan Xiaowei Wang T, Bu CH, Behrendt CL, Zeng M, Wang Y, Misawa T, Li X, Tang M, Zhan X, Scott L, Hildebrand S, Murray AR, Moresco MY, Hooper LV, Beutler B (2017) IgD class switching is initiated by microbiota and limited to mucosa-associated lymphoid tissue in mice. Proc Natl Acad Sci U S A 114:E1196–E1204. https://doi.org/10.1073/pnas.1621258114
Clem LW (1971) Phylogeny of immunoglobulin structure and function. IV. Immunoglobulins of the giant grouper Epinephelus itaira. J Biol Chem 246:9–15
Clem LW, McLean WE (1975) Phylogeny of immunoglobulin structure and function. VII. Monomeric and tetrameric immunoglobulins of the margate a marine teleost fish. Immunol Commun 29:791–799
Cooper MD, Alder MN (2006) The evolution of adaptive immune systems. Cell 124:815–822. https://doi.org/10.1016/j.cell.2006.02.001
Coscia MR, Varriale S, De Santi C, Giacomelli S, Oreste U (2010) Evolution of the Antarctic teleost immunoglobulin heavy chain gene. Mol Phylogenet Evol 55:226–233. https://doi.org/10.1016/j.ympev.2009.09.033
Criscitiello MF, Flajnik MF (2007) Four primordial immunoglobulin light chain isotypes including λ and κ identified in the most primitive living jawed vertebrates. Eur J Immunol 37:2683–2694. https://doi.org/10.1002/eji.200737263
Dacanay A, Bentley BE, Brown LL, Roberts AJ, Johnson SC (2006) Unique multimeric immunoglobulin crosslinking in four species from the family Gadidiae. Fish Shellfish Immunol 21:215–219. https://doi.org/10.1016/j.fsi.2005.11.004
Daggfeldt A, Bengtén E, Pilström L (1993) A cluster type organization of the loci of the immunoglobulin light chain in Atlantic cod (Gadus morhua L.) and rainbow trout (Oncorhynchus mykiss Walbaum) indicated by nucleotide sequences of cDNAs and hybridization analysis. Immunogenetics 38:199–209. https://doi.org/10.1007/BF00211520
Danilova N, Bussmann J, Jekosch K, Steiner LA (2005) The immunoglobulin heavy-chain locus in zebrafish: identification and expression of a previously unknown isotype immunoglobulin Z. Nat Immunol 6:295–302. https://doi.org/10.1038/ni1166
Das S, Mohamedy U, Hirano M, Nei M, Nikolaidis N (2010) Analysis of the immunoglobulin light chain Genes in zebra finch: evolutionary implications. Mol Biol Evol 27:113–120. https://doi.org/10.1093/molbev/msp212
Du CC, Mashoof SM, Criscitiello MF (2012) Oral immunization of the African clawed frog (Xenopus laevis) upregulates the mucosal immunoglobulin IgX. Vet Immunol Immunopathol 145:493–498. https://doi.org/10.1016/j.vetimm.2011.10.019
Edelman GM, Poulik MD (1961) Studies on structural units of the gamma-globulins. J Exp Med 113:861–884. https://doi.org/10.1084/jem.113.5.861
Edholm ES, Wilson M, Sahoo M, Miller NW, Pilström L, Wermenstam NE, Bengtén E (2009) Identification of Igσ and Igλ in channel catfish Ictalurus punctatus and Igλ in Atlantic cod Gadus morhua. Immunogenetics 61:353–370. https://doi.org/10.1007/s00251-009-0365-z
Edholm ES, Bengtén E, Stafford JL, Sahoo M, Taylor EB, Miller NW, Wilson M (2010) Identification of two IgD + B cell populations in channel catfish Ictalurus punctatus. J Immunol 185:4082–4094. https://doi.org/10.4049/jimmunol.1000631
Edholm ES, Wilson M, Bengten E (2011) Immunoglobulin light (IgL) chains in ectothermic vertebrates. Dev Comp Immunol 35:906–915. https://doi.org/10.1016/j.dci.2011.01.012
Elcombe BM, Chang RJ, Taves CJ, Winkelhake JL (1985) Evolution of antibody structure and effector functions: comparative hemolytic activities of monomeric and tetrameric IgM from rainbow trout Salmo gairdnerii. Comp Biochem Physiol Part B Comp Biochemist 80:697–706. https://doi.org/10.1016/0305-0491(85)90448-1
Fillatreau S, Six A, Magadan S, Castro R, Sunyer JO, Boudinot P (2013) The astonishing diversity of Ig classes and B-cell repertoires in teleost fish. Front Immunol 4:28. https://doi.org/10.3389/fimmu.2013.00028
Flajnik MF (2002) Comparative analyses of immunoglobulin genes: surprises and portents. Nat Rev Immunol 2:688–698. https://doi.org/10.1038/nri889
Flajnik MF (2018) A cold-blooded view of adaptive immunity. Nat Rev Immunol 18:435–453. https://doi.org/10.1038/s41577-018-0003-9
Flajnik MF, Du Pasquier L (2004) Evolution of innate and adaptive immunity: Can we draw a line? Trends Immunol 25:640–644. https://doi.org/10.1016/j.it.2004.10.001
Fricke R, Eschmeyer WN, van der Laan R (eds) (2020) Eschmeyer’s catalog of fishes: genera, species, references. http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp
Fu X, Zhang H, Tan E, Watabe S, Asakawa S (2015) Characterization of the torafugu (Takifugu rubripes) immunoglobulin heavy chain gene locus. Immunogenetics 67:179–193. https://doi.org/10.1007/s00251-014-0824-z
Fu X, Zhang F, Watabe S, Asakawa S (2017) Immunoglobulin light chain (IGL) genes in torafugu: genomic organization and identification of a third teleost IGL isotype. Sci Rep 7:40416. https://doi.org/10.1038/srep40416
Fu X, Sun J, Tan E, Shimizu K, Reza MS, Watabe S, Asakawa S (2018) High-throughput sequencing of the expressed Torafugu (Takifugu rubripes) antibody sequences distinguishes IgM and IgT repertoires and reveals evidence of convergent evolution. Front Immunol 9. https://doi.org/10.3389/fimmu.2018.00251
Galson JD, Trück J, Fowler A, Münz M, Cerundolo V, Pollard AJ, Lunter G, Kelly DF (2015) In-depth assessment of within-individual and inter-individual variation in the B-cell receptor repertoire. Front Immunol 6:531. https://doi.org/10.3389/fimmu.2015.00531
Gambón-Deza F, Sánchez-Espinel C, Magadán-Mompó S (2010) Presence of an unique IgT on the IGH locus in three-spined stickleback fish (Gasterosteus aculeatus) and the very recent generation of a repertoire of VH genes. Dev Comp Immunol 34:114–122. https://doi.org/10.1016/j.dci.2009.08.011
Gambón-Deza F, Sánchez-Espinel C, Mirete-Bachiller S, Magadán-Mompó S (2012) Snakes antibodies. Dev Comp Immunol 38:1–9. https://doi.org/10.1016/j.dci.2012.03.001
Ghaffari SH, Lobb CJ (1993) Structure and genomic organization of immunoglobulin light chain in the channel catfish: An unusual genomic organizational pattern of segmental genes. J Immunol 151:6900–6912
Ghaffari SH, Lobb CJ (1999) Structure and genomic organization of a second cluster of immunoglobulin heavy chain gene segments in the channel catfish. J Immunol 162:1519–15129
Guselnikov SV, Baranov KO, Najakshin AM, Mechetina LV, Chikaev NA, Makunin AI, Kulemzin SV, Andreyushkova DA, Stöck M, Wuertz S, Gessner J, Warren WC, Schartl M, Trifonov VA, Taranin AV (2018) Diversity of immunoglobulin light chain genes in non-teleost ray-finned fish uncovers IgL subdivision into five ancient isotypes. Front Immunol 9:1079. https://doi.org/10.3389/fimmu.2018.01079
Gutzeit C, Chen K, Cerutti A (2018) The enigmatic function of IgD: some answers at last. Eur J Immunol 48:1101–1113. https://doi.org/10.1002/eji.201646547
Hansen JD, Landis ED, Phillips RB (2005) Discovery of a unique Ig heavy-chain (IgT) in rainbow trout: implications for a distinctive B-cell developmental pathway in teleost fish. Proc Natl Acad Sci U S A 102:6919–6924. https://doi.org/10.1073/pnas.0500027102
Hikima J, Jung TS, Aoki T (2011) Immunoglobulin genes and their transcriptional control in teleosts. Dev Comp Immunol 35:924–936. https://doi.org/10.1016/j.dci.2010.10.011
Hirono I, Nam BH, Enomoto J, Uchino K, Aoki T (2003) Cloning and characterisation of a cDNA encoding Japanese flounder Paralichthys olivaceus IgD. Fish Shellfish Immunol 15:63–70. https://doi.org/10.1016/S1050-4648(02)00139-0
Holtmeier W (2000) IgA and IgM VH repertoires in human colon: evidence for clonally expanded B cells that are widely disseminated. Gastroenterology 119:1253–1266. https://doi.org/10.1053/gast.2000.20219
Hordvik I (2002) Identification of a novel immunoglobulin δ transcript and comparative analysis of the genes encoding IgD in Atlantic salmon and Atlantic halibut. Mol Immunol 39:85–91. https://doi.org/10.1016/S0161-5890(02)00043-3
Hordvik I, Thevarajan J, Samdal I, Bastani N, Krossøy B (1999) Molecular cloning and phylogenetic analysis of the Atlantic salmon immunoglobulin D gene. Scand J Immunol 50:202–210. https://doi.org/10.1046/j.1365-3083.1999.00583.x
Hozumi N, Tonegawa S (1976) Evidence for somatic rearrangement of immunoglobulin genes coding for variable and constant regions. Proc Natl Acad Sci U S A 73:3628–3632. https://doi.org/10.1073/pnas.73.10.3628
Hu YL, Zhu LY, Xiang LX, Shao JZ (2011) Discovery of an unusual alternative splicing pathway of the immunoglobulin heavy chain in a teleost fish Danio rerio. Dev Comp Immunol 35:253–257. https://doi.org/10.1016/j.dci.2010.10.009
Jiang N, Weinstein JA, Penland L, White RA, Fisher DS, Quake SR (2011) Determinism and stochasticity during maturation of the zebrafish antibody repertoire. Proc Natl Acad Sci 108:5348–5353. https://doi.org/10.1073/pnas.1014277108
Kaattari S, Evans D, Klemer J, Kaattari S, Evans D, Kieraer J (1998) Varied redox forms of teleost IgM: an alternative to isotypic diversity? Immunol Rev 166:133–142. https://doi.org/10.1111/j.1600-065X.1998.tb01258.x
Kaattari SL, Zhang HL, Khor IW, Kaattari IM, Shapiro DA (2002) Affinity maturation in trout: clonal dominance of high affinity antibodies late in the immune response. Dev Comp Immunol 26:191–200. https://doi.org/10.1016/S0145-305X(01)00064-7
Krasnov A, Jørgensen SM, Afanasyev S (2017) Ig-seq: deep sequencing of the variable region of Atlantic salmon IgM heavy chain transcripts. Mol Immunol 88:99–105. https://doi.org/10.1016/j.molimm.2017.06.022
Lindner C, Wahl B, Föhse L, Suerbaum S, Macpherson AJ, Prinz I, Pabst O (2012) Age microbiota and T cells shape diverse individual IgA repertoires in the intestine. J Exp Med 209:365–377. https://doi.org/10.1084/jem.20111980
Lindner C, Thomsen I, Wahl B, Ugur M, Sethi MK, Friedrichsen M, Smoczek A, Ott S, Baumann U, Suerbaum S, Schreiber S, Bleich A, Gaboriau-Routhiau V, Cerf-Bensussan N, Hazanov H, Mehr R, Boysen P, Rosenstiel P, Pabst O (2015) Diversification of memory B cells drives the continuous adaptation of secretory antibodies to gut microbiota. Nat Immunol 16:880–888. https://doi.org/10.1038/ni.3213
Lobb CJ, Clem LW (1981) Phylogeny of immunoglobulin structure and function. XI. Secretory immunoglobulins in the cutaneous mucus of the sheepshead Archosargus probatocephalus. Dev Comp Immunol 5:587–596. https://doi.org/10.1016/S0145-305X(81)80033-X
Lobb CJ, Hayman JR (1989) Activation of complement by different immunoglobulin heavy chain isotypes of the channel catfish (Ictalurus punctatus). Mol Immunol 26:457–465. https://doi.org/10.1016/0161-5890(89)90105-3
Lobb CJ, William Clem L (1981) Phylogeny of immunoglobulin structure and function-XII. Secretory immunoglobulins in the bile of the marine teleost Archosargus probatocephalus. Mol Immunol 18:615–619. https://doi.org/10.1016/0161-5890(81)90032-8
Lundqvist ML, Middleton DL, Radford C, Warr GW, Magor KE (2006) Immunoglobulins of the non-galliform birds: Antibody expression and repertoire in the duck. Dev Comp Immunol 30:93–100. https://doi.org/10.1016/j.dci.2005.06.019
Magadan S, Sunyer OJ, Boudinot P (2015) Unique features of fish immune repertoires: particularities of adaptive immunity within the largest group of vertebrates. Results Probl Cell Differ 57:235–264. https://doi.org/10.1007/978-3-319-20819-0-10
Magadan S, Jouneau L, Puelma Touzel M, Marillet S, Chara W, Six A, Quillet E, Mora T, Walczak AM, Cazals F, Sunyer O, Fillatreau S, Boudinot P (2018) Origin of public memory B-cell clones in fish after antiviral vaccination. Front Immunol 9:2115. https://doi.org/10.3389/fimmu.2018.02115
Magadan S, Jouneau L, Boudinot P, Salinas I (2019a) Nasal Vaccination drives modifications of nasal and systemic antibody repertoires in rainbow trout. J Immunol 203:1480–1492. https://doi.org/10.4049/jimmunol.1900157
Magadan S, Krasnov A, Hadi-Saljoqi S, Afanasyev S, Mondot S, Lallias D, Castro R, Salinas I, Sunyer O, Hansen J, Koop BF, Lefranc MP, Boudinot P (2019b) Standardized IMGT® nomenclature of salmonidae IGH genes the paradigm of Atlantic salmon and rainbow trout: from genomics to repertoires. Front Immunol 10:2541. https://doi.org/10.3389/fimmu.2019.02541
Magadán-Mompó S, Sánchez-Espinel C, Gambón-Deza F (2011) Immunoglobulin heavy chains in medaka (Oryzias latipes). BMC Evol Biol 11:165. https://doi.org/10.1186/1471-2148-11-165
Magadán-Mompó S, Zimmerman AM, Sánchez-Espinel C, Gambón-Deza F (2013) Immunoglobulin light chains in medaka (Oryzias latipes). Immunogenetics 65:387–396. https://doi.org/10.1007/s00251-013-0678-9
Magor BG (2015) Antibody affinity maturation in fishes—our current understanding. Biology (Basel) 4:512–524. https://doi.org/10.3390/biology4030512
Malmstrøm M, Matschiner M, Tørresen OK, Star B, Snipen LG, Hansen TF, Baalsrud HT, Nederbragt AJ, Hanel R, Salzburger W, Stenseth NC, Jakobsen KS, Jentoft S (2016) Evolution of the immune system influences speciation rates in teleost fishes. Nat Genet 48:1204–1210. https://doi.org/10.1038/ng.3645
Mashoof S, Criscitiello MF (2016) Fish immunoglobulins. Biology (Basel) 5:45. https://doi.org/10.3390/biology5040045
Morrison RN, Nowak BF (2001) Affinity purification and partial characterisation of systemic immunoglobulin of the snapper (Pagrus auratus). Aquaculture 201:1–17. https://doi.org/10.1016/S0044-8486(01)00566-X
Mountz JD, Mushinski JF, Owens JD, Finkelman FD (1990) The in vivo generation of murine IgD-secreting cells is accompanied by deletion of the C mu gene and occasional deletion of the gene for the C delta 1 domain. J Immunol 145:1583–1591
Mussmann R, Du Pasquier L, Hsu E (1996) Is xenopus IgX an analog of IgA? Eur J Immunol 26:2823–2830. https://doi.org/10.1002/eji.1830261205
Navelsaker S, Magadan S, Jouneau L, Quillet E, Olesen NJ, Munang’andu HM, Boudinot P, Evensen Ø (2019) Sequential immunization with heterologous viruses does not result in attrition of the B-cell memory in rainbow trout. Front Immunol 10:2687. https://doi.org/10.3389/fimmu.2019.02687
Ohta Y, Flajnik M (2006) IgD like IgM is a primordial immunoglobulin class perpetuated in most jawed vertebrates. Proc Natl Acad Sci U S A 103:10723–10728. https://doi.org/10.1073/pnas.0601407103
Pabst O, Hazanov H, Mehr R (2015) Old questions new tools: Does next-generation sequencing hold the key to unraveling intestinal B-cell responses? Mucosal Immunol 8:29–37. https://doi.org/10.1038/mi.2014.103
Partula S, Schwager J, Timmusk S, Pilström L, Charlemagne J (1996) A second immunoglobulin light chain isotype in the rainbow trout. Immunogenetics 45:44–51. https://doi.org/10.1007/s002510050165
Ramirez-Gomez F, Greene W, Rego K, Hansen JD, Costa G, Kataria P, Bromage ES (2012) Discovery and characterization of secretory IgD in rainbow trout: secretory IgD is produced through a novel splicing mechanism. J Immunol 188:1341–1349. https://doi.org/10.4049/jimmunol.1101938
Rego K, Bengtén E, Wilson M, Hansen JD, Bromage ES (2020a) Characterization of immunoglobulin light chain utilization and variable family diversity in rainbow trout. Dev Comp Immunol 104:103566. https://doi.org/10.1016/j.dci.2019.103566
Rego K, Hansen JD, Bromage ES (2020b) Genomic architecture and repertoire of the rainbow trout immunoglobulin light chain genes. Dev Comp Immunol 113:103776. https://doi.org/10.1016/j.dci.2020.103776
Salinas I, Zhang YA, Sunyer JO (2011) Mucosal immunoglobulins and B cells of teleost fish. Dev Comp Immunol 35:1346–1365. https://doi.org/10.1016/j.dci.2011.11.009
Savan R, Aman A, Nakao M, Watanuki H, Sakai M (2005) Discovery of a novel immunoglobulin heavy chain gene chimera from common carp (Cyprinus carpio L.). Immunogenetics 57:458–463. https://doi.org/10.1007/s00251-005-0015-z
Sepahi A, Salinas I (2016) The evolution of nasal immune systems in vertebrates. Mol Immunol 69:131–138. https://doi.org/10.1016/j.molimm.2015.09.008
Shan Y, Gras R (2011) 43 genes support the lungfish-coelacanth grouping related to the closest living relative of tetrapods with the Bayesian method under the coalescence model. BMC Res Notes 4:49. https://doi.org/10.1186/1756-0500-4-49
Shen L, Stuge TB, Evenhuis JP, Bengtén E, Wilson M, Chinchar VG, Clem LW, Miller NW (2003) Channel catfish NK-like cells are armed with IgM via a putative FcμR. Dev Comp Immunol 27:699–714. https://doi.org/10.1016/S0145-305X(03)00042-9
Stenvik J, Jørgensen T (2000) Immunoglobulin D (IgD) of Atlantic cod has a unique structure. Immunogenetics 51:452–461. https://doi.org/10.1007/s002510050644
Sun Y, Huang T, Hammarstrom L, Zhao Y (2020) The immunoglobulins: New insights implications and applications. Annu Rev Anim Biosci 8:145–169. https://doi.org/10.1146/annurev-animal-021419-083720
Tacchi L, Musharrafieh R, Larragoite ET, Crossey K, Erhardt EB, Martin S, Lapatra SE, Salinas I (2014) Nasal immunity is an ancient arm of the mucosal immune system of vertebrates. Nat Commun 5:1–11. https://doi.org/10.1038/ncomms6205
Tonegawa S (1983) Somatic generation of antibody diversity. Nature 302:575–581. https://doi.org/10.1038/302575a0
Turchaninova MA, Davydov A, Britanova OV, Shugay M, Bikos V, Egorov ES, Kirgizova VI, Merzlyak EM, Staroverov DB, Bolotin DA, Mamedov IZ, Izraelson M, Logacheva MD, Kladova O, Plevova K, Pospisilova S, Chudakov DM (2016) High-quality full-length immunoglobulin profiling with unique molecular barcoding. Nat Protoc 11:1599–1616. https://doi.org/10.1038/nprot.2016.093
Uchida D, Hirose H, Chang PK, Aranishi F, Hirayabu E, Mano N, Mitsuya T, Prayitno SB, Natori M (2000) Characterization of Japanese eel immunoglobulin M and its level in serum. Comp Biochem Physiol Part B Biochem Mol Biol 127:525–532. https://doi.org/10.1016/S0305-0491(00)00290-X
Wang B, Wang P, Wu ZH, Lu YS, Wang ZL, Jian JC (2016) Molecular cloning and expression analysis of IgD in nile tilapia (Oreochromis niloticus) in response to streptococcus agalactiae stimulus. Int J Mol Sci 17:348. https://doi.org/10.3390/ijms17030348
Weinstein JA, Jiang N, White RA, Fisher DS, Quake SR (2009) High-throughput sequencing of the zebrafish antibody repertoire. Science 324(5928):807–810. https://doi.org/10.1126/science.1170020
Wilson M, Bengtén E, Miller NW, Clem LW, Du Pasquier L, Warr GW (1997) A novel chimeric Ig heavy chain from a teleost fish shares similarities to IgD. Proc Natl Acad Sci U S A 94:4593–4597. https://doi.org/10.1073/pnas.94.9.4593
Xu Z, Parra D, Gómez D, Salinas I, Zhang YA, von Gersdorff JL, Heinecke RD, Buchmann K, LaPatra S, Sunyer JO (2013) Teleost skin an ancient mucosal surface that elicits gut-like immune responses. Proc Natl Acad Sci U S A 110:13097–13102. https://doi.org/10.1073/pnas.1304319110
Xu Z, Takizawa F, Parra D, Gómez D, Von Gersdorff JL, Lapatra SE, Sunyer JO (2016) Mucosal immunoglobulins at respiratory surfaces mark an ancient association that predates the emergence of tetrapods. Nat Commun 7:10728. https://doi.org/10.1038/ncomms10728
Xu Z, Takizawa F, Casadei E, Shibasaki Y, Ding Y, Sauters JC, Yu Y, Salinas I, Sunyer JO (2020) Specialization of mucosal immunoglobulins in pathogen control and microbiota homeostasis occurred early in vertebrate evolution. Sci Immunol 5:eaay 3254. https://doi.org/10.1126/sciimmunoLAay3254
Yasuike M, de Boer J, von Schalburg KR, Cooper GA, McKinnel L, Messmer A, So S, Davidson WS, Koop BF (2010) Evolution of duplicated IgH loci in Atlantic salmon Salmo salar. BMC Genomics 11:486. https://doi.org/10.1186/1471-2164-11-486
Ye J, Bromage ES, Kaattari SL (2010) The strength of B-cell interaction with antigen determines the degree of IgM polymerization. J Immunol 184:844–850. https://doi.org/10.4049/jimmunol.0902364
Ye J, Bromage E, Kaattari I, Kaattari S (2011) Transduction of binding affinity by B lymphocytes: a new dimension in immunological regulation. Dev Comp Immunol 35:982–990. https://doi.org/10.1016/j.dci.2011.01.015
Ye J, Kaattari IM, Ma C, Kaattari S (2013) The teleost humoral immune response. Fish Shellfish Immunol 35:1719–1728. https://doi.org/10.1016/j.fsi.(2013).10.015
Yel L (2010) Selective IgA deficiency. J Clin Immunol 30:10–16. https://doi.org/10.1007/s10875-009-9357-x
Yu Y, Kong W, Yin YX, Dong F, Huang ZY, Yin GM, Dong S, Salinas I, Zhang YA, Xu Z (2018) Mucosal immunoglobulins protect the olfactory organ of teleost fish against parasitic infection. PLoS Pathog 14:1–24. https://doi.org/10.1371/journal.ppat.1007251
Zhang YA, Salinas I, Li J, Parra D, Bjork S, Xu Z, Lapatra SE, Bartholomew J, Sunyer JO (2010) IgT a primitive immunoglobulin class specialized in mucosal immunity. Nat Immunol 11:827–835. https://doi.org/10.1038/ni.1913
Zhang T, Tacchi L, Wei Z, Zhao Y, Salinas I (2014) Intraclass diversification of immunoglobulin heavy chain genes in the African lungfish. Immunogenetics 66:335–351. https://doi.org/10.1007/s00251-014-0769-2
Zhang N, Zhang XJ, Chen DD, Sunyer JO, Zhang YA (2017) Molecular characterization and expression analysis of three subclasses of IgT in rainbow trout (Oncorhynchus mykiss). Dev Comp Immunol 70:94–105. https://doi.org/10.1016/j.dci.2017.01.001
Zhao Y, Kacskovics I, Pan Q, Liberles DA, Geli J, Davis SK, Rabbani H, Hammarstrom L (2002) Artiodactyl IgD: the missing link. J Immunol 169:4408–4416. https://doi.org/10.4049/jimmunol.169.8.4408
Zhao Y, Pan-Hammarström Q, Kacskovics I, Hammarström L (2003) The porcine Ig δ gene: unique chimeric splicing of the first constant region domain in its heavy chain transcripts. J Immunol 171:1312–1318. https://doi.org/10.4049/jimmunol.171.3.1312
Zimmerman AM, Yeo G, Howe K, Maddox BJ, Steiner LA (2008) Immunoglobulin light chain (IgL) genes in zebrafish: genomic configurations and inversional rearrangements between (VL-JL-CL) gene clusters. Dev Comp Immunol 32:421–434. https://doi.org/10.1016/j.dci.2007.08.005
Zimmerman AM, Romanowski KE, Maddox BJ (2011) Targeted annotation of immunoglobulin light chain (IgL) genes in zebrafish from BAC clones reveals kappa-like recombining/deleting elements within IgL constant regions. Fish Shellfish Immunol 31:697–703. https://doi.org/10.1016/j.fsi.2010.09.015
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Mendoza, M., Magadán, S. (2022). Immunoglobulins in Teleost. In: Buchmann, K., Secombes, C.J. (eds) Principles of Fish Immunology . Springer, Cham. https://doi.org/10.1007/978-3-030-85420-1_7
Download citation
DOI: https://doi.org/10.1007/978-3-030-85420-1_7
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-85419-5
Online ISBN: 978-3-030-85420-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)