Abstract
Segmentation of medical images is typically one of the first and most critical steps in medical image analysis. Manual segmentation of volumetric images is labour-intensive and prone to error. Automated segmentation of images mitigates such issues. Here, we compare the more conventional registration-based multi-atlas segmentation technique with recent deep-learning approaches. Previously, 2D U-Nets have commonly been thought of as more appealing than their 3D versions; however, recent advances in GPU processing power, memory, and availability have enabled deeper 3D networks with larger input sizes. We evaluate methods by comparing automated liver segmentations with gold standard manual annotations, in volumetric MRI images. Specifically, 20 expert-labelled ground truth liver labels were compared with their automated counterparts. The data used is from a liver cancer study, HepaT1ca, and as such, presents an opportunity to work with a varied and challenging dataset, consisting of subjects with large anatomical variations responding from different tumours and resections. Deep-learning methods (3D and 2D U-Nets) proved to be significantly more effective at obtaining an accurate delineation of the liver than the multi-atlas implementation. 3D U-Net was the most successful of the methods, achieving a median Dice score of 0.970. 2D U-Net and multi-atlas based segmentation achieved median Dice scores of 0.957 and 0.931, respectively. Multi-atlas segmentation tended to overestimate total liver volume when compared with the ground truth, while U-Net approaches tended to slightly underestimate the liver volume. Both U-Net approaches were also much quicker, taking around one minute, compared with close to one hour for the multi-atlas approach.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Over the last few decades, the rapid development of non-invasive imaging technologies has given rise to large amounts of data; analysis of such large datasets has become an increasingly complex task for clinicians. For example, in abdominal magnetic resonance imaging (MRI), image segmentation can be used for measuring and visualising internal structures, analysing changes, surgical planning, and extracting quantitative metrics. The high variability of location, size, and shape of abdominal organs makes segmentation a challenging problem. Segmentation of medical images is often one of the first and most critical steps in medical image analysis. Manual segmentation of volumetric medical images is a labour-intensive task that is prone to inter-rater and intra-rater variability. There is a need to automate the process to increase both efficiency and reproducibility and decrease subjectivity. Developing a robust automated segmentation method using deep learning has been an area of intense research in recent years [1].
Popular automated segmentation techniques include: using statistical models [2], image registration, classical machine learning algorithms [3] and, most recently, deep learning. Statistical models and classical machine learning algorithms do not generalise as well. Classical machine learning algorithms require careful feature engineering which is a time consuming and complex process. In this paper we compare ‘feature engineering free’ state-of-the-art multi-atlas based segmentation to recent deep-learning approaches.
Multi-atlas segmentation (MAS) was first introduced and popularised by Rohlfing et al. in 2004 [4]. Since then, substantial progress has been made [5]. MAS is a process that warps a number an expert-labelled atlas images (the moving images) into the coordinate space of a target image, via non-linear image registration. The iterative process of image registration involves optimising some similarity metric, such as cross-correlation, between the warped and target image by means of deformation of the ‘moving’ image [5]. Atlas labels are then propagated onto the target image and fused together in a way that can use the relationship between the target image and registered atlases, alongside the propagated labels [6]. MAS has the ability to capture anatomical variation much better than a model-based average or registration to one chosen case [4]. MAS is a computationally intensive process and has consequently grown in popularity due to an increase in computational resources [5].
Deep learning refers to neural networks with numerous layers that are capable of automatically extracting features [1]. This self-learning capability has a significant advantage over traditional machine learning algorithms, namely that features do not have to be hand-crafted. Deep learning has been applied to numerous fields [1]; image analysis tasks, such as object recognition, is one such. Convolutional neural networks (CNNs) were first introduced in 1989 [7] but have only recently become popular after a breakthrough in 2012 [9], along with rapid increases in GPU power, memory and accessibility. CNNs are currently by far the most popular approach for image analysis tasks [8].
Advancements in algorithms, and GPU technology, along with increased availability of training sets, has enabled larger, more robust networks. CNNs can automatically learn representations of image data with increasing levels of abstraction via convolutional layers [1]. These convolutional layers drastically reduce the number of parameters, when compared with traditional ‘fully connected’ neural networks, as weights are shared among convolutional layers [9].
U-Net was first introduced in 2015 by Ronneberger et al. as a deep CNN architecture geared towards biomedical image segmentation [10]. U-Net is a development of the fully convolutional network architecture [11]. A contracting encoder, which analyses the full image, is followed by an expanding decoder to provide the final segmentation; shallower layers in the network capture local information while deeper layers, whose retrospective field is much larger, capture global information [10]. The expanding decoder aims to recover a full-resolution pixel-to-pixel label map, from the different feature maps created in the contracting layers. Previously, 2D U-Net architectures have been thought of as more appealing than their 3D versions due to limitations in computational cost and GPU memory [12]. This said, current advancements in GPU memory and accessibility (cloud services such as Amazon Web Services) has enabled deeper 3D networks, with larger input sizes.
Here we compare the performance of a state-of-the-art multi-atlas segmentation approach with more recent 2D and 3D U-Net approaches. We evaluate the performance of each method by comparing manual ground truth liver labels with their automated counterparts, in challenging volumetric MRI images. Segmenting the liver is a process often used in surgical planning [13].
2 Materials
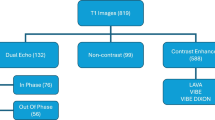
Data for the evaluation of methods was from ‘HepaT1ca’ a liver cancer study. T1-weighted 3D-SPGR images of the abdomen were collected according to the HepaT1ca study protocol [14]. HepaT1ca data implies working with a varied and challenging dataset, consisting of subjects with large anatomical variations responding from different tumours and resections. The different segmentation methods should ideally be robust to such dramatic changes between different images, and complicates both testing and training.
135 livers were labelled by a trained analyst. Subjects in the dataset have various tumours and resections of different shapes and sizes. The dataset was split into: 115 training cases and 20 test cases (Fig. 1).
3 Methods
3.1 Multi-atlas Segmentation Method
For the multi-atlas segmentation, 45 subjects were chosen at random from the ‘training’ set. A number of random atlases were used in order to capture anatomical variation within a population (one of the underlying principles of MAS). For each subject in the test set, the 45 random atlases were non-linearly registered to the test image.
The registration step was divided into two parts: affine registration (scaling, translation, rotation and shear mapping), followed by non-linear registration. We used the affine transform from ANTs (advanced normalisation tools) package [15]. We chose the DEEDS (dense displacement sampling) algorithm [16] for the non-linear part. DEEDS has been shown to yield the best registration performance in abdominal imaging [17], when compared with other common registration algorithms. Image grids are represented as a minimum spanning tree; a global optimum of the cost function is found using discrete optimisation. After all of the atlases (and their corresponding labels) had been registered to the target image, we used STEPS [6], as a template selection and label fusion algorithm, to produce the final liver segmentation.
3.2 Deep Learning Segmentation Methods
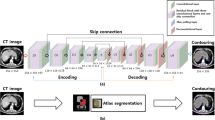
For the deep learning methods, we used slightly different model architectures for the 3D and 2D U-Net implementations. Figure 2 illustrates architecture of the 2D U-Net implementation; an expansion of the network previously used for quantitative liver segmentation [18].
Like the original 2D U-Net, our implementation has a contracting and expanding path. The input size of the network is a \(288 \times 224\) image with 1 channel (black and white image). Images were padded to ensure the output size of feature maps were the same size as the input image. We used batch normalisation (BN), after each convolution, to improve performance and stability of the network [19]. BN was followed by a rectified linear unit (ReLu) activation function. As suggested in [20], in the contracting path we doubled the number of channels, prior to max pooling, to avoid any bottlenecks in the network. We also applied this same principle in the expanding path. An addition of two max pooling (downsampling) layers, that further reduce the dimensionality of the image, resulted in better localisation, and as such, improve final segmentation performance of the network.
The 3D U-Net implementation was essentially identical to the 2D network, but with 3D operations instead, e.g. a \(3 \times 3 \times 3\) convolution instead of a \(3 \times 3\) convolution. The input size to the network was \(224 \times 192 \times 64\) voxels. We also used one fewer max pooling layer; the lowest resolution/ highest dimensional representation of an image was \(14 \times 12 \times 4\). This allowed for better depth localisation. If we still had 2 additional max pooling layers there would only have been 2 layers in the depth dimension.
Each model was implemented using the Keras framework, with Tensorflow as the backend.
Pre-processing. Each image underwent some pre-processing before being fed into the network. First, we applied 3 rounds of N4 Bias Field Correction [21] to remove any image contrast variations due to magnetic field inhomogeneity. All intensity values were then normalised between a standard reference scale (between 0 and 100). We also winsorised images, by thresholding the maximum intensity value to the 98th percentile; a heuristic that gives a reasonable balance between the reduction of high signal artefacts and image contrast.
For the 2D U-Net network, volumes were split into their respective 2D slices in the axial plane. Slices could then be reassembled into their respective volumes after a liver segmentation had been predicted by the network.
Training and Data Augmentation. Both networks were trained on NVIDIA Tesla V100 GPUs for 100 epochs, with a learning rate of 0.00005. We used a batch size of 10 and 1 for the 2D and 3D networks, respectively. During training we employed strategies to prevent overfitting; an important process that ensures that true features of images are learnt, instead of specific features that only exist in the training set. In addition to batch normalisation, when training the 2D network, slices were randomly shuffled between all subjects and batches for each epoch. Batch order was also randomized during each epoch when training the 3D network. Anatomically plausible data augmentation was applied ‘on-the-fly’ to further reduce the risk of overfitting. We applied small affine transformations with 5 degrees of rotation, 10% scaling and 10% translation. Both networks used Adam optimisation [22] with binary cross-entropy as the loss function. Each network took around 6 h to train.
After each network was trained, liver masks were predicted for each volumetric image in the test set.
3.3 Evaluation
Methods were evaluated by comparing the automated liver segmentations, produced by the automated segmentation methods, with the expert-labelled ground truth image. The first comparison metric we used was the Dice overlap score.
Dice measures the number of voxels that overlap between the ground truth segmentation (X) and the automated segmentation (Y). A score of 1 represents a perfect overlap between two 3D segmentations, while 0 represents no overlap between segmentations. In addition to Dice, we also measured performance by calculating the percentage difference in volume between the ground truth and automated segmentations. This highlighted if methods tended to underestimate or overestimate total liver volume.
V1 represents the liver volume of the ground truth segmentation. V2 represents the liver volume of the automated segmentation. The time taken for each segmentation method to run was also recorded for each test image.
We then used a paired t-test, to evaluate the differences in mean and variance between Dice metrics and volume percentage differences, between the segmentations produced by each method.
4 Results
Figure 3 shows a boxplot of the Dice overlap scores between the automated liver segmentations and ground truth annotations, for all images in the test set.
The multi-atlas approach was found to perform significantly worse than both the 3D U-Net (t = 3.397, p = 0.003)Footnote 1 and 2D U-Net (t = 2.628, p = 0.017)Footnote 2 approaches, while between the U-Net approaches, the 3D version slightly outperforms the 2D version (t = 2.016, p = 0.051) (Fig. 4).
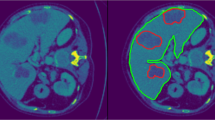
Figures 5 and 6 show examples of a single slice from different volumetric images, their corresponding automated liver segmentations and the ground truth liver segmentations. Figure 5 shows a more challenging case in the test set, whereby the subject has had a previous liver resection and is missing a substantial part of the liver. Figure 6 highlights an image with an exemplar liver.
Boxplot of dice scores for each segmentation method. The box extends from the lower to upper quartile values of the data, with a line at the median. The whiskers extend from the box to show the range of the data. Flier points are those past the end of the whiskers. The outlier seen in the multi-atlas dice scores is 3.8 standard deviations away from the mean.
The multi-atlas approach tended to overestimate the overall volume of the liver. The percentage differences in volume, of the automated multi-atlas segmentations with the ground truth, were significantly different than differences in volume for the 2D U-Net (t = 3.432, p = 0.003)Footnote 3 and the 3D U-Net (t = 3.812, p = 0.001)Footnote 4. The 3D U-Net tended to underestimate the volume (median = −3.01%) more the then 2D U-Net (median = −0.15%). The distributions in volume differences between the 2D U-Net and 3D U-Net are different (t = 3.824, p = 0.001).
The multi-atlas based approach took close to 1 h to compute a liver segmentation. The deep-learning based approaches, once trained, took around 1 min to compute a liver segmentation (Fig. 7).
Boxplot of the percentage differences in volumes for all test cases for each segmentation method. The box extends from the lower to upper quartile values of the data, with a line at the median. The whiskers extend from the box to show the range of the data. Flier points are those past the end of the whiskers. The outlier seen in the multi-atlas volume percentage difference scores is 3.8 standard deviations away from the mean.
5 Discussion
Both deep learning models significantly outperformed the multi-atlas based approach, with the 3D U-Net achieving slightly better performance, in terms of overlap with the ground truth, than the 2D U-Net. This could be due to spatial encoding in the 3D U-Net; inputs to the 2D network are completely independent slices that have no information about where, within the volume, the slice was located.
A disadvantage of fully convolutional 3D networks is that they have much larger computational cost and GPU memory requirements. Previously, this has limited the depth of a network and the filters’ field-of-view, two key factors for performance gains, resulting in better performance from 2D networks. More complex network architectures have been developed to avoid some of these drawbacks by using 2D networks with encoded spatial information [12]. However, recently state-of-the-art GPUs are now easily accessible on Cloud services, such as Amazon Web Services, and have increasingly larger amounts of GPU processing power and memory, allowing for deeper networks and larger inputs. We believe utilising these state-of-the-art GPUs was a contributing factor to the superior performance from the 3D network. We did not have to employ patch-based methods in order to effectively use a 3D network. Input images were only slightly downsampled (to around 90% of the original dimension) to fit the 3D model in GPU memory. This slight downsampling could be a reason why the 3D U-Net underestimated liver volume more than the 2D U-Net. 2D U-Nets could be more useful for automated liver volumetry; however, this does not mean 2D U-Nets are the best for other applications such as surgical planning and extracting quantitative metrics.
The deep learning approaches were several orders of magnitude faster than the multi-atlas based approach. Although, in a clinical workflow for fully-automated segmentation, this may not be a limiting factor, faster segmentation time does provide significant advantages when analysing larger datasets. Inter-observer variability is a factor to consider when assessing the performance of an automated segmentation. However, here ground-truth delineations were provided by a single annotater which was appropriate given the tests were of how close the methods resembled the annotations they were trained on.
When using the multi-atlas segmentation method, we saw a much larger variation in segmentation accuracy when compared with the deep learning approaches (Fig. 3). The probabilistic multi-atlas approach did not generalize well when compared with the deep learning approaches, this could be due to insensitivity to biologically-relevant variance (as seen in Fig. 5). Variance could be more apparent in this dataset due to a larger variation in liver shapes and sizes between subjects, from tumours and previous liver resections. That being said, although not studied in depth here, the number of tumours within a liver did not seem to alter the segmentation performance of any of the automated techniques. There are more advance atlas selection techniques [23] which could reduce the variance; however, it does not alleviate the computational time drawback of a multi-atlas technique.
In conclusion, the U-Net approaches were much more effective at automated liver delineation (once trained), both in terms of time and accuracy, than the multi-atlas segmentation approach.
Notes
- 1.
(t = 4.886, p = 0.0001) excluding the MAS outlier.
- 2.
(t = 3.499, p = 0.003) excluding the MAS outlier.
- 3.
(t = 4.906, p = 0.0001) excluding the MAS outlier.
- 4.
(t = 5.381, p = 0.00004) excluding the MAS outlier.
References
Lecun, Y., Bengio, Y., Hinton, G.: Deep learning. Nature 521, 436–444 (2015)
Heimann, T., Meinzer, H.P.: Statistical shape models for 3D medical image segmentation: a review. Med. Image Anal. 13, 543–563 (2009)
Fritscher, K., Magna, S., Magna, S.: Machine-learning based image segmentation using Manifold Learning and Random Patch Forests. In: Imaging and Computer Assistance in Radiation Therapy (ICART) Workshop, MICCAI 2015, pp. 1–8 (2015)
Rohlfing, T., Russakoff, D.B., Maurer Jr., C.R.: An expectation maximization-like algorithm for multi-atlas multi-label segmentation. In: Proceedings of the Bildverarbeitung frdie Medizin, pp. 348–352 (2004)
Iglesias, J.E., Sabuncu, M.R.: Multi-atlas segmentation of biomedical images: a survey. Med. Image Anal. 24, 205–219 (2015)
Jorge Cardoso, M., et al.: STEPS: similarity and truth estimation for propagated segmentations and its application to hippocampal segmentation and brain parcelation. Med. Image Anal. 17, 671–684 (2013)
Lecun, Y., Jackel, L.D., Boser, B., Denker, J.S., Gral, H., Guyon, I.: Handwritten digit recognition. IEEE Commun. Mag. 27 (1989)
Zhao, Z.-Q., Zheng, P., Xu, S., Wu, X.: Object detection with deep learning: a review. IEEE Trans. Neural Netw. Learn. Syst. (2019)
Krizhevsky, A., Sutskever, I., Hinton, G.: ImageNet classification with deep convolutional neural networks. In: Advances in Neural Information Processing Systems, vol. 2, pp. 1097–1105 (2012)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Long, J., Shelhamer, E., Darrell, T.: Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 3431–3440 (2015)
Li, X., Chen, H., Qi, X., Dou, Q., Fu, C., Heng, P.: H-DenseUNet: hybrid densely connected UNet for liver and tumor segmentation from CT volumes. IEEE Trans. Med. Imaging 37, 2663–2674 (2018)
Gotra, A., et al.: Liver segmentation: indications, techniques and future directions. Insights Imaging 8, 377–392 (2017)
Mole, D.J., et al.: Study protocol: HepaT1ca, an observational clinical cohort study to quantify liver health in surgical candidates for liver malignancies. BMC Cancer 18, 890 (2018)
Avants, B.B., Tustison, N.J., Song, G., Cook, P.A., Klein, A., Gee, J.C.: A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage 54(3), 2033–2044 (2010)
Heinrich, M.P., Jenkinson, M., Brady, S.M., Schnabel, J.A.: Globally optimal deformable registration on a minimum spanning tree using dense displacement sampling. In: Ayache, N., Delingette, H., Golland, P., Mori, K. (eds.) MICCAI 2012. LNCS, vol. 7512, pp. 115–122. Springer, Heidelberg (2012). https://doi.org/10.1007/978-3-642-33454-2_15
Xu, Z., et al.: Evaluation of six registration methods for the human abdomen on clinically acquired CT. IEEE Trans. Biomed. Eng. 63, 1563–1572 (2016)
Irving, B., et al.: Deep quantitative liver segmentation and vessel exclusion to assist in liver assessment. In: Valdés Hernández, M., González-Castro, V. (eds.) MIUA 2017. CCIS, vol. 723, pp. 663–673. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-60964-5_58
Ioffe, S., Szegedy, C.: Batch normalization: accelerating deep network training by reducing internal covariate shift (2015)
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., Wojna, Z.: Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2818–2826 (2016)
Tustison, N.J., et al.: N4ITK: improved N3 bias correction. IEEE Trans. Med. Imaging 29, 1310–1320 (2010)
Kingma, D.P., Ba, J.: Adam: a method for stochastic optimization. arXiv Preprint arXiv:1412.6980 (2014)
Antonelli, M., et al.: GAS: a genetic atlas selection strategy in multi-atlas segmentation framework. Med. Image Anal. 52, 97–108 (2019)
Acknowledgements
Portions of this work were funded by a Technology Strategy Board/Innovate UK grant (ref 102844). The funding body had no direct role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Owler, J., Irving, B., Ridgeway, G., Wojciechowska, M., McGonigle, J., Brady, S.M. (2020). Comparison of Multi-atlas Segmentation and U-Net Approaches for Automated 3D Liver Delineation in MRI. In: Zheng, Y., Williams, B., Chen, K. (eds) Medical Image Understanding and Analysis. MIUA 2019. Communications in Computer and Information Science, vol 1065. Springer, Cham. https://doi.org/10.1007/978-3-030-39343-4_41
Download citation
DOI: https://doi.org/10.1007/978-3-030-39343-4_41
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-39342-7
Online ISBN: 978-3-030-39343-4
eBook Packages: Computer ScienceComputer Science (R0)