Abstract
Accurate liver and lesion segmentation plays a crucial role in the clinical assessment and therapeutic planning of hepatic diseases. The segmentation of the liver and lesions using automated techniques is a crucial undertaking that holds the potential to facilitate the early detection of malignancies and the effective management of patients’ treatment requirements by medical professionals. This research presents the Generalized U-Net (G-Unet), a unique hybrid model designed for segmentation tasks. The G-Unet model is capable of incorporating other models such as convolutional neural networks (CNN), residual networks (ResNets), and densely connected convolutional neural networks (DenseNet) into the general U-Net framework. The G-Unet model, which consists of three distinct configurations, was assessed using the LiTS dataset. The results indicate that G-Unet demonstrated a high level of accuracy in segmenting the data. Specifically, the G-Unet model, configured with DenseNet architecture, produced a liver tumor segmentation accuracy of 72.9% dice global score. This performance is comparable to the existing state-of-the-art methodologies. The study also showcases the influence of different preprocessing and postprocessing techniques on the accuracy of segmentation. The utilization of Hounsfield Unit (HU) windowing and histogram equalization as preprocessing approaches, together with the implementation of conditional random fields as postprocessing techniques, resulted in a notable enhancement of 3.35% in the accuracy of tumor segmentation.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
In 2020, 905,700 individuals worldwide were diagnosed with liver cancer, and 830,200 died from it [30]. Liver cancer is one of the top three causes of cancer deaths in 46 countries. The number of new cases and fatalities from liver cancer might rise by more than 55% by 2040. Many malignancies are detected in their advanced stages, when there are few therapeutic treatments and the prognosis is poor. Early cancer identification leads to more effective therapy and much improved survival rates. However, 50% of cancers are still detected after they have progressed significantly. Improved early cancer detection has the potential to considerably increase survival rates. Recent advances in early detection have undoubtedly saved lives, but more advances and the development of early cancer detection systems are essential.
Medical professionals commonly employ contrast-enhanced computed tomography (CT) as a diagnostic modality for the detection of cancer. Abdominal CT scans are employed in cases of hepatocellular carcinoma. The manual segmentation of liver tumors from CT images in the early stages poses significant challenges due to the subtle distinctions between normal and malignant tissues. There are numerous sizes and configurations of liver tumors, and the appearance of each case can vary considerably. The density and contrast enhancement patterns of liver tumors may resemble those of the adjacent healthy liver tissue. The aforementioned resemblance poses a visual impediment in differentiating the tumor from the healthy liver parenchyma, especially when utilizing early-phase or non-contrast CT images. An additional consequence of employing contrast in CT images is the amplification of noise levels. The utilization of manual demarcation may introduce human limitations that compromise the accuracy of segmentation. Expert judgment, on the other hand, relies on the skill and experience of the individuals involved. The presented visual representation in Fig. 1 depicts a cross-sectional view of a CT image, wherein the liver region is demarcated using the color green, while the liver tumors are demarcated using the color red.
In order to tackle these challenges, software tools and computer-assisted techniques have been devised to aid radiologists in the delineation of hepatic tumors. These instruments can segment tumors based on their density, texture, and other characteristics using sophisticated algorithms, producing more objective and consistent results. Moreover, by reducing the likelihood of human error and accelerating the procedure, these automated methods can improve the precision of liver tumor detection in CT images.
Deep learning-based algorithms have made substantial progress in liver tumor segmentation from CT scans by improving accuracy, consistency, and efficiency. Deep learning models utilizing convolutional neural networks (CNNs) have demonstrated the ability to learn about intricate patterns and characteristics found in CT images, which traditional image processing approaches have struggled with. Operating in real-time or surgical environments, they furnish surgeons with instantaneous feedback throughout the course of procedures. Models capable of deep learning are adaptable to image quality, patient anatomical, and tumor-specific variations. Their capacity to acquire knowledge from a variety of datasets renders them practical in an extensive spectrum of clinical situations. To mitigate the requirement for large annotated datasets, deep learning methodologies, including semi-supervised and weakly supervised learning, have been implemented. When labeled data is insufficient, deep learning models for hepatic tumor segmentation can be trained more easily using this method. As additional data is acquired and algorithms progress, it is possible to consistently enhance and update the models. This adaptability guarantees that the models remain current with the most recent insights and developments. Deep learning has fueled research in liver tumor segmentation, resulting in the development of new architectures and approaches that continue to push the field forward. Potentially, these developments will enhance liver cancer patient care, treatment planning, and prognoses.
Despite the extensive body of research undertaken in this domain employing many deep learning architectures, the bulk of these models exhibit complexity, necessitating substantial computing resources and prolonged training durations. There exists a significant possibility for enhancement in terms of precision and accuracy. The applications of this study include a wide range of medical modalities, including MRI, ultrasound, X-ray, PET, etc. This research will be useful for applications that need similar image segmentation.
Numerous investigations have been conducted over the last few decades to explore the application of automated and semi-automatic techniques for liver tumor segmentation using CT images. Prior research has indicated that conventional image processing methods, such as statistical form models, region-growing models, graph-cut models, and similar approaches, exhibit limited efficacy when used to segment CT images. In recent times, there has been a notable emergence of deep learning models that rely on fully convolutional neural networks (FCNs) for various tasks, including classification and segmentation. These models, such as AlexNet, SegNet, U-Net, Residual Networks (ResNets), and Densely Connected Convolutional Neural Networks (DenseNets), have demonstrated significant potential and yielded promising outcomes in their respective domains.
Most traditional segmentation techniques, which depend on image processing and basic machine learning algorithms, require prior knowledge and information on tissue qualities. Consequently, these strategies exhibit a lack of reliability in achieving precise segmentation. A wide range of deep learning models, including AlexNet, SegNet, U-Net, ResNet, DenseNets, FCNs, DenseUnets, MA-Net, and their derivatives, have been used for liver tumor segmentation. The segmentation accuracy of some models does not meet the expected standard. Some models exhibit a multitude of layers, resulting in a complicated network structure that involves a significant number of parameters. This necessitates a significant amount of training time and computing capacity. Therefore, an effective model that is parameter-efficient and demands a tolerable amount of processing power is still needed for autonomously segmenting liver tumors from abdominal CT images.
This work employs a combination of successful models to develop a hybrid model capable of precisely and autonomously segmenting liver cancers. The proposed G-Unet model has the capability to accommodate U-Net, ResNet, and DenseNet models with few adjustments.
The key contributions of the investigation are as follows:
-
In this study, we provide a novel and versatile model known as the G-Unet model, which has been meticulously developed with a modular approach. With a few adjustments, the G-Unet has the capability to be set as a conventional Convolutional Neural Network (CNN), ResNet, or DenseNet. The architectural structure will stay consistent with that of a typical Convolutional Neural Network (CNN). The ResNet architecture incorporates identity mapping by utilizing shortcut connections that perform element-wise addition of features. Each layer in DenseNet gets feature maps from all the preceding layers and adds features element by element. Since each layer contributes to the same set of channels, the feature map size stays constant since summation is utilized rather than concatenation. The model that is obtained exhibits both parameter and computational efficiency.
-
We experiment with the LiTS dataset to evaluate the performance of different G-Unet setups with CNN, ResNet, and DenseNet.
-
In this study, we examine the impact of various preprocessing and postprocessing approaches on the performance of the G-Unet architecture.
This paper is organized as follows: In Section 2, a comprehensive review of the existing literature is presented, focusing on the deep learning methodologies as well as the preprocessing and postprocessing approaches employed in the context of liver tumor segmentation. Section 3 gives a detailed discussion of the proposed approach. Section 4 discusses the experimental plan and findings. The conclusion is presented in Section 5.
2 Literature review
Image segmentation tasks have become extremely efficient because of advances in machine learning techniques, the emergence of convolutional neural networks, and CNN-based deep learning models. Deep learning algorithms have also gained prominence as a result of publicly available datasets. ISBI 2017 conducted a liver lesion segmentation competition named LITS in conjunction with MICCAI 2017. The majority of current research on liver lesion segmentation is based on the LITS challenge. This section describes the various deep learning techniques employed for liver tumor segmentation, as well as the preprocessing and postprocessing techniques used to enhance segmentation accuracy.
2.1 Deep learning-based segmentation methods
Some of the earliest segmentation techniques were based on convolutional neural networks and fully convolutional neural networks (FCNs). The authors in [10] presented the most significant earlier research on liver tumor segmentation, which relied on cascaded fully convolutional neural networks (CFCN) for segmenting liver tumors from CT images. The U-Net architecture [29], which is appropriate for pixel-wise prediction, was implemented at two levels. As a postprocessing phase, dense 3D conditional random fields with spatial coherence are used to enhance segmentation accuracy. Using a similar technique, [11] segmented liver tumors from MRI images. Both of these approaches utilized Hounsfield unit adjustment and contrast enhancement as preprocessing steps.
The U-Net [29] rose to prominence in biomedical image segmentation applications. Numerous researchers have used variants of the U-Net model to segment CT images. The authors of [9] proposed a technique based on a two-dimensional U-Net architecture and a random forest classifier as a postprocessing step. The technique was able to obtain 0.65 dice coefficient accuracy. To prevent the obscuring of image features caused by U-Net’s skip connection, Seo et al. [31] proposed modifying U-Net to include a residual path in the skip connection. Another similar model based on U-Net was proposed in [3], which used class balancing to improve segmentation accuracy by removing slices without lesions. Li et al. [21] proposed a U-Net that focuses specifically on bottleneck feature vectors to prevent information loss during training. Another architecture based on U-Net and 3D FCN was proposed in [39]. Additionally, they employed the level set method and fuzzy C-means clustering to enhance the segmentation accuracy. Tran et al. [36] Un-net is another U-Net-based model in which the convolution block is redesigned to retrieve more features and employs dilated convolution to detect wider output features while preserving spatial information. Kushnure and Talbar [19] HFRU Net is also based on the U-Net design with modified skip connections to improve high-level functionality. In the skip connections, squeeze and excitation networks are used to improve feature extraction. The study in [33] presents a U-Net-based level set model that aims to automatically separate the liver from CT images. A level-set framework receives the segmented images transmitted by the initial U-Net. The proposed multi-phase level set formulation successfully segmented large and intricate images in a shorter amount of time. Song et al. [34] proposed a model based on spatial and temporal convolutional neural networks that aids in the exploration of contextual data from CT images. The method was only capable of segmenting tumors with an accuracy of 0.688 Dice coefficient. The primary drawbacks of these methods are the lengthy training period and less precise segmentation.
Residual networks have demonstrated high efficiency in applications related to image recognition and segmentation. In their study, the authors [15] introduced a novel deep convolutional neural network architecture that combines the U-Net and ResNet [16]. The utilization of long-range U-Net connections and ResNets’ skip connections resulted in an enhancement in the model’s performance. These techniques had prolonged training periods and poor precision. Cascaded deep residual networks were also utilized by Bi et al. [5] to enhance learning; however, segmentation accuracy was quite poor.
In [28], the researchers employed a hybrid approach by integrating the U-Net and ResNet models. Meanwhile, in [2], the ResNet-50 network was utilized in conjunction with the YOLOV3 model. However, the sample size in both investigations was insufficient to yield a generalizable outcome. Eres-UNet++ is a residual network-based model that builds upon the UNet++ architecture as its foundation. It enhances feature extraction and addresses uneven sample distribution by incorporating a channel attention module within the residual block. However, the model has an undersegmentation issue.
The architecture of the Residual Multi-Scale Attention U-Net proposed in [18] consists of two main components: a residual block and a multiscale attention block. The utilization of multiscale attention blocks facilitates the effective capture of both multiscale and spatial elements. Tumor segmentation accuracy was 0.76 dice score using the approach. SAA-Net [38] is an architectural framework that leverages the U-Net model as its foundational structure and incorporates scale attention and axis attention techniques to enhance the precision of segmentation. In [17], a 3D Fully Convolutional Network (FCN) was introduced, utilizing the Attention Hybrid Connection Network (AHCN) architecture. The approach demonstrates the utilization of attention processes in order to localize the liver and tumor. The model demonstrated a reasonable level of accuracy in segmenting tumors, as indicated by a dice global value of 0.591.
RIU-NET [25] is a network that has been created by integrating U-Net, ResNet, and InceptionV3. This approach works in 2.5D to identify spatial information and employs Inception convolution to reduce the number of parameters. The model’s dice accuracy for tumor segmentation was 73.79%. The approach employed in the study was based on multichannel fully convolutional networks [35]. The three distinct stages of contrast-enhanced CT images underwent processing using three separate Fully Convolutional Networks (FCNs) that were trained with different settings. Additionally, a feature fusion layer was employed to merge the extracted features. In their study, the authors [6] employ a methodology that involves the utilization of two consecutive deep encoder-decoder convolutional neural networks. These networks are specifically designed for the purpose of detecting liver and liver cancers and are built around the SegNet model. The performance of the model was not able to get a level of accuracy that is equivalent in the task of liver tumor segmentation. In a previous study [1], a comparable approach utilizing SegNet was investigated, but with minimal enhancements in terms of accuracy.
The architectural design proposed in [4] comprises a 3D U-Net model utilized for liver segmentation, a fractal residual structure employed for liver tumor segmentation from CT images, and a multi-scale candidate generation technique employed for creating tumor candidates. The model demonstrated a modest accuracy of 0.67 dice. Fan et al. [14] The multi-scale attention network employed position-wise and multi-scale attention blocks to effectively identify spatial and channel-wise information, representing a unique approach in this domain. The liver tumor segmentation model achieved a dice score of 0.749.
In [23] the model proposed has a high-resolution backbone composed of deep convolutional layers, which are responsible for extracting multiscale information and then fusing them. Additionally, the model has a self-attention module to effectively capture and consider long-range contextual data. The liver tumor segmentation dice score attained by the model on the LiTS dataset was 58.49+-27.83.
Different versions of DenseNet have demonstrated encouraging outcomes in the domain of semantic segmentation challenges. Chi et al. [8] The X-Net model is a variant of the DenseUnet architecture that incorporates an additional deconvolution operation. This modification enhances the effectiveness of tumor segmentation while improving efficiency. Additionally, a loss function based on active contours is employed to compare contour areas. The model successfully attained a dice score of 76.4% for tumor segmentation. Li et al. [22] The H-DenseUNet architecture is a hybrid composition consisting of the U-Net and DenseNet designs. Additionally, the model has a hybrid feature fusion layer that takes into account both intra-slice and inter-slice information while performing the segmentation process. In a previous study, a comparable model known as U-ADenseNet was introduced [40]. This model incorporates U-Net as a first step, followed by the use of ADenseNet for the purpose of fine segmentation. Despite the high accuracy exhibited by both models, the training process was time-consuming.
Chen et al. [7] TKiU-NeXt is an enhanced version of the 3D KiUNet framework, comprising two distinct components known as TK-Net and UNext. TK-Net is a kite-net that uses a transformer-based architecture to acquire knowledge from compact structures, whereas UNext is a model that specializes in capturing the structural characteristics. The complexity of the model demands a substantial amount of time for training. The TD-Net architecture, as described in [12], is constructed by combining a U-shaped convolutional neural network (CNN) with a Transformer module. This design allows for the retrieval of comprehensive global contextual information. The segmentation performance did not meet the expected standard. In [24], the authors put forward a network based on Generative Adversarial Networks (GANs) with the aim of creating synthetic lesions. Incorporating the created synthetic lesions into the training dataset increased segmentation performance by 4-5
In our earlier investigation, the liver and liver tumor were segmented on CT images using a modified version of the U-Net architecture [13]. The methodology employed in this study consisted of employing two distinct U-Nets for the purpose of performing segmentation tasks. The findings of the study demonstrate that the U-Net model, despite its straightforward design, is capable of achieving exceptional levels of accuracy in the task of segmentation.
2.2 Preprocessing and postprocessing techniques
In addition to employing deep learning architectures, numerous studies have incorporated preprocessing and postprocessing techniques as a means to enhance the accuracy of liver tumor segmentation. Hounsfield Unit (HU) windowing is the preprocessing method that is most frequently employed. The process involves altering the Hounsfield Unit (HU) values to selectively display the liver and its associated tissues. Histogram equalization is a frequently employed preprocessing method [6, 10, 11, 27] that aims to enhance the contrast of a image by redistributing the intensity values of its pixels.
Several studies [17, 18, 20] have employed data augmentation methods, such as geometric transformations (e.g., clipping, shifting, flipping, scaling) on CT slices, to enrich the training data and enhance the network’s ability to generalize. The MinMax normalization procedure, also known as feature scaling or min-max scaling, is a commonly used technique for transforming the pixel intensity values of CT scans to a predefined range [17, 21]. The application of this normalization technique enhances the ability to compare pixel values across different images.
A Gaussian filter has been employed in some research [40] to minimize image noise and smooth out minute variations in pixel values. Several studies [15, 37] have utilized statistical methods to preprocess data by removing the mean energy. This strategy aims to mitigate the inconsistencies observed in the dataset. Morphological filters are commonly employed for the purpose of filling gaps or low-intensity regions within a image [5]. The study in [32] introduces an adaptive local variance-based level set (ALVLS) framework for the segmentation of medical images. The ALVLS model is utilized to differentiate between noise points and object edges, thereby enhancing the accuracy of segmenting medical images that exhibit intensity inhomogeneity and noise.
The application of many post-processing approaches can enhance the segmentation outcomes obtained by diverse deep learning algorithms. It has been discovered in many studies [10, 11, 40] that conditional random fields (CRFs) are very effective in incorporating geographical context and encouraging smoothness in segmentation results. Active contour models, often known as snakes, are widely utilized in the domain of image segmentation to precisely delineate object boundaries inside images [4]. The utilization of these methods is of great importance in the task of segmenting objects that display irregular shapes and do not have well-defined boundaries. Consequently, they play a crucial role in improving the segmentation of CT images. The segmentation results obtained from previous segmentation approaches may be improved and refined by including a Random Forest classifier [9, 40] as a post-processing tool in the context of segmenting CT images.
According to the findings of the literature review, U-Net, ResNet, and DenseNet architectures have demonstrated notable efficacy in the context of medical image segmentation tasks. Many of these models’ modifications have produced cutting-edge outcomes, but their design is quite complicated, training takes a long time, and they use a lot of resources. Additionally, the models exhibit a significant constraint in terms of their ability to generalize. The fundamental objective of this research is to provide a framework for CT image segmentation that can modularly accommodate different models without requiring a redesign of the architecture. The work also aims to investigate the impact of preprocessing and postprocessing methods on segmentation.
3 Methodology
The overall structure comprises a liver segmentation and lesion segmentation network, which is built using the LiTS dataset and a conventional U-Net architecture. The network has been structured in a modular fashion to facilitate the implementation of various network designs. Due to its modular architecture, any convolution block inside the pipeline may be employed as a conventional convolutional network, a residual network, or a dense convolution neural network.
3.1 Preprocessing
The training dataset comprises 200 abdominal CT scans in the NIfTI (Neuroimaging Informatics Technology Initiative) format, obtained from the Liver Tumor Segmentation Challenge (LiTS). The NIfTI file format is widely utilized in the storage of neuroimaging data, including magnetic resonance imaging (MRI) and CT scans. The Python library known as nibabel is utilized for the purposes of reading, writing, and manipulating NIfTI files. Subsequently, a NumPy array file is generated for each CT slice. Each slice is transformed into a 512 by 512 pixel image and transmitted to the framework.
Based on a review of the literature and experiments, Hounsfield Unit (HU) windowing and histogram equalization were discovered to be particularly successful in emphasizing features and boosting segmentation accuracy. As a result, HU windowing was performed as a preprocessing step before submitting the CT images to the G-Unet, followed by Historgram equalization.
Within the area of CT imaging, the practice of HU windowing, sometimes referred to as CT windowing or just windowing, entails the manipulation of the visual representation of CT images in order to effectively visualize designated intervals of Hounsfield Unit values. The Hounsfield unit is a quantifiable metric employed in CT imaging for the purpose of expressing the radiodensity of various tissues or substances present in the human body. The scale is established using water (HU = 0) and air (HU = -1000) as reference points, with other substances positioned along this scale according to their radiodensities. The radiodensity of the liver is often characterized by a certain range of HU values. By correctly changing the window width (WW) and window level (WL), it is possible to precisely highlight the liver.
The window width (WW) refers to the range of HU values that are visually represented on the grayscale image. A WW covering the HU values related to the hepatic tissues is selected in order to see the liver. The WW needs to be large enough to encompass not just the liver but also its blood vessels and other liver-related components. The selection of the appropriate WW for liver imaging is dependent upon several factors, including the individual CT scanner model, patient dimensions, and acquisition settings. However, it is generally observed that a WW range of around 200 to 400 HU is commonly employed in this context.
The middle gray on the display is represented by the HU value, known as the WL. For liver imaging, the WL is commonly set to the average HU value of the liver tissue. This technique guarantees that the liver is shown as a shade of gray that is neither too light nor too dark in the image, hence facilitating its differentiation from surrounding anatomical components. Since liver tissue often falls between 40 and 60 HU, the WL for liver imaging is typically set in this range.
Through the implementation of these specific window settings, the liver tissues will be effectively emphasized and exhibited with optimal contrast, hence facilitating the deep learning framework in accurately discerning hepatic structures as well as detecting any potential anomalies or lesions. Although the surrounding tissues and organs may appear in various tones of gray, they won’t be as noticeable as the liver.
Histogram equalization is a widely employed method in image processing that aims to improve the contrast of images. It can be challenging to discern between key details or characteristics when using CT imaging since the data that is acquired may have a broad range of intensity values, and certain portions of the image may look overly dark or bright. Histogram equalization can solve this issue and improve image quality by shifting intensity levels in a way that increases contrast.
The initial procedure involves calculating the histogram of the CT image. The histogram provides a visual representation of the frequency distribution of intensity values over the entirety of the image. The data shown illustrates the number of pixels associated with each respective intensity level. The Cumulative Distribution Function (CDF) is obtained by deriving it from the histogram and serves as a representation of the cumulative probability associated with each intensity value. The value is derived by aggregating the probability of pixel occurrences across all intensity levels. The subsequent phase involves the development of a transformation function. In order to create a more even and contrast-enhancing image, this function essentially redistributes the pixel values by mapping the original intensity values to new intensity values. The transformation function is systematically applied to each individual pixel within the CT image, resulting in the conversion of the initial intensity values to their respective new values. This method expands the spectrum of intensities, leading to an image that exhibits enhanced contrast. After histogram equalization, normalization is carried out to make sure that the improved image’s intensity values fall within the acceptable range.
The histogram equalization procedure improves contrast by maximizing the available intensity range. The lower range is increased to accommodate dark places, while the higher range is expanded to accommodate bright areas. Consequently, the visibility and differentiation of features in both dark and bright regions are improved, resulting in an image that exhibits higher contrast as well as improved visual quality.
Equation 1 is used to create the transformation function that maps the initial intensity values to the new intensity values, where L represents the maximum intensity level, CDF(Original_Intensity) denotes the cumulative distribution function value associated with the original intensity value, M represents the total number of rows in this image, and N represents the total number of columns in the image. The CT slices that have been preprocessed are used to train the proposed G-Unet framework.
3.2 G-Unet framework
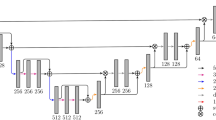
G-Unet is a variant of the conventional U-Net design. The modular design of the G-Unet architecture enables it to incorporate regular convolution neural networks, residual networks, and densely connected convolution neural networks with minimal modification. G-Unet can therefore be adjusted to operate with various settings and compare the results based on the type of input data. Figure 2 illustrates the G-Unet architecture.
Overall, the G-Unet architecture is comparable to the U-Net architecture. It comprises a network of encoders and decoders. The encoder network is accountable for capturing the context and characteristics of the input image while simultaneously reducing its spatial dimensions. It is termed the “contracting path” because the spatial resolution progressively decreases while the number of feature channels increases. After each convolutional block, the number of feature channels is doubled, allowing the network to capture increasingly intricate features as it proceeds deeper. The goal of the decoder network is to generate a segmentation mask by progressively upsampling the feature maps to the size of the original image. Convolutions are used to accomplish the upsampling. These procedures increase spatial resolution while decreasing the number of feature channels. To preserve spatial information, the feature maps from the contracting path (encoder) are concatenated with their corresponding upsampled feature maps in the decoder. Between the contracting path and the expanding path, the architecture includes skip connections. These connections enable the decoder to access low-level feature information from the encoder, enabling the upsampling process to preserve fine-grained details.
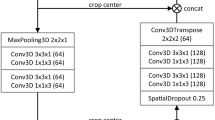
Modularity is provided by convolutional modules. As shown in Fig. 3, each convolution block can be configured to operate as a conventional convolutional neural network, residual network, or dense network. Each residual block in ResNets comprises a shortcut link that adds the input to the output, thereby learning the residual difference between the input and the intended output. The residual connections include the element-wise addition of a residual block’s input and output. In DenseNets, each layer receives additional inputs from all preceding levels and transmits its own feature maps to all subsequent layers in a feedforward manner. DenseNets are also designed to employ element-wise addition of previous layer feature maps rather than concatenation to minimize model complexity.
Each convolution block in Fig. 2 consists of a group of four convolution layers connected by a batch normalization layer, a feature dropout layer, and a leaky ReLU activation function.
The process operates on two distinct levels with the same architecture but distinct hyperparameter configurations. The first level segments the liver from the input CT image, and the second level segments the liver tumor from the first level’s masked CT image.
3.3 Convolution block as normal convolution neural network
A convolution block may be set up to function like a typical CNN. In conventional CNN design, a convolutional block often denotes the fundamental unit employed in both the encoder and decoder components. This block usually includes two or more convolutional layers, followed by normalization layers and activation functions. The primary function within a convolutional block involves the implementation of one or more convolutional layers. Each layer of the convolutional neural network is responsible for learning and extracting distinct feature mappings from the input data. The feature maps in question serve to collect a multitude of patterns and information that are inherent within the input image. The application of Leaky ReLU activation functions is performed in an element-wise manner on the feature maps. Activation functions play a crucial role in introducing non-linear characteristics to the model, hence enabling it to acquire a deeper understanding of intricate interactions between the input and output variables. Batch normalization is used to speed up and stabilize the training process. Batch normalization is a technique that standardizes the activations of the preceding layer, resulting in enhanced network stability and less sensitivity to the initial weight selection. Instead of maxpooling, strided convolution is employed.
3.4 Convolution block as ResNets
In G-Unet, a convolution block may alternatively be set up as a ResNet. ResNets may be used efficiently for liver tumor segmentation tasks to improve model accuracy and handle complicated and intricate tumor shapes. ResNets are capable of addressing challenges such as the presence of small or irregular tumors, variations in tumor morphology and intensity, and the need for precise pixel-level segmentation. ResNets excel at learning hierarchical and abstract characteristics from CT scans. In ResNet, every layer is designed to acquire more features, and the inclusion of skip connections enables the seamless propagation of gradient information, hence enabling the successful training of extremely deep neural networks. This feature facilitates the model’s ability to grasp the delicate intricacies and subtle patterns present in liver tumor images.
ResNets leverage the inclusion of residual blocks to enable the development of deep neural networks while mitigating the adverse effects of the vanishing gradient problem. This depth helps the model learn intricate tumor representations, increasing segmentation precision. The skip connections in ResNets permit the direct propagation of features from one layer to the next, thereby creating shortcuts for gradient flow during backpropagation. This helps to maintain minute details while also preventing data loss as it travels through the network. Liver tumors can manifest diverse characteristics, including differences in their form, size, and visual attributes. ResNets provide the capability to effectively adapt to the inherent unpredictability in data by simultaneously collecting high-level global context and low-level local information. The residual blocks allow the model to concentrate on discovering the disparities, or residuals, between tumor regions and the adjacent healthy liver tissue.
Let X represent the input CT image and Y denote the ground truth segmentation mask for liver tumors. A residual block is composed of a sequence of convolutional layers and skip connections. The calculation of the residual block can be formally stated as shown in (2).
In this context, the symbol Z denotes the intermediate feature map that is derived subsequent to the application of convolutional layers and activation functions within the residual block. The convolution process is denoted by “Conv”, the learnable parameters are represented by “\(W_{i}\)”, the Leaky Relu activation function is denoted as “Activation”, and batch normalization is represented as “BatchNorm”.
The difference between the input and the intermediate feature map is computed by the residual function \(F(X,W_{i})\) as shown in (3).
The residual block’s output, which establishes a shortcut connection, is the sum of its input and residual function as given in (4).
The residual block’s output is represented by \({Y_{residual}}\). ResNets allow the network to learn the residual mapping between the input image and the required segmentation mask, allowing for enhanced feature learning, adaptability to variances, and accurate liver tumor segmentation.
3.5 Convolution block as DenseNets
The convolution block inside the G-Unet architecture may also be customized to function as a DenseNet. DenseNet is a form of neural network in which each neuron in a given layer is connected to each neuron in the succeeding layer. In contrast to convolutional or recurrent neural networks, which have skipped connections, the connections are dense in this network. During the process of forward propagation, the input data is sequentially sent across the many layers of the neural network. The input values are multiplied by the relevant weights assigned to each connection, and the resulting products are aggregated to generate an intermediate value for each neuron. Subsequently, a non-linear activation function is employed to introduce non-linearity, resulting in the output of the activation function serving as the output of the given neuron. To add non-linearity, element-wise leaky Relu activation functions are applied to the intermediate values in the neurons. This non-linearity enables the network to represent complicated relationships in data and to learn and approximate non-linear functions. The learnable parameters of the dense network are the weights and biases associated with the connections between neurons. Throughout the training procedure, the weights undergo updates using gradient descent in order to minimize a loss function that measures the disparity between the anticipated output and the true target output. DenseNets create a more complex connection structure that iteratively concatenates all feature outputs in a feedforward manner. Consequently, the output of the layer l is as shown in (5).
Here, the concatenation of feature maps is represented by [...]. In this particular case, the architecture of H comprises a batch normalization layer, a rectified linear unit (ReLU) activation function, a convolutional layer, and a dropout layer. The aforementioned connection topology facilitates the reutilization of features and guarantees that all levels of the architecture get supervisory signals directly. The output dimension of each layer has k feature maps. K, commonly referred to as the growth rate, is frequently set to a small value. Hence, the number of feature mappings in DenseNets exhibits a direct correlation with the network’s depth. Thus, after l layers, the input \( [\ x_{l-1},\ x_{l-2},\ \ldots ,\ x_0]\) will have l x k feature maps.
The DenseNet architecture incorporates the concatenation of feature maps from previous layers, resulting in a substantial increase in parameters and the formation of a complex network structure. In this model, the operation of concatenation is replaced with summation, specifically element-wise addition. This modification enhances the model’s efficiency and accelerates the learning process.
Furthermore, DenseNets offer improved training capabilities because of their increased parameter efficiency as well as enhanced information and gradient flow. The gradients of the loss function and the initial input signal are directly accessible to each layer, resulting in automatic deep supervision. This facilitates the training of network topologies that exhibit higher levels of complexity.
3.6 Hyperparameter settings
The same G-Unet model is employed at two different levels: liver segmentation and liver tumor segmentation. However, the hyperparameter values vary for each level. The hyperparameters employed in this model exhibit uniqueness in the context of a hybrid model of this nature and have the potential to enhance the accuracy of CT image segmentation.
Leaky ReLu is preferred to ReLU because it is computationally cheaper, rapidly converges, and activates the network sparsely. By allowing the contextual feature information gained in the encoder blocks to be utilized, skip connections help in the construction of the segmentation map. The purpose is to generate a feature map from the encoder blocks’ high-resolution features. The use of a very modest slope parameter \(\alpha \) that takes into account negative value information, as seen in (6), prevents negative values from being sent to zero.
The initial level employs a 3X3 filter size. Initialization of the weights is performed via Kaiming Initialization [16]. Kaiming devised a robust initialization technique through meticulous simulation of the non-linearity shown by Rectified Linear Units (ReLUs), enabling the convergence of models with significant depth. This methodology effectively mitigates the issue of vanishing and inflating gradients by ensuring that gradients do not become too small or huge.
The output of each convolution layer can be described using the (7).
\(y_l\) , \(x_l\) and \(b_l\) are vectors. \(x_l\) is a, \(n_l\) * 1 vector of activations from the previous layer \(y_l\) -1 using activation function f represented as \(x_l\) = f (\(y_l\) -1). Here \(n_l\) represents the number of activations of layer l. \(W_l\) is a \(d_l\) * \(n_l\) weight matrix between \(l-1\) and l layers. \(d_l\) is the number of channels. Layer l biases are represented by \(b_l\).
Equation 8 suggests layer l weights to be initialized with zero mean gaussian distribution and a standard deviation of \(\sqrt{\frac{2}{n_l}}\) .
Where Variance is represented by Var. \(w_l\) represents each weight element in \(W_l\). Kaiming Initialization gives the initialized weights as shown in (9).
To prevent overfitting, the first-level configuration has an L2 regularization value of 0.00001. When the complexity of the model increases, regularization effectively increases the penalty. The regularization term lambda (\(\lambda \)) penalizes all parameters besides the intercept, ensuring that the model adequately generalizes the data and does not overfit. L2 regularization is useful when there are interdependent features. L2 regularization (R) is represented by (10). The weight matrix is represented by W with i and j as indexes.
In order to mitigate the issue of overfitting, the initial level also incorporates a dropout rate of 0.2. The dropout technique is a form of regularization in neural networks that aids in reducing interdependencies among neurons during the learning process.
The segmentation model’s performance is measured using multi-class cross-entropy loss, which returns a probability value between 0 and 1. The cross-entropy loss grows when the anticipated probability and actual label diverge. The multi-class cross-entropy loss is represented by L in (11). y is the target value, \(\hat{y}\) is the projected value, and \(y^{\left( k\right) }\) is a value between 0 and 1 depending on the correct prediction.
The utilization of the batch normalization layer facilitates the independent functioning of each network layer. The purpose of this component is to normalize the output generated by the preceding levels. The learning rate is set at 0.00003 in the first level, with 90% of the dataset being used for training and 10% being used for validation.
The second level also uses an L2 regularization value of 0.00004 and a dropout rate of 0.2. The employed loss function consists of the multiclass cross-entropy loss, which is subsequently followed by the multiclass dice loss. In order to mitigate the issue of overfitting, the experimental setup utilizes a batch size of four along with the implementation of batch normalization. The training dataset comprises 90% of the CT scans, while the remaining 10% is allocated for testing purposes. Various data augmentation techniques, including vertical flipping, horizontal flipping, rotation, and magnification, are employed to expand the size of the training dataset.
3.7 Post processing using CRF
Based on a comprehensive review of relevant literature and empirical investigations, it has been established that Conditional Random Fields (CRFs) exhibit a high level of efficacy in enhancing the precision of segmentation outcomes, particularly in cases where there exists a discernible correlation among adjacent pixels within CT images. CRFs have demonstrated notable efficacy in integrating spatial context and promoting smoothness in the outputs of segmentation. This capability is advantageous as it aids in mitigating segmentation artifacts and enhancing the overall accuracy of segmentation.
The present work employs CRFs as a postprocessing methodology to enhance the original tumor segmentation acquired using the G-Unet framework. The segmentation output produced by the G-Unet model is utilized as the input for the CRF.
For each pixel in the CT image, intensity values are extracted as features and is represented as X. For each pixel in the CT image, the label y indicates whether the pixel belongs to the tumor region \((y=1)\) or the background \((y=0)\).
The energy function of the CRF is formulated as shown in (12).
Where Y is the set of label variables for all pixels \({y_1, y_2, ..., y_N}\) and X is the set of input feature variables for all pixels X1, X2, ..., XN. \(\sum _{i}{\psi _u\left( y_i,X_i\right) }\ \ \) is the unary potential function that represents the compatibility between the label \(y_i\) and the input features \(X_i\) for pixel i.\( \sum _{ij}{\psi _p\left( y_i,y_j\right) }\ \) is the pairwise potential function that models the interactions between neighboring pixels i and j. It captures the contextual information and encourages smoothness in the segmentation.
The unary potential function \( \sum _{i}{\psi _u\left( y_i,X_i\right) }\ \ \) measures the compatibility of label \(y_i\) with the observed features \(X_i\) of pixel i as shown in (13). It is determined by the degree of resemblance between the features and tumor patterns.
Here,\( \ P\left( y_i\ \vert X_i\right) \ \) represents the probability of assigning label \(y_i\) to pixel i given its features \(X_i\). The negative log-likelihood is used to convert the probability into an energy term.
The pairwise potential function \( \psi _p\left( y_i,y_j\right) \) models the interactions between neighboring pixels i and j as shown in (14). It discourages rapid label changes and promotes nearby pixels with similar features to share the same label. A common choice is the Potts model:
Here, \(\lambda \) is a weight parameter controlling the influence of the pairwise potential, and \( \delta \left( y_i,y_j\right) \) is the Kronecker delta function that equals 1 when \(y_i = y_j\) (same label for neighboring pixels) and 0 otherwise.
During the inference stage, probabilistic inference is used to discover the most likely label configuration Y that minimizes the energy function E(Y, X). The result of the CRF postprocessing is the refined segmentation in which the pixel labels have been modified based on the learned pairwise potentials and the input features.
4 Experimental setup and results
4.1 Dataset used for training and evaluation
The model was evaluated using 200 NIfTI-format abdominal CT images from the MICCAI 2017 and LiTS challenges (CodaLab Competition). The approach was evaluated using 200 CT images, 130 of which were utilized for training with ground truth masks and the remaining 70 without masks. 117 of the 130 samples were used for training, while 13 were used for validation.
4.2 Metrics used for performance evaluation
Competition organizers typically give ground truths to compare with segmented CT scans. Many criteria are used to evaluate segmentation accuracy and performance. The most widely used performance statistic is discussed in this section.
The following are the definitions of the metrics, where S represents the segmented region and R represents the ground truth.
The computation of Volumetric Overlap Error (VOE) involves the comparison of the intersection and union of two sets of segmentations. Values that are close to zero indicate efficient segmentation, whereas larger scores indicate discrepancies among segmented images. The formula for calculating the variable of interest (VOE) is shown in (15).
The dice similarity coefficient is a metric used to measure the level of pixel segmentation inside a certain region of interest. A DSC (Dice Similarity Coefficient) value of 1 signifies perfect segmentation, whereas values approaching 0 suggest more discrepancies. The (16) provides the mathematical expression for computing the DSC.
The Relative Volume Difference (RVD) is calculated as the percentage obtained by dividing the total volume of a segmented region by the total volume present in the ground truth. Insufficient segmentation is expected to yield negative results, while excessive segmentation is expected to yield positive values. The formula presented in (17) is utilized for the calculation of Relative Volume Difference (RVD).
The average symmetric surface distance (ASSD) refers to the mean value obtained by calculating the distances between points located on the border of the machine-segmented region and the corresponding boundary of the ground truth. ASSD is computed as the mean of all the distances measured using the Euclidean distance formula. A measurement of 0 mm indicates optimal segmentation.
The Root Mean Square Symmetric Surface Distance (RMSD) is a variant of the Average Symmetric Surface Distance (ASSD) that incorporates the square root of the ASSD value. A measurement of 0 mm indicates optimal segmentation. The maximum Symmetric Surface Distance (MSD) is a modified version of the Average Symmetric Surface Distance (ASSD) metric. MSD measures the greatest distance between the boundary voxels of a segmented region and the corresponding boundary voxels in the ground truth. A measurement of 0 mm indicates optimal segmentation.
Precision and recall are often employed criteria for assessing the accuracy of segmentation models. Precision is a metric that quantifies the proportion of accurately predicted positive cases out of all expected positive instances. In the context of segmentation, positive examples refer to the pixels or voxels that have been categorized as targets by the model. Precision is calculated using (18).
The metric of recall, sometimes referred to as sensitivity or true positive rate, quantifies the proportion of properly predicted positive cases out of all actual positive instances by the model. Recall is calculated using (19).
True positives (TP) refer to the pixels or voxels that have been accurately categorized by the model as belonging to the target class. False positives (FP) refer to the pixels or voxels that have been erroneously categorized as the target by the model. False negatives (FN) refer to the pixels or voxels that have been erroneously overlooked by the model and inaccurately categorized as not pertaining to the intended class.
The Intersection over Union (IoU), also known as the Jaccard Index, is a significant statistic used to assess the accuracy of segmentation. It quantifies the degree of overlap between the determined segmentation mask and the ground truth mask. Equation 20 is used to calculate IoU.
IoU has a range of 0 to 1, with 1 denoting a perfect match between the anticipated mask and the ground truth mask.
The evaluation metric known as precision at 50% overlap (IOU) is commonly employed in the assessment of segmentation models. This metric is particularly useful in tasks involving the dense annotation of object instances inside images, since it provides a measure of accuracy. The proposed metric is a modified version of the conventional precision metric, which incorporates the consideration of the intersection between the predicted segmentation masks and the ground truth masks. Precision at 50% IOU is a specific case of precision, where a threshold of 50% IoU is considered. The metric quantifies the ratio of accurately detected object instances (true positives) to the overall predicted instances within the regions where the predicted and ground truth masks exhibit an overlap of 50% or more. The formula for Precision at 50% IoU is given in (21).
Where, (Number TPs with IoU \(\ge 0.5\)) is the number of correctly identified instances for which the IoU with the corresponding ground truth instance is 50% or higher. Number of Predicted Instances (P) is the total number of instances predicted by the model. Recall at 50% IoU is a specific case of recall, where a threshold of 50% IoU is considered. The metric quantifies the ratio of accurately detected object instances (true positives) in relation to the overall number of ground truth instances inside the regions where the predicted and ground truth masks exhibit an overlap of 50% or more. The formula for Recall at 50% IoU is given in (22).
Where, (Number of True Positives (TP) with \(\ge 0.5\)) is the number of correctly identified instances for which the IoU with the corresponding ground truth instance is 50% or higher. Number of Ground Truth Instances (G) is the total number of instances (object instances) present in the ground truth annotations.
4.3 Training setup
The training procedure was conducted on a Dell R430-E5 server equipped with 128 GB of RAM and a Tesla P100 16GB GPU. During the training process, the central processing unit (CPU) usage reached a level of 20 gigabytes (GB), while around 10 GB of graphics processing unit (GPU) RAM was utilized.
4.4 Liver segmentation training and results
Three alternative configurations, namely Unet, ResNet, and DenseNet, were utilized in the experimentation of G-Unet. Initially, the training process for liver segmentation was conducted over a span of 55 epochs. Figure 4 displays the training losses associated with various epochs. Figure 5 displays the accuracy on the training dataset as a function of the dice coefficient for various epochs. Figure 6 displays the accuracy on the validation dataset as a function of the dice coefficient for various epochs. The maximum validation dice for the G-Unet setup as a typical CNN was 0.9598. The maximum validation dice for the G-Unet configured as ResNet was 0.9663. The maximum validation dice for the G-Unet with DensNet configuration was 0.9786.
4.5 Liver tumor segmentation training and results
The liver CT slices that have been masked during the first phase of liver segmentation are employed as training data for tumor segmentation. This is accomplished by utilizing G-Unet, which incorporates various configurations of regular CNN, ResNet, and DenseNet. The training for liver tumor segmentation was similarly conducted over a span of 55 epochs. Figure 7 displays the training loss values associated with various epochs. Figure 8 displays the accuracy on the training dataset as a function of the dice coefficient for various epochs. Figure 9 displays the accuracy of the validation dataset for various epochs in terms of the dice coefficient. The G-Unet, when built as a conventional Convolutional Neural Network (CNN), demonstrated a peak validation dice coefficient of 0.7272. The G-Unet, when set as ResNet, demonstrated a peak validation dice coefficient of 0.7349. The G-Unet, when configured as DensNet, demonstrated a validation dice coefficient of 0.7429 at its peak performance.
4.6 LiTS competition results
The architectural design was assessed by utilizing the set of 70 CT images supplied by the LiTS competition. The created lesion masks (segmentation output) were uploaded to the website for the LiTS Competition and received excellent results. Three experimental trials were conducted using varying configurations of the G-Unet. The liver segmentation findings are presented in Table 1, whereas Table 2 displays the liver tumor segmentation results.
The findings indicate that the G-Unet model, specifically configured with DenseNet, produced the most favorable outcomes when compared to the other two setups.
4.7 Preprocessing and postprocessing impact
The utilization of preprocessing and postprocessing techniques has the potential to enhance the accuracy of segmentation. Based on the findings of the literature research, it was determined that the most appropriate preprocessing approaches for CT image processing encompass Hounsfield Unit (HU) windowing, histogram equalization, min-max normalization, Gaussian filters, and morphological filters. Numerous trials and a multitude of studies have demonstrated that the utilization of HU windowing and histogram equalization techniques yields superior results in enhancing CT images and facilitates improved feature extraction in deep learning frameworks. Therefore, HU-windowing and histogram equalization techniques were used to preprocess the raw CT images from the LiTS dataset.
Active contour models, random forest classifiers, and conditional random fields (CRFs) are the most often utilized post-processing methods for CT image processing. Dense Conditional Random Fields (CRFs) have been found to be highly successful at integrating spatial context and promoting smoothness in the outputs of image segmentation. This capability is crucial in mitigating segmentation artifacts and enhancing the overall accuracy of the segmentation process.
To evaluate the effect of preprocessing and postprocessing techniques, 130 CT images with ground truths from the LiTS dataset were utilized. 117 of the 130 samples were used for training, while 13 were used for validation. Table 3 presents the impact of preprocessing and postprocessing methodologies on the accuracy of segmentation achieved through the utilization of the G-Unet framework. As a result of using HU-windowing and histogram equalization, the segmentation accuracy of the liver and liver tumor was both improved by 0.71% and 1.44%, according to the results. The utilization of Conditional Random Fields (CRF) as a postprocessing methodology has resulted in a notable enhancement in the accuracy of liver segmentation by 1.23% and liver tumor segmentation by 1.91%. The use of combined preprocessing and postprocessing strategies substantially boosted the accuracy of liver tumor segmentation by 3.35%.
4.8 Performance comparison with related work
A comparison of the effectiveness of liver tumor segmentation across several studies using recognized metrics is shown in Table 4. Furthermore, our previous investigation [13] only employed the U-Net framework, which yielded a Dice global score of 73%. The current model outperformed our prior model, earning a Dice global score of 79.2%. Table 4 demonstrates that, within the context of research publications, the suggested technique demonstrates a greater degree of performance and accuracy across the majority of criteria. In comparison to the algorithms currently employed, which involve several layers and need significant training duration, the model proposed in this research demonstrates the ability to achieve outstanding outcomes with a very short training period. The validation results of the LiTS competition have been made available on the CodaLab website under the username Deepak [39].
Figure 10 depicts the outcomes of liver segmentation, whereas Fig. 11 depicts the results of lesion segmentation. The first column represents a CT slice, the second a ground truth slice, and the third a segmented CT slice. The segmentation results in Fig. 11 show that the model can distinguish even the smallest tumor tissues.
5 Conclusions and future work
In this paper, we present the G-Unet modular framework for segmenting liver and liver lesions from abdominal CT images. G-Unet is a hybrid model constructed with the U-Net model as its backbone that can easily incorporate standard CNN, ResNet, and DenseNet with minimal configuration changes. G-Unet provides a modular architecture that is readily reconfigurable to accommodate different types of datasets and applications. The LiTS dataset is used to evaluate the various G-Unet configurations. G-Unet implemented as a conventional CNN is highly efficient at learning hierarchical features, retaining spatial data, and adapting to varying tumor sizes. G-Unet configured as ResNet acquires incremental features, and skip connections facilitate the flow of gradient data without interruption. This facilitates the model in obtaining images of the liver that contain intricate details and subtle patterns. G-Unet configured as DenseNet provides numerous benefits, such as dense connection, feature reuse, gradient stability, parameter efficiency, and the ability to incorporate both local and global contextual information. The performance of the different configurations of G-Unet was evaluated using the LiTS dataset, and the results indicate that G-Unet with the DenseNet configuration was able to achieve an accuracy of 79.2% dice global score for liver tumor segmentation, which is comparable to the current state-of-the-art techniques. In addition, the impact of preprocessing techniques such as HU-Windowing and histogram equalization, as well as postprocessing techniques such as conditional random fields, on the segmentation performance of G-Unet was evaluated. The application of preprocessing and postprocessing techniques resulted in a 3.35 % improvement in segmentation accuracy.
The fact that G-Unet with the DenseNet configuration achieved the highest segmentation accuracy for the LiTS dataset does not preclude the use of the other two configurations. Depending on the application and data set type, any of the three G-Unet configurations may be utilized. Consequently, G-Unet can be expanded to incorporate additional medical modalities and computer vision applications. The proposed methodology is applicable for feature extraction in diverse computer vision applications and may be implemented across numerous medical modalities, including MRI, ultrasound, X-ray, PET, and others. Future research endeavors may delve into various 3D neural network topologies, multi-label methodologies, and post-processing techniques in order to enhance the accuracy of lesion segmentation.
The huge number of parameters in the G-Unet with the DenseNet model may increase training time, particularly when working with large datasets. When the size of the training dataset is restricted, the model’s extensive parameter count may additionally increase the likelihood of overfitting. The potential scope of G-Unet and DenseNet’s efficacy in segmentation tasks might be restricted to particular domains or data types. Addressing these constraints frequently requires striking a careful balance between model complexity, processing power, and characteristics of the segmentation task at hand.
Data Availability
Dataset is freely available and can be downloaded from [41].
References
Almotairi S, Kareem G, Aouf M, Almutairi B, Salem MA-M (2020) Liver Tumor Segmentation in CT Scans Using Modified SegNet. Sensors (Basel) 20:1516. https://doi.org/10.3390/s20051516
Amin J, Anjum MA, Sharif M, Kadry S, Nadeem A, Ahmad SF (2022) Liver Tumor Localization Based on YOLOv3 and 3D-Semantic Segmentation Using Deep Neural Networks. Diagnostics 12:823. https://doi.org/10.3390/diagnostics12040823
Ayalew YA, Fante KA, Mohammed MA (2021) Modified U-Net for liver cancer segmentation from computed tomography images with a new class balancing method. BMC Biomed Eng 3:4. https://doi.org/10.1186/s42490-021-00050-y
Bai Z, Jiang H, Li S, Yao Y-D (2019) Liver Tumor Segmentation Based on Multi-Scale Candidate Generation and Fractal Residual Network. IEEE Access 7:82122–82133. https://doi.org/10.1109/ACCESS.2019.2923218
Bi L, Kim J, Kumar A, Feng D (2017) Automatic liver lesion detection using cascaded deep residual networks. arXiv:1704.02703 [cs]
Budak Ü, Guo Y, Tanyildizi E, Şengür A (2020) Cascaded deep convolutional encoder-decoder neural networks for efficient liver tumor segmentation. Med Hypotheses 134:109431. https://doi.org/10.1016/j.mehy.2019.109431
Chen G, Li Z, Wang J, Wang J, Du S, Zhou J, Shi J, Zhou Y (2023) An improved 3D KiU-Net for segmentation of liver tumor. Comput Biol Med 160:107006. https://doi.org/10.1016/j.compbiomed.2023.107006
Chi J, Han X, Wu C, Wang H, Ji P (2021) X-Net: Multi-branch UNet-like network for liver and tumor segmentation from 3D abdominal CT scans. Neurocomputing 459:81–96. https://doi.org/10.1016/j.neucom.2021.06.021
Chlebus G, Meine H, Moltz JH, Schenk A (2017). Neural network-based automatic liver tumor segmentation with random forest-based candidate filtering. https://doi.org/10.48550/arXiv.1706.00842
Christ PF, Elshaer MEA, Ettlinger F, Tatavarty S, Bickel M, Bilic P, Rempfler M, Armbruster M, Hofmann F, D’Anastasi M, Sommer WH, Ahmadi S-A, Menze BH (2016) Automatic liver and lesion segmentation in CT using cascaded fully convolutional neural networks and 3D conditional random fields. In: Ourselin S, Joskowicz L, Sabuncu MR, Unal G, Wells W (eds) Medical Image computing and computer-assisted intervention – MICCAI 2016. Springer International Publishing, Cham, pp 415–423
Christ PF, Ettlinger F, Grün F, Elshaera MEA, Lipkova J, Schlecht S, Ahmaddy F, Tatavarty S, Bickel M, Bilic P, Rempfler M, Hofmann F, Anastasi MD, Ahmadi S-A, Kaissis G, Holch J, Sommer W, Braren R, Heinemann V, Menze B (2017) Automatic liver and tumor segmentation of CT and MRI volumes using cascaded fully convolutional neural networks. arXiv:1702.05970 [cs]
Di S, Zhao Y-Q, Liao M, Zhang F, Li X (2023) TD-Net: A Hybrid End-to-End Network for Automatic Liver Tumor Segmentation From CT Images. IEEE J Biomed Health Inform 27:1163–1172. https://doi.org/10.1109/JBHI.2022.3181974
Doggalli D, Sunil Kumar BS (2022) The Efficacy of U-Net in Segmenting Liver Tumors from Abdominal CT Images. IJIES 15:151–161. https://doi.org/10.22266/ijies2022.1031.14
Fan T, Wang G, Li Y, Wang H (2020) MA-Net: A Multi-Scale Attention Network for Liver and Tumor Segmentation. IEEE Access 8:179656–179665. https://doi.org/10.1109/ACCESS.2020.3025372
Han X (2017) Automatic Liver Lesion Segmentation Using A Deep Convolutional Neural Network Method. Med Phys 44:1408–1419. https://doi.org/10.1002/mp.12155
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: 2016 IEEE conference on computer vision and pattern recognition (CVPR). pp 770–778
Jiang H, Shi T, Bai Z, Huang L (2019) AHCNet: an application of attention mechanism and hybrid connection for liver tumor segmentation in CT volumes. IEEE Access 7:24898–24909. https://doi.org/10.1109/ACCESS.2019.2899608
Jiang L, Ou J, Liu R, Zou Y, Xie T, Xiao H, Bai T (2023) RMAU-Net: Residual Multi-Scale Attention U-Net For liver and tumor segmentation in CT images. Comput Biol Med 158:106838. https://doi.org/10.1016/j.compbiomed.2023.106838
Kushnure DT, Talbar SN (2022) HFRU-Net: High-Level Feature Fusion and Recalibration UNet for Automatic Liver and Tumor Segmentation in CT Images. Comput Methods Programs Biomed 213:106501. https://doi.org/10.1016/j.cmpb.2021.106501
Li J, Liu K, Hu Y, Zhang H, Heidari AA, Chen H, Zhang W, Algarni AD, Elmannai H (2023) Eres-UNet++: Liver CT image segmentation based on high-efficiency channel attention and Res-UNet++. Comput Biol Med 158:106501. https://doi.org/10.1016/j.compbiomed.2022.106501
Li S, Tso GKF, He K (2020) Bottleneck feature supervised U-Net for pixel-wise liver and tumor segmentation. Expert Syst Appl 145:113131. https://doi.org/10.1016/j.eswa.2019.113131
Li X, Chen H, Qi X, Dou Q, Fu C-W, Heng P-A (2018) H-DenseUNet: Hybrid Densely Connected UNet for Liver and Tumor Segmentation From CT Volumes. IEEE Trans Med Imaging 37:2663–2674. https://doi.org/10.1109/TMI.2018.2845918
Li Y, Zou B, Liu Q (2021) A deep attention network via high-resolution representation for liver and liver tumor segmentation. Biocybern Biomed Eng 41:1518–1532. https://doi.org/10.1016/j.bbe.2021.08.010
Liu Y, Yang F, Yang Y (2023) A partial convolution generative adversarial network for lesion synthesis and enhanced liver tumor segmentation. J Appl Clin Med Phys 24:e13927. https://doi.org/10.1002/acm2.13927
Lv P, Wang J, Wang H (2022) 2.5D lightweight RIU-Net for automatic liver and tumor segmentation from CT. Biomed Signal Process Control 75:103567. https://doi.org/10.1016/j.bspc.2022.103567
Manjunath RV, Kwadiki K (2022) Modified U-NET on CT images for automatic segmentation of liver and its tumor. Biomed Eng Adv 4:100043. https://doi.org/10.1016/j.bea.2022.100043
Moghbel M, Mashohor S, Mahmud R, Saripan MIB (2018) Review of liver segmentation and computer assisted detection/diagnosis methods in computed tomography. Artif Intell Rev 50:497–537. https://doi.org/10.1007/s10462-017-9550-x
Rahman H, Bukht TFN, Imran A, Tariq J, Tu S, Alzahrani A (2022) A Deep Learning Approach for Liver and Tumor Segmentation in CT Images Using ResUNet. Bioengineering 9:368. https://doi.org/10.3390/bioengineering9080368
Ronneberger O, Fischer P, Brox T (2015) U-Net: convolutional networks for biomedical image segmentation. https://doi.org/10.48550/ARXIV.1505.04597
Rumgay H, Arnold M, Ferlay J, Lesi O, Cabasag CJ, Vignat J, Laversanne M, McGlynn KA, Soerjomataram I (2022) Global burden of primary liver cancer in 2020 and predictions to 2040. J Hepatol 77:1598–1606. https://doi.org/10.1016/j.jhep.2022.08.021
Seo H, Huang C, Bassenne M, Xiao R, Xing L (2020) Modified U-Net (mU-Net) with Incorporation of Object-Dependent High Level Features for Improved Liver and Liver-Tumor Segmentation in CT Images. IEEE Trans Med Imaging 39:1316–1325. https://doi.org/10.1109/TMI.2019.2948320
Shu X, Yang Y, Liu J, Chang X, Wu B (2023) ALVLS: Adaptive local variances-Based levelset framework for medical images segmentation. Pattern Recognit 136:109257. https://doi.org/10.1016/j.patcog.2022.109257
Shu X, Yang Y, Wu B (2021) Adaptive segmentation model for liver CT images based on neural network and level set method. Neurocomputing 453:438–452. https://doi.org/10.1016/j.neucom.2021.01.081
Song L, Wang H, Wang ZJ (2021) Bridging the Gap Between 2D and 3D Contexts in CT Volume for Liver and Tumor Segmentation. IEEE J Biomed Health Inform 25:3450–3459. https://doi.org/10.1109/JBHI.2021.3075752
Sun C, Guo S, Zhang H, Li J, Chen M, Ma S, Jin L, Liu X, Li X, Qian X (2017) Automatic segmentation of liver tumors from multiphase contrast-enhanced CT images based on FCNs. Artif Intell Med 83:58–66. https://doi.org/10.1016/j.artmed.2017.03.008
Tran S-T, Cheng C-H, Liu D-G (2021) A multiple Layer U-Net, U n -Net, for liver and liver tumor segmentation in CT. IEEE Access 9:3752–3764. https://doi.org/10.1109/ACCESS.2020.3047861
Zhang C, Hua Q, Chu Y, Wang P (2021) Liver tumor segmentation using 2.5D UV-Net with multi-scale convolution. Comput Biol Med 133:104424. https://doi.org/10.1016/j.compbiomed.2021.104424
Zhang C, Lu J, Hua Q, Li C, Wang P (2022) SAA-Net: U-shaped network with Scale-Axis-Attention for liver tumor segmentation. Biomed Signal Process Control 73:103460. https://doi.org/10.1016/j.bspc.2021.103460
Zhang Y, Jiang B, Wu J, Ji D, Liu Y, Chen Y, Wu EX, Tang X (2020) Deep Learning Initialized and Gradient Enhanced Level-Set Based Segmentation for Liver Tumor From CT Images. IEEE Access 8:76056–76068. https://doi.org/10.1109/ACCESS.2020.2988647
Zhu Y, Yu A, Rong H, Wang D, Song Y, Liu Z, Sheng VS (2021) Multi-Resolution Image Segmentation Based on a Cascaded U-ADenseNet for the Liver and Tumors. JPM 11:1044. https://doi.org/10.3390/jpm11101044
CodaLab - Competition. https://competitions.codalab.org/competitions/17094
Funding
No Funding was obtained for this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors have no conflict of interests on the manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
D J, D., B S, S.K. Liver tumor segmentation using G-Unet and the impact of preprocessing and postprocessing methods. Multimed Tools Appl (2024). https://doi.org/10.1007/s11042-024-18759-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11042-024-18759-y