Abstract
A simple method, for image encryption using 1D discrete chaos and DNA encoding, is proposed in this article. Here, chaotic maps are used to encode the secret image as DNA sequence and also to diffuse the encoded image with DNA operations and finally a cipher image is obtained. Additionally, hash function SHA-256 is employed to plain image and this makes the current method more robust. The result of the present method is studied and analyzed to established the aptness of the proposed method.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Information was always considered as a valuable possession. After the age of the internet dawned upon us, the value of information has surpassed any other material in existence. Information is also susceptible to corruption, theft and illegal exposure and images are no different in this regard. So, it is utmost important that the information should be protected so that an authorized person only has the permission to access that information. With respect to images, the process of making meaningless data out of a plain image is called image encryption. Conventional encryption algorithms like, DES [5] and AES [14] they don’t work well in image domain. Usually, an image encryption algorithm has two phases, namely, confusion and diffusion. The position of the pixels is altered in the first phase and intensity value of the pixels is modified in the last phase. Chaotic system is well used for encryption purposes due to its sensitivity, pseudo-randomness, ergodicity and reproduction [15] to initial value of the parameters. In recent years, because of these strengths of chaotic systems, many studies have implemented in image encryption algorithms [3, 9, 20, 22, 24]. In [21], S-boxes are formulated using chaotic systems for encryption. A method for encrypting an image is proposed in [12] where a 5D multiwing hyperchaotic system has employed. In [17] hyperchaos-based image encryption technique is implemented that uses a 6-dimensional hyperchaotic structure.
Besides the chaos based image encryption many other techniques are also studied for image encryption. In [7], different sine waves have constructed and using these random sequences are generated to confusion and diffusion phases. In [4], a polynomial interval bisection method is applied to obtain random sequences and these are used for arranging pixels of rows/columns of the plain image. To change the pixels value, in the diffusion phase, value Substitution and iterative XOR operation are applied. Another non-chaotic cryptosystem is developed using the cyclic group in [11]. Here, the cyclic group is employed to get permutation sequences and pixels and bits of the original image are permuted with the help of these permutation sequences. In [8], to get permutation some points are selected using certain distance on the perimeter of a defined circle with user defined radius depending the image size. In both the phases, the generated permutation is employed to get an encrypted image. In [13], a new technique for image encryption is proposed and for this purpose fibonacci transform is used. The pixels are permuted and also the pixel values are changed using this fibonacci transform.
Due to the some features of DNA encoding like parallelism of coding, consumption of low power and high density of information, it is attracted by various researchers in image encryption domain. For enhancing the security of the encryption method, many researchers have studied hybrid techniques using DNA encoding and computing with chaotic system. A chaotic map with DNA sequence based image encryption method is designed in [19], where bitwise XOR is executed among the pixels of the plain image and then DNA encoding is applied on the confused image and finally row/column are permuted to obtain the encrypted image. Using DNA permutation another scheme is proposed in [18] for color image encryption. In this scheme Lorenz map is also applied for random sequences generation and this step is used to increase the security level. A cryptosystem [23] is developed using 2D Hénon-Sine map (2D-HSM) and the DNA encoding. Here, only DNA XOR operation is defined to diffused image pixels. A hybrid model is designed in [10]. The model is composed of DNA coding and computing, SHA-2 and Lorenz map. Here, the DNA coding and operation and the Lorenz chaotic system improve the security of the model. In [16], a new image encryption system is introduced with permutations at pixel-level and bit-level followed by DNA coding. Here, a hyperchaotic (5D) map is applied for random sequence generation that is used in confusion. Then, DNA coding, XOR operation is employed to diffused the permuted image. Based on chaotic system and DNA coding another encryption technique is suggested in [2]. It is composed of two rounds. In the first round keys are generated and the permutation, substitution and diffusion is carried out in the last round. Additionally, to generate initial parameters for the chaotic map SHA-256 system is used.
In this article, a new method for image encryption, using 1D chaotic map and DNA computing, is proposed. The parameter(s) of these maps are initialized with the hash value of original image. To compute hash value, SHA-256 is used. The present method is a private key crypto-system. The Renyi map is applied for generating a key image (same size as the secret image). In this work, both images are encoded as DNA sequence using Tent map and then these encoded image are diffused to generate a diffused image. The proposed method has the provision to iterate the DNA encoding and diffusion a number of times. From the diffused image, an encrypted image is earned through DNA decoding. The presence of 1D maps makes the present method useable in terms of power consumption and computation aspects. The current method having no confusion step, it has only diffusion process. Now, we describe the structure of the rest part of this article. The basics of tools used in this method are discussed in Sect. 2. In Sect. 3 is dedicated for the proposed image encryption technique. The performance and its analysis of the proposed method are presented in Sect. 4. Section 5 highlights the conclusion of the present work.
2 Preliminaries
In the proposed image encryption algorithm, we have used 1D Tent map, 1D Renyi map and DNA encoding technique to encrypt a plain image.
2.1 Renyi Map and Tent Map
Tent map is applied to choose i) the rule of DNA coding and ii) the DNA operation. On the other hand Renyi map is employed for key image generation. According to [1] the randomness of the 1D Renyi system is very good. The 1D Renyi Map is defined in Eq. (1).
where \(a,~b \in [0, 10]\) and \(\alpha ~ \in (0, 12345]\). We use the Renyi map as the randomness of this map is very uniform and it has good bifurcation behavior. The bifurcation diagram of the Renyi map is shown in Fig. 1.
The Tent map is simple and extensively applied in image encryption algorithms. The expression of Tent map is offered in Eq. (2).
where \(x_0\) is the initial value, \(x_i \in [0, 1]\) and \(r \in [0, 2]\). The bifurcation diagram of the Tent map is presented in Fig. 2. From the Fig. 2 it is clear that for \(r \in [1.4,2]\), the randomness is at its highest level.
2.2 DNA Coding and DNA Operations
DNA is a molecule containing the genetic information of every living organism and various viruses to evolve, create, function, and reproduce. In DNA sequencing, the basic block of genetic code is built upon four bases i.e. Adenine (A), Thymine (T), Guanine (G) and Cytosine (C). According to the Watson–Crick [6] principle, A and T are complement of each other (i.e., A \(\overleftrightarrow {complement}\) T), similarly C and G are also complement of each other (i.e., C \(\overleftrightarrow {complement}\) G). Generally, these four DNA bases are encoded by 2-bit binary numbers ‘00’, ‘11’, ‘01’, and ‘10’. So, there are 24 (= 4!) rules to encode these four DNA bases in binary representation.
Suppose, ‘A’ is represented by ‘00’ and then due to complement relation ‘11’ is the code of ‘T’. For ‘C’, there are two options: i) ‘C’ =‘10’ (accordingly ‘G’=‘01’) or ii) ‘C’=‘01’ (and ‘G’=‘10’). So, for the particular choice of ‘A’ we have i) ‘A’ \(\sim \) ‘00’, ‘T’ \(\sim \) ‘11’, ‘C’ \(\sim \) ‘01’ and ‘G’ \(\sim \) ‘10’ or ii) ‘A’ \(\sim \) ‘00’, ‘T’ \(\sim \) ‘11’, ‘C’ \(\sim \) ‘10’ and ‘G’ \(\sim \) ‘01’. Again, ‘A’ can be represented by any of the four options {‘00’, ‘11’, ‘01’, ‘10’}. So, only eight rules out of 24 encoding rules satisfy the complementary relationship within the bases. Inverse of the encoding rules are the DNA decoding rules. The encoding rules for the DNA encoder is described in Table 1. For example, 177 is gray value of a particular pixel of an image and its binary representation is “10110001”. With respect to fifth encoding rule, the pixel will be encoded as “ACGT”.
Binary operations like addition, subtraction and XOR are also applied between DNA sequences to obtained an intermediate image. These operations are actually executed on binary strings and here, the operands and result are labeled by bases \(\{A, B, C, D\}\). The result of labeling (i.e., binary string \(\longleftrightarrow \) DNA sequence) depends on the DNA encoding rule. Hence, the DNA operations are defined with an encoding rule. For example, addition and subtraction operations with the help of first encoding rule are given in the Table 2 and Table 3, respectively. The XOR operation using fourth encoding rule is illustrated in the Table 4.
3 Proposed Image Encryption Method
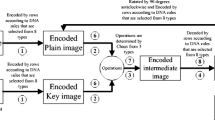
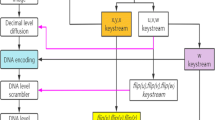
In this article, we have implemented a new approach for image encryption with the use of DNA encoder and two linear chaotic systems. The encryption algorithm is presented in Fig. 3. In the present method, unlike two conventional phases confusion and diffusion, we have implemented only diffusion phase. To carried out the diffusion step, first the plain image and key image are encoded by DNA encoder and then these two images are operated by DNA operations. To perform the proposed technique, we have certain steps which are as follows:
3.1 Generation of DNA Encoded Images
From Fig. 3, it is clear that the current method handles two DNA encoded images: i) one generated from the given plain image and ii) the other one is derived from the key image. Now, we are describing the process of key image generation and then the DNA encoder is presented. To generate the key image Renyi map is used and the DNA encoder is defined with the help of the Tent map. For any chaotic map, initial parameters are required to get the random sequences. The initial parameters can be selected arbitrary, but should be within the domain. In this method, we have generate the initial parameters from the plain image. For this purpose SHA-256 function is applied on the plain image. The reason to use the hash function is discussed later. The initial part of the sequence generated using chaotic map may not be random in nature. For shake of simplicity, we ignore first thousands elements of the sequence and then the rest part is considered as the sequence (which will be used in the experiment) generated by the map. In this implementation it is assumed that when a part of a sequence is used for a particular purpose, then this part will be removed from the sequence and remaining part will be used for future requirement.
Key Image Generation: In this step, we have constructed a key image (\(KI_{M \times N}\)) using Renyi chaotic map. The size of KI and plain image are same. A sequence \(\{S_r\}\) generated using the Renyi map and elements of the sequence is multiplied by a constant ‘\(Fact_1\)’ which is large enough so that new value is grater than thousand and so. Then, KI computed from new sequence using the following algorithm.

where ‘int (u, v)’ returns q when \(u = q.v +r\).
DNA Encoder: In this section we have converted the plain and key image into DNA encoded images. To convert gray scale value of image into DNA sequence we have to select a rule from the eight DNA encoding rules (which is already illustrated in Sect. 2.2). In this work, each time we have randomly selected a rule using sequences generated by the Ten map. Let, the sequence \(\{T_m(r)\}\) is generated using Tent map. Based upon the sequence value \(T_m(r)\), \(i^{th}\) encoding rule is selected where \(i= int (T_m(x) \times Fact_2, 8) + 1\). In the encoding process, we choose pixel-wise encoding rule or row-wise/column-wise encoding rule. For simplicity, in this method, we have fixed the same encoding rule for all pixels of a row. Suppose, the encoding rules (given in Table 1) is stored in the matrix \(Rule_{4 \times 8}\). Also assume that a pixel ‘p’ is represented by ‘\(p_1p_2p_3p_4p_5p_6p_7p_8\)’ (binary representation). Respect to \(t^{th}\) rule (\(1 \le t \le 8\)), a particular pixel ‘p’ is encoded as ‘\(X_1X_2X_3X_4\)’ where \(X_i= Rule(dec(p_{(i-1)*2+1}p_{i*2}), t)\), for \(1 \le i \le 4\) and ‘dec()’ is decimal to binary converter. The algorithmic flow of the used DNA encoder is as given below:

3.2 Diffusion Using DNA Operation
From previous step, we have two DNA encoded images in hand and in the present step these two images are diffused (see the Fig. 3). To diffuse the encoded images, we have used base-wise (symbol-wise) addition, subtraction or XOR operations as discussed in Sect. 2.2. In this work, three operations (addition, subtraction or XOR) are defined and result of these operations are also depending on the the encoding rule. Like previous, we can fix the encoding rule as well as the operation pixel-wise, row/column wise (same for all pixels in that row/column). Here, we fix them row-wise, i.e., for each pixel of the row same encoding rule and same operation have used. Henceforth, we use the term ‘DNA Operation’ to represent the diffusion of DNA encoded images with selection of encoding rule and binary operation. In this step, sequence of the Tent map helps to find the diffused image. Suppose, \(Oper_{8 \times 3 \times 4 \times 4}\) is used to store the ‘DNA Operation’, eight rules, three operations and \(4 \times 4\) operation table as given in Table 2, 3 and 4. The algorithmic structure of the DNA operation is described as below.

3.3 Generation of Encrypted Image
This is the last step of the proposed algorithm. In this step, diffused image is coded into binary form using DNA decoder. The DNA decoder is the inverse process of DNA encoder and may algorithmically presented as Algorithm 4: DNA Decoder(). Due to the space limitation, this algorithm is not presented here. Like DNA encoder, the DNA decoder needs the encoding rule convert the encoded image into binary form. This binary image may be consider as the (final) cipher image or may be consider as an intermediate image and iterated through the DNA encoder, DNA Operation and DNA Decoder as shown as a loop in Fig. 3. The proposed image encryption technique is algorithmically discussed below.

The current method is a private key encryption system and the original image can be retrieved by following the steps in reverse order with inverse operations using same key.
4 Experimental Result and Security Analysis
Here, we have reported the performance of the presented method and also analyze the security of the present method. This method is implemented in MATLAB environment.
4.1 Results
To demonstration the performance of this method, we have considered standard grayscale images ‘Lena’, ‘Girlface’, ‘Barbara’ and ‘Cameraman’. Size of each image is \(512 \times 512\). The parameters of the proposed method are set up as: the value of the parameter ‘itr’ set to 10, ‘\(Fact_1\)’ set to 2399 (a prime number), ‘\(Fact_2\)’ is set to 151 (a prime number). To define the keys, we take the hash value of given image and odd positions bit is considered as the \(key_1\) and even positions bit form \(key_2\). The original images are displayed in Fig. 4(a) and the cipher images shown in Fig. 4(b). In Fig. 4(c) and (d), the decrypted images with exact key and different key are illustrated. We have noticed that decryption with incorrect key gives meaningless image and so the achievement of this method is quite good and hence the proposed method can be applied in different applications.
4.2 Security Analysis
Here, the robustness of the proposed method under different different attacks, like differential plaintext attack, statistical attack, brute-force attack etc., have studied.
Histogram Analysis: Histogram analysis measures the defense against statistical attacks. An ideal encryption algorithm will distribute the pixel intensities evenly. From an evenly distributed histogram of encrypted images in Fig. 5, we can claim that our proposed algorithm will withstand against statistical attacks.
Information Entropy: The information entropy, which is expressed in bits for given encryption algorithm, is ideally 8 for grayscale images. An encryption algorithm with better information entropy will divulge lesser information to the world. Basically, this is the factor which illustrates the randomness. In case of an 8-bits gray-scale image (I) entropy is calculated with equation \(H(I)=-\sum _{p=0}^{255}f(p)log_2 f(p)\) bit, where the f(p) denotes the normalized frequency of occurrences of the intensity p. The comparative analysis of the performance of the proposed method in terms of entropy is reported in Table 5. From this table, it is observed that the present method achieved almost ideal value and hence rate of information disclosure is practically nil. So, any type of entropy attack on encrypted images will get no information.
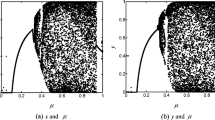
Correlation Analysis: Correlation between a pair of adjacent pixels hugely positive in plain images. Due to this positive correlation, an attacker can easily guess the pixel intensity of an adjacent pixel. During encryption, this correlation must be reduced. We are expecting zero (0) correlation value (between two immediate neighbor pixels) for an encrypted image. In Table 6, we have reported the correlation values generated before and after encryption. In case of adjacency, we have taken horizontal, vertical and diagonal adjacent pixels. It is evident from the table that there is very low amount of correlation exists between two adjoined pixels. In implementation, randomly we have chosen 5000 pairs of adjacent pixels and respective distribution diagrams are shown in Fig. 6. From the scattered diagram, we can see that plain image has a huge amount of positive correlation, because of which values generated are alighted at \(45^{\circ }\). In contrast to the scattered diagram of original images, it is observed that in case of cipher images points are well distributed all over the area. This implies that two neighboring pixels are not related.
4.3 Key Space Analysis
The proposed method is a symmetric key encryption system and the security of the proposed method depends on the initial parameters of Renyi and Tent maps. Also note that to select the encoding rule or DNA operation, we have used sequence \(\{T_m\}\) and parameters ‘\(Fact_1\)’ and ‘\(Fact_2\)’. Someone may consider these two parameters as secret information. In this work, we have taken these as public data. Another important paragenetic is ‘itr’, which denoted the number iterations the diffusion of the intermediate image will be executed. This can be consider as a private key and it takes some moderate value of size, say, \(2^{10}\). To initialize the parameters of the chaotic maps, we use SHA-256(I). The space defined by the hash value is \(2^{256}\) and from this value two keys \(key_1\) and \(key_2\) are derived. Therefore, the size of the key space of the present method is (\(2^{266}\)), which is notably high and present method can withstand brute-force attacks.
4.4 Key Sensitivity
As per our previous description, the pair (hash, itr) represents the key of the proposed method. To study the robustness, in this experiment, two different keys are used to encrypt the same image. For example, in this implementation, an image is encrypted by (hash, itr), \((hash', itr)\) and \((hash, itr-1)\). Here, \(hash'\) is obtained from hash by complementing the LSB of hash. Let these keys give the cipher images \(Enc_1\), \(Enc_2\) and \(Enc_3\). If cipher images are heavily different from other, then it ensures that current method is sensitive to key. The UACI and NPCR parameters (see Eq. (3)) have used to measure the dissimilarity between two encrypted images. Theoretical optimum value of UACI and NPCR are 33.33% and 100%, respectively. From Table 7, it may be concluded that performance of the current method is acceptable and may be applicable to different applications.
Differential Attacks: An encryption method should have power to resist the differential attack, i.e., encryption should return totally dissimilar encrypted image while a very little modification is done in the plain image. In this implementation, we have used two methods: i) where the LSB of the first pixel is complemented and ii) LSB of the last pixel is complemented. Let the encrypted images obtained from no change in plain image, change at first pixel and change at last pixel are denoted as \(Enc_1\), \(Enc_2\) and \(Enc_3\), respectively. Then, the difference in encrypted images are evaluated with UACI and NPCR. These parameters are used to test the strength against the differential attack. UACI and NPCR are calculated in Eq. (3).
The result of the proposed method against the differential attack is presented in Table 8, which shows that UACI and NPCR indexes are very similar to the theoretical values, so it reveals that the encryption algorithm is stable against the differential attack. The keys \(key_1\) and \(key_2\) are obtained from ‘hash’ of the given image, so for a very small difference in secret image and we have completely different hash value and hence, we have different keys. Different keys means different chaotic sequences. So, different encoding rule, different operation will be selected and as a result we obtained significantly dissimilar encrypted images even if there is a very little modification in the original image.
5 Conclusions
Here, a symmetric key image encryption algorithm is proposed. The current method uses Renyi map, Tent map and DNA encoding. Here, the initial parameters are constructed from the secret image itself with the help of SHA-256 hash function for better security. From the performance and analysis of the results it may be noted that the present method is robust against brute force attack, differential attack and statistical attack. Our future target is to modify and extend the current method to make it robust against the plain image attack.
References
Alzaidi, A., Ahmad, M., Doja, M., Solami, E., Beg, M.: A new 1D chaotic map and \(\beta \)-hill climbing for generating substitution-boxes. IEEE Access 6, 55405–55418 (2018)
Belazi, A., Talha, M., Kharbech, S., Xiang, W.: Novel medical image encryption scheme based on chaos and DNA encoding. IEEE Access 7, 36667–36681 (2019)
Biswas, P., Kandar, S., Dhara, B.C.: A novel image encryption technique using one dimensional chaotic map and circular shift technique. In: Proceedings of the 6th International Conference on Software and Computer Applications, pp. 112–116 (2017)
Biswas, P., Kandar, S., Dhara, B.C.: An image encryption scheme using sequence generated by interval bisection of polynomial function. Multimed. Tools Appl. 79, 1–24 (2020)
Blakley, G., Borosh, I.: Rivest-Shamir-Adleman public key cryptosystems do not always conceal messages. Comput. Math. Appl. 5(3), 169–178 (1979)
Cisse, I.I., Kim, H., Ha, T.: A rule of seven in Watson-Crick base-pairing of mismatched sequences. Nat. Struct. Mol. Biol. 19(6), 623 (2012)
Das, S.K., Dhara, B.C.: An image encryption technique using sine curve. In: 2017 Ninth International Conference on Advances in Pattern Recognition (ICAPR), pp. 1–6. IEEE (2017)
Das, S.K., Dhara, B.C.: A new image encryption method using circle. In: 2017 8th International Conference on Computing, Communication and Networking Technologies (ICCCNT), pp. 1–6. IEEE (2017)
El Assad, S., Farajallah, M.: A new chaos-based image encryption system. Sig. Process.: Image Commun. 41, 144–157 (2016)
Guesmi, R., Farah, M., Kachouri, A., Samet, M.: A novel chaos-based image encryption using DNA sequence operation and secure hash algorithm SHA-2. Nonlinear Dyn. 83(3), 1123–1136 (2016). https://doi.org/10.1007/s11071-015-2392-7
Kandar, S., Chaudhuri, D., Bhattacharjee, A., Dhara, B.C.: Image encryption using sequence generated by cyclic group. J. Inf. Secur. Appl. 44, 117–129 (2019)
Li, Y., Wang, C., Chen, H.: A hyper-chaos-based image encryption algorithm using pixel-level permutation and bit-level permutation. Opt. Lasers Eng. 90, 238–246 (2017)
Maiti, C., Dhara, B.C.: Image encryption with a new Fibonacci transform. In: 2018 Fifth International Conference on Emerging Applications of Information Technology (EAIT), pp. 1–4. IEEE (2018)
NIST FIPS PUB 197: Advanced Encryption Standard (AES) (2001)
Ravichandran, D., Praveenkumar, P., Rayappan, J.B.B., Amirtharajan, R.: Chaos based crossover and mutation for securing DICOM image. Comput. Biol. Med. 72, 170–184 (2016)
Sun, S.: A novel hyperchaotic image encryption scheme based on DNA encoding, pixel-level scrambling and bit-level scrambling. IEEE Photon. J. 10(2), 1–14 (2018)
Wang, T., Wang, M.H.: Hyperchaotic image encryption algorithm based on bit-level permutation and DNA encoding. Opt. Laser Technol. 132, 106355 (2020)
Wang, X.Y., Li, P., Zhang, Y.Q., Liu, L.Y., Zhang, H., Wang, X.: A novel color image encryption scheme using DNA permutation based on the Lorenz system. Multimed. Tools Appl. 77(5), 6243–6265 (2018)
Wang, X.Y., Zhang, Y.Q., Bao, X.M.: A novel chaotic image encryption scheme using DNA sequence operations. Opt. Lasers Eng. 73, 53–61 (2015)
Wang, X., Guan, N., Zhao, H., Wang, S., Zhang, Y.: A new image encryption scheme based on coupling map lattices with mixed multi-chaos. Sci. Rep. 10(1), 1–15 (2020)
Wang, X., Wang, Q.: A novel image encryption algorithm based on dynamic s-boxes constructed by chaos. Nonlinear Dyn. 75(3), 567–576 (2014). https://doi.org/10.1007/s11071-013-1086-2
Wang, X., Zhang, H.: A color image encryption with heterogeneous bit-permutation and correlated chaos. Opt. Commun. 342, 51–60 (2015)
Wu, J., Liao, X., Yang, B.: Image encryption using 2D Hénon-Sine map and DNA approach. Sig. Process. 153, 11–23 (2018)
Zhang, Y.: The unified image encryption algorithm based on chaos and cubic S-box. Inf. Sci. 450, 361–377 (2018)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Biswas, M., Das, S.K., Dhara, B.C. (2021). An Image Encryption Method Using Chaos and DNA Encoding. In: Dutta, P., Mandal, J.K., Mukhopadhyay, S. (eds) Computational Intelligence in Communications and Business Analytics. CICBA 2021. Communications in Computer and Information Science, vol 1406. Springer, Cham. https://doi.org/10.1007/978-3-030-75529-4_4
Download citation
DOI: https://doi.org/10.1007/978-3-030-75529-4_4
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-75528-7
Online ISBN: 978-3-030-75529-4
eBook Packages: Computer ScienceComputer Science (R0)