Abstract
After several decades studying different acto-myosin complexes at lower and intermediate resolution – limited by the electron microscope instrumentation available then – recent advances in imaging technology have been crucial for obtaining a number of excellent high-resolution 3D reconstructions from cryo electron microscopy. The resolution level reached now is about 3–4 Å, which allows unambiguous model building of filamentous actin on its own as well as that of actin filaments decorated with strongly bound myosin variants. The interface between actin and the myosin motor domain can now be described in detail, and the function of parts of the interface (such as, e.g., the cardiomyopathy loop) can be understood in a mechanistical way. Most recently, reconstructions of actin filaments decorated with different myosins, which show a strongly bound acto-myosin complex also in the presence of the nucleotide ADP, have become available. The comparison of these structures with the nucleotide-free Rigor state provide the first mechanistic description of force sensing. An open question is still the initial interaction of the motor domain of myosin with the actin filament. Such weakly interacting states have so far not been the subject of microscopical studies, even though high-resolution structures would be needed to shed light on the initial steps of phosphate release and power stroke initiation.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
4.1 Introduction
When considering “the structure of acto-myosin” two seemingly different meanings come to mind: in the first place we think about the mechanism of force generation and motility via the actin/myosin molecular motor system. The notion that the structure of the interacting complex of the motor protein myosin and its track, filamentous actin, will define function is evident, and ever since electron microscopists produced the first visualisation of the interactions of myosin with actin in a muscle sarcomere (cf. Fig. 4.1; Huxley 1957) structural biologists carried out work to understand different states of the crossbridge cycle at finer and finer detail.
Electron micrograph of an ultrathin section of chemically fixed, heavy metal stained, and resin embedded muscle sample. Note the sarcomere Z-lines and the area of overlapping thick and thin filaments (red circle). In the area one can clearly see the myosin motor domains as “crossbridges” between the filaments. Such micrographs represent the typical first visualization of the acto-myosin complex and are considered recently again – in their 3D tomogram flavour – a valuable source for information about crossbridge states (cf. Figs. 4.11, 4.12 and text Sect. 4.5). (Image adapted from Huxley 1957)
On the other hand, the phrase “acto-myosin” has for a long time been used by electron microscopists when talking about “decorated F-actin”. This means purified filaments of either actin on its own, or sometimes as reconstituted or even native thin filaments with their tropomyosin and troponin components “decorated” with myosin sub-fragments where myosin binds strongly to the actin in its nucleotide-free Rigor state. This protein complex was for many years the only state of the crossbridge cycle accessible until crystals of myosin could finally be grown (Rayment et al. 1993a; Houdusse et al. 2000). “Decorated actin” was thus excessively studied, first by conventional electron microscopy via the so-called negative-staining technique, then since the 1990s by cryo electron microscopy. Typical reconstructions are shown in Fig. 4.2 illustrating some very early examples of this work (Moore et al. 1970; Milligan and Flicker 1987; Flicker et al. 1991).
Typical micrographs and 3D reconstructions from early myosin decorated F-actin in their nucleotide-free strong binding state, contrasted with negative stain (a), or embedded in a vitrified ice layer (b). In the case of the cryo electron microscopy sample (b), tropomyosin (blue in the 3D reconstruction) was present. (Images in (a) adapted from Moore et al. (1970), in (b) from Flicker et al. (1991))
As has happened often in structural biology, high-resolution structures from protein X-ray crystallography and not from electron microscopy have dominated the progress in our understanding of molecular functions. The common wisdom was that X-ray structures intrinsically have higher resolution and will give much more information than 3D reconstructions from electron micrographs. Acto-myosin has not been an exception from this rule, and X-ray crystallographers have produced a very large number of structures using nucleotide analogues, myosin mutants and/or unconventional myosins to produce a detailed picture of the possible conformational space of the motor domain on its own. What has been lacking for a long time was detailed information about the motor domain interacting with its track F-actin. Despite numerous attempts to form a small enough acto-myosin complex, which might be in the correct binding conformation and able to form crystals (Dawson et al. 2003; Qu et al. 2018), to date there is not yet any crystallographic interaction structure.
This leaves the field still wide open for electron microscopy, which in recent years has developed from a medium-resolution technique, providing typical 1-nm resolution data, to a truly structural biology tool, contributing data at the 3–4-Å resolution level at present. Over the last 25 years advances in sample preparation, image processing, as well as development of new microscopes, detectors, and automated data collection have boosted the field (cf. review by Schröder 2015) resulting in the awarding of Nobel prizes to Dubochet, Frank, and Henderson (Cressey and Callaway 2017). In the last 2 years high-resolution structures of F-actin (3.1 Å, Chou and Pollard 2019) and myosin-decorated F-actin (3.7 Å, Mentes et al. 2018) have been published. They show very clearly that electron microscopy can now be used to study the mechanism of the actin-myosin motor system.
Figure 4.3 illustrates the crossbridge cycle of actin and myosin, indicating also which structural biology technique (X-ray crystallography or electron microscopy) might be most useful to obtain additional information in the future. In this chapter we will briefly look back at some earlier work, which already pointed out interesting questions, such as the mechanism for the actin-activation of myosin ATP hydrolysis or the coupling between nucleotide state and lever arm movement, also known as the power stroke. It is also interesting to see how the increase in resolution developed and fostered a completely new understanding of how the conformational changes connect to function. This will lead to an in-depth discussion of the highest resolution structure of decorated F-actin to date and its implications for the detailed description of the strongly interacting acto-myosin states. Here, we will not repeat the crystallographic results. They are described in detail in Chap. 2 by Sweeney and colleagues and also in a very recent review by Robert-Paganin et al. 2019. We will end the chapter by exploring some ideas regarding future microscopy studies, which should close the blind spots of both X-ray crystallography and static electron microscopy.
Schematics of the acto-myosin crossbridge cycle. The background pie segments assign the states in the cycle to either X-ray crystallography (blue) or cryo electron microscopy (orange) as that technique, which is most commonly used to obtain structural information about that particular acto-myosin state. The two orange ellipses denote states, which might become accessible for cryo electron microscopy in the future (cf. also Sect. 4.5). (Image of the crossbridge cycle adapted from Wulf et al. 2016)
4.2 Studying the Canonical Acto-myosin State – The Strongly Bound Motor
As discussed above, electron microscopy was the natural technique to investigate the strongly bound states of myosin to F-actin (cf. Fig. 4.3, light orange background). At times when only skeletal myosins were examined, the nucleotide-free myosin state (Rigor) was studied, as no difference between an ADP-bound or nucleotide-free state could be observed at the resolution attainable then. Early work was satisfied with visualising the binding state with a first attempt to dock the quasi-molecular crystallographic structure into the reconstructed density, which was at a resolution level of about 20–30 Å (Flicker et al. 1991; Rayment et al. 1993b; Schröder et al. 1993). These models were in hindsight not very helpful as their definition of the actin-myosin interface was not good enough to draw detailed conclusions.
This situation changed when myosins other than skeletal muscle myosin 2 were explored. For example, two states were reported for smooth muscle myosin 2 (Whittacker et al. 1995) shown in Fig. 4.4a and for brush border myosin 1 (now myosin 1A; Myo1a) (Jontes et al. 1995). Besides the conventional Rigor state, the ADP state was described, i.e., the strongly bound myosin where the nucleotide ADP is still bound. Such a nucleotide state can be populated for a number of myosins, e.g., myosin 5 (Wulf et al. 2016) and myosin 1B (Mentes et al. 2018) as will be discussed later. Interestingly, the reconstructions showed a clear difference in the direction the lever arm is pointing. In the Rigor state it swings slightly downwards towards the barbed end of the decorated filament, while the ADP state reveals a more upwards directed lever arm. These two states were also subsequently found in single-molecule mechanics experiments, identified as the two states in displacement graphs from optical trap studies (Veigel et al. 1999; Sellers and Veigel 2006).
(a) Low resolution helical reconstruction of F-actin decorated with the smooth muscle myosin 1 motor domain and its light chain, left in the nucleotide-free Rigor state, right in AM.ADP state. Images adapted from Whittacker et al. (Whittaker et al. 1995). Note the difference in the direction of the light chains pointing away from the F-actin axis. Together with work on brush border myosin 1 (Jontes et al. 1995), these were the first structural data indicating that the motor domain and its lever arm are in an intermediate power stroke position when ADP is still bound. Cf. Sect. 4.3 and the high-resolution work by Mentes et al. (2018), also Fig. 4.9. (b) Closure of the cleft in the upper 50 kDa motor domain upon strong binding to actin. Image from Holmes et al. (2003). The reconstruction of the acto-myosin complex at medium resolution provided a well-defined molecular envelope, which then allowed molecular fitting of the crystallographic model of skeletal myosin 2. To fit into the envelope the crystal structure had to be modified. The upper row shows the unmodified structure with the lower 50 kDa domain (grey) already docked to the actin surface. The lower row shows the fit with the closed cleft and both, upper and lower 50 kDa domain in a position forming the actin-myosin interface for the strongly bound complex
in the middle of the 1990’s it was already well anticipated from these experiments that the “strongly bound” state should come in different flavours and should be more populated in some myosins than others. This also indicates a correlation between existence of the ADP state and motor function. The existence of such a long-lived ADP state is today understood as a stopping point in the crossbridge cycle. In this state the myosin can sense force, e.g., strain pulling at the lever arm (cf. e.g., Chap. 8 by Sellers and Takagi as well as the discussion of the ADP high-resolution reconstructions below).
Another flavour of the strongly bound state is its function as a trigger for the power stroke. Rephrased in a more mechanistical way, the transition from a weakly to a strongly bound myosin must be the signal for the motor to start with the power stroke. This order of events is mandatory, as otherwise – in a decoupled scenario – part of the power stroke’s effect would be lost. Myosin’s sensing of its binding coordination upon binding to F-actin has been investigated in numerous studies, again using electron microscopy, but also myosin mutants, spectroscopic probes, and elaborate biochemical state preparations for X-ray crystallography. One of the first visualisations of a conformational change upon strong binding is illustrated in Fig. 4.4b (Holmes et al. 2003) where the closure of the cleft between the upper and lower 50-kDa domains was observed when trying to fit the original crystallographic skeletal myosin 2 model into a 3D reconstruction of decorated actin at about 14-Å resolution.
Unfortunately, this level of resolution was still not good enough to derive a detailed model of the molecular mechanisms, but it pointed to a number of hotspots in the myosin structure which were then studied with a number of different approaches.
4.3 Changing the Game – Cryo Electron Microscopy 2.0 and High-Resolution Reconstructions of the Nucleotide-Free Rigor State and ADP States
4.3.1 Pushing the Limits – Advances in Instrumentation and Image Processing
In the years from about 2010 new ideas and technical advances have changed electron microscopy. While progress in many details such as instrument stability and automated data collection had happened in secrecy, the appearance of novel reconstruction software (Scheres 2012) coincided with the availability and application of direct electron detectors (Campbell et al. 2012; Clough et al. 2014; McMullan et al. 2016) and put a spotlight on the possibility of obtaining high-resolution structures in the 3–4-Å resolution range by cryo electron microscopy (Kühlbrandt 2014).
This development is reflected in the boost of resolution for many reconstructions of F-actin and myosin-decorated F-actin or thin filaments over the years. Figs. 4.5 and 4.6 reflect the change in appearance and information content of the 3D reconstructions obtained. For myosin-decorated actin filaments (Fig. 4.5) representative reconstructions were published by Behrmann et al. (2012, 7.4 Å resolution, data found as EMD-1988 in EMDataResource.org), Fujii and Namba (2017, 5.2 Å, EMD-6664), and van der Ecken et al. (2016, 3.9 Å, EMD-8165), which illustrate also the limits of resolution imposed, e.g., by the electron detectors used. Maybe the best comparison and most obvious effect is seen by the direct juxtaposition of a by then state-of-the-art “pre-revolution” F-actin reconstruction (Fujii et al. 2010) and the at present best published F-actin cryo electron microscopy reconstruction at about 3.1 Å resolution (Chou and Pollard 2019) as shown in Fig. 4.6.
Recent 3D helical reconstructions of actin filaments decorated with myosin subfragment-1 at increasing resolution: (a) 7.4 Å from Behrmann et al. (2012) including tropomyosin, data found as EMD-1988 in EMDataResource.org; (b) 5.2 Å from Fujii and Namba (2017) as pure F-actin, EMD-6664; (c) 3.9 Å from von der Ecken et al. (2016) including tropomyosin, EMD-8165. Note the different appearance of details with increasing resolution but also the likeness of the general fit of the binding motor domain. The red circle marks the N-terminal beta-barrel of myosin. (All maps are visualized using the graphics software USCF Chimera (Pettersen et al. 2004))
Comparison of 3D reconstructions of filamentous actin before (a) and after (b) the “resolution revolution” originating from the use of movie correction made possible by the fast frame rates of Direct Electron Detectors. (a) from Fuji et al. (2010), data found as EMD-5168 in EMDataResource.org; (b) from Chou and Pollard (2019), EMD-7936. This direct comparison of the two reconstructed 3D maps illustrates the difference in information content and obvious detail when highest resolution (here 3.1 Å) is available. (All maps are visualized using the graphics software USCF Chimera (Pettersen et al. 2004))
That latter study included an interesting comparison of F-actin structures published over the years derived by different techniques ranging from X-ray diffraction on ordered gels to cryo electron microscopy. Again, the boost in resolution for microscopy is apparent, but also the differences in resolution, which correlate with details of data collection and data processing. Here, technologies proved a mixed bag, e.g., the use of electron energy filtering microscopy, which has been around for a long time but has been belittled, seems to improve resolution, while the recently hyped physical phase plates in their current implementation seem to deteriorate highest resolution. It will be interesting to see where cryo electron microscopy goes from there and which standard will emerge in the next years.
The newest reconstructions of F-actin provide a detailed analysis of the actin polymerization mechanism derived from the comparison of three reconstructions (AMPPNP, ADPPi, ADP), all in the resolution range of 3.1–3.7 Å (Chou and Pollard 2019; cf. also Bradshaw and Paul 2019).
4.3.2 The 50-kDa Domain in Its Strongly Bound State and the Acto-myosin Interface
Quite a few high-resolution reconstructions have by now described the Rigor state of the myosin motor domain bound to F-actin alone (Mentes et al. 2018) or thin filaments without troponin (Behrmann et al. 2012; van der Ecken et al. 2016). In the overall structure of the Rigor complex, i.e., the orientation of myosin’s motor domain at the surface of F-actin, there have been very few changes to earlier medium resolution data (cf. Fig. 4.4b, Holmes et al., 2003). In particular, the details about the closure of the deep cleft in the 50-kDa domain of myosin still hold. This closure changes the orientation of the so-called switch-1 with respect to the bound nucleotide (cf. Fig. 4.10 and also Chap. 2 on myosin structure by Sweeney and colleagues), thus weakening its binding interaction. The detailed amino acid interactions of this coordination vary slightly among the different reconstructions, but the general concept that the transition from a weakly to a strongly bound state of the acto-myosin interaction will affect nucleotide coordination and binding strength is still correct. Interestingly, the dissociation of the motor domain from the filament upon ATP binding must – in principle – reverse this step. That is, by recruiting switch-1 for nucleotide coordination, the cleft in the 50-kDa domain is forced to open and the motor domain dissociates from F-actin. As is described below, the weakly bound state upon formation of the acto-myosin binding complex and also the structures when myosin dissociates from the strongly bound complex are not known at present, and thus a molecular mechanism for this is not really known. But we can assume that the detailed interactions in myosin are not “symmetric”, i.e., the sequence of steps when going through the crossbridge cycle is reflected in an order of structural changes not freely interchangeable. An example of such a directed change could be the coordination of the N-terminal extension of myosin with the lever arm now seen upon ADP release (cf. Fig. 4.9, Mentes et al. 2018). It has to be seen which other structures of the crossbridge cycle will be available in the future.
It should be mentioned that the structures by Behrmann et al. (2012) and van der Ecken et al. (2016) show additional details about the interactions of amino acids in the interface between myosin and actin. This gives new information regarding the effects of mutations in surface loops of actin and myosin interrupting the interface between myosin and actin and thus cause pathological effects, such as, e.g., familial cardiomyopathies.
4.3.3 The ADP-State of Myosin Strongly Bound to Actin – And What It Tells Us About Force Sensing
Two studies, albeit at different levels of resolution, lately compared the nucleotide-free myosin Rigor model with the conformation where the nucleotide ADP is bound. Wulf et al. (2016) used myosin 5 (myo5) for their reconstructions (ending up at about 6-8 Å resolution), while Mentes et al. (2018) used myosin 1b (Myo1b) reaching resolutions in the order of 3.2–4.1 Å. Figure 4.7 shows the general appearance of the reconstructions and focusses also on the central beta-sheet (the so-called transducer; Coureux et al. 2004). It was long speculated that this beta-sheet – together with other structural elements, in particular the N-terminal domain, which includes part of the beta-sheet – couples the lever arm to the nucleotide-binding site. And indeed, one of the more obvious changes between the ADP and Rigor state is the position of the lever arm and the bending and twisting of the transducer domain. In simple terms, pulling at the lever arm in opposite direction to its power stroke will also pull at the central beta-sheet (and other structural elements, see below) which leads to a lower probability of the ADP leaving the nucleotide-binding site. Conversely, when allowed to swing freely in its genuine power stroke direction, myosin changes its conformation such that ADP can rapidly leave the molecule.
Comparison of 3D reconstructions of F-actin decorated with myosin 5 (a) from Wulf et al. (2016) and F-actin 1B decorated with myosin (b) from Mentes et al. (2018). Note the difference in resolution (about 6 Å for (a), about 3.3 Å for (b)), equivalent to the situation for F-actin alone illustrated in Fig. 4.6. Both reconstructions are from samples in their AM.ADP states. (Pictures copyright by PNAS/United States National Academy of Science (US))
This simple scheme can provide a first mechanistical model, e.g., for the coordination of heads in the processive walking of myosin 5 as well as the force sensing of a number of different myosins.
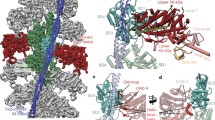
Mentes et al. (2018) in fact contribute a very detailed mechanistical model of how mechanical load prevents MgADP release and thus obstructs the transition from an intermediate acto-myosin-ADP state to the final Rigor state after the power stroke. Figure 4.8 (adapted from their original publication) illustrates the detailed changes of the molecular conformation when transitioning from (i) an AM.ADP state (denoted AM.ADPA) with the lever arm not yet in a Rigor-like position to (ii) a second AM.ADP state which still has ADP bound, but the lever arm has already moved (AM.ADPB), to (iii) the Rigor state. The definition of the second AM.ADPB state with its moved lever arm resulted from the classification of reconstructed data, i.e., while the majority of acto-myosin complexes were in the state AM.ADPA, about 15% of the complexes included in the reconstruction had the lever arm almost in the Rigor position, but ADP was still present. This succession of states and their changes regarding the coordination of loop 5 and the N-terminal subdomain, as illustrated in Fig. 4.9, can be put together as a working hypothesis in the following mechanistical model: The swinging of the lever arm is coupled to the nucleotide, which is bound in the motor domain by the now so-called N-terminal subdomain (which also includes part of the central beta-sheet). In the individual small figures in row D this subdomain is shown in blue, also including the N-terminal extension. While the latter coordinates with the subdomain in the AM.ADP state (aka AM.ADPA, cf. Fig. 4.9 row C, note the different binding of the extension coloured in magenta), it starts to interact with the lever arm in the intermediate state (AM.ADPB). Future studies (e.g., Molecular Dynamics calculations driving a transition among these three structures) may show what the energy landscape of these three structures looks like and why the conformational changes happen in the sequence observed now. It may be envisaged that pulling at the lever arm may allow the N-terminal extension to change its binding position with a higher probability and thus to weaken the otherwise stabilized AM.ADPA state. In the rigor state this extension is then again coordinated to the lever arm. The important result of the work is the visualization of an existing intermediate state, which shows the changes of the coordination of the N-terminal extension.
The conformations of myosin’s central beta-sheet in the nucleotide-free Rigor state and the AM.ADP state – where ADP is still bound in the nucleotide binding pocket (upper two rows; top row from Wulf et al. (2016) – beta strands in the sheet are not resolved at 6 Å resolution, while the strands are very well resolved in the reconstruction from Mentes et al. (2018) at 3.3 Å resolution. Note the distinct difference in the bending of the beta-sheet for the two different nucleotide states. The difference between Rigor and AM.ADP is also reflected in the direction of the lever arm, which is illustrated for the data from Mentes et al. (2018) (bottom row). (Pictures adapted from images in the cited publications in PNAS/United States National Academy of Science (US))
Figure from Mentes et al. (2018) illustrating the detailed changes in lever arm direction and coordination of the N-terminal subdomain/N-terminal extension in the different states AM.ADPA, AM.ADPB, and Rigor (AM). The two ADP states were found in the ensemble of recorded ADP acto-myosin filaments, with the majority (85%) of complexes in the AM.ADPA form. AM.ADPB is supposed to be an intermediate with the lever arm already swinging into the usual direction of the Rigor state. (a) shows the general conformation of nucleotide binding pocket, central beta-sheet, N-terminal domain (blue) and lever arm (red). (b) shows details of the nucleotide binding site, and (c) illustrates the corresponding conformations for the N-terminal extension (magenta) interacting either with the N-terminal domain or the lever arm. These changes are combined with a large change of the coordination of loop-5 and the swinging of the lever arm into its Rigor direction. (d) Schematic models for the three discussed states. (Picture copyright by PNAS/United States National Academy of Science (US))
It should be mentioned here that for myosin 6 (Wells et al. 1999) mechanisms for force sensing were discussed as well (Gurel et al. 2017), but they were not based on any structural data of acto-myosin at that level of resolution.
4.4 Putting All the Information Together – The Crossbridge Cycle
By combining all the crystallographic, spectroscopic and microscopic data, one can compile a detailed description of states and transitions of the motor’s crossbridge cycle. The best compilation to date is given in Fig. 4.10, adapted from a recent review by Robert-Paganin et al. (2019; cf. also Chap. 2 on myosin structure by Sweeney and colleagues).
Schematic of the crossbridge cycle proposing a detailed structural and mechanistic model for the different states of the myosin motor domain. This detailed model has very recently been proposed in Robert-Paganin et al. (2019), cf. also chapter 2 on the structure of myosin in this book. (Picture copyright by American Chemical Society (US))
Two details of this crossbridge cycle scheme are noteworthy: At present we have no experimental evidence for a defined structure after the first, non-stereospecific initial association of the ADP.Pi-loaded pre-power stroke myosin. However, there must be a transition to a specific binding of the lower and upper 50-kDa domain, which initiates a sequence of conformational changes leading to the power stroke. The second point then concerns the structure of this specific binding, which can be imagined as a somewhat open conformation as proposed by Robert-Paganin et al. (2019) or already with a further closed cleft as proposed e.g., by Behrmann et al. (2012). Experimental evidence for such details is not overwhelming as all the modelling is based on crystallographic states of myosin on its own. It is most likely that these conformations – which a priori only describe possible states in free myosin’s conformational space – will also be used by myosin when interacting with actin. But we have to remember that there are no interpretable structural data for any weakly bound acto-myosin states (cf. below). Considering the evidence for other states and transitions in the crossbridge cycle, in particular our detailed understanding of the transition between AM.ADP and the Rigor state, it seems not irrelevant to wait for more structural data of such weakly bound complexes. Whether this will be crystallographic data or reconstructions from cryo electron microscopy has to be seen, but certainly both will help to shed more light onto the details of the initial binding of myosin to actin, phosphate release and the initiation of the lever arm swing with its transition to the above discussed AM.ADPA state.
4.5 The Future of the Field – Chasing the Weakly Bound Acto-myosin States
Very little progress has been reported on crystallizing any acto-myosin complex, and the small steps, e.g., efforts to stop actin from polymerizing resulting in formation of a small, F-actin-like oligomer (Qu et al. 2018; Dawson et al. 2003), should not be overinterpreted. However, after the success of cryo electron microscopy described so far in analyzing more or less ordered complexes – and also watching the advances of classification and sorting of states applied in other fields of biomedical research – one should reconsider the possibilities of advanced sample preparation.
As is well known from stop-flow kinetic experiments, rapid mixing is one possible way to populate transient states. In the case of electron microscopy this could be translated into a rapid mixing and freeze experiment. In this experiment a myosin suspension with gold nanoparticles is sprayed onto an electron microscopy grid that is already covered by a thin film of a suspension of actin filaments. The grid is quick-frozen milliseconds after spraying, so that the mixing time is very short. The gold nanoparticles in the myosin solution are used as markers that the myosin spray has actually hit the sample. Figure 4.11a from Walker et al. (1999) shows a vitrified ice layer with two such nanoparticles and two F-actin filaments, which do not show the typical arrowhead appearance of well-ordered myosin decorated F-actin filaments (cf. Figs. 4.2 and 4.3), but look like myosin randomly interacting with the filament. This indicates that myosin might not yet be bound in a stereospecific way, which makes it likely that such myosins are attached in a weakly binding state – as desired.
Capturing dynamic states of the acto-myosin complex, which are intermediates in the crossbridge cycle (cf. Fig. 4.3, states marked by orange ellipses). (a) nucleotide-free myosin subfragment-S1 sprayed onto a suspension of F-actin and then quick-frozen to stop binding reaction in an early/intermediate weak binding state. Picture from Walker et al. (1999), copyright by PNAS/United States National Academy of Science (US). Note the disordered appearance of the myosin molecules interacting with F-actin. Conventionally, i.e. some more time after mixing, myosin would form nicely ordered “arrowheads” as shown in Fig. 4.2. The data are consistent with the hypothesis that the initial coordination of myosin molecules upon actin binding is non-stereospecific. However, it should be noted that the experiments shown here are in the absence of nucleotide, i.e., they are not identical to the initial binding of myosin in an ADP.Pi state. (b) Time-resolved dissociation of myosin subfragment-1 from F-actin (Ménétret et al. 1991). In this experiment flash-photolysis of caged-AMPPNP was used, which slows down nucleotide binding and complex dissociation and thus allows to freeze intermediate states. Freezing delay after flashing (left to right): no flashing, 20 ms, 30 ms, 80 ms, 2 s. Scale bar 35.5 nm. Note the curly appearance of the intermediates at 30 ms delay time, which may indicate breaking of the helical symmetry of the F-actin filament by the still interacting myosin molecules. (Picture from Ménétret et al. (1991), copyright by Elsevier)
The lack of symmetry, which is in contrast to what would be expected from a well-ordered decoration, makes image processing complicated for such samples. In principle, a single-particle approach could be applied if myosin molecules were only sparsely attached to the filament. Then, the F-actin backbone could be used for aligning the molecules, while classification might sort a wider distribution of motor domain attachment geometries. If enough molecules are selected even averaging the molecules to higher resolution might be feasible. Such an approach needs to be repeated with modern microscopes and direct electron detectors. It may help to visualize the first interactions of the motor domain with F-actin and to find out what conformation of the 50-kDa cleft is adopted (cf. Fig. 4.9 and discussion above). An additional interesting detail is exposed by Fig. 4.10a: While a cooperative effect for strong binding of myosin to actin has been described many times (e.g., Prochniewicz et al. 1996), it seems as if the weakly bound states could prime the filament for motor domain binding, too. While two actin filaments seem to be covered by myosin to a rather high degree, another filament is still completely bare. This is a surprising observation, which might indicate that already for non-stereospecific interactions myosin molecules can link adjacent actin monomers, e.g., bridge them with different attachment geometries of the lower and upper 50-kDa domain with a still reasonably open cleft. This is all speculation at this point, but this discussion should awaken interest in more dynamic sample preparation experiments and high-resolution image processing of micrographs from such samples.
Another experiment exploring a dynamic sample preparation technique is illustrated in Fig. 4.11b, which shows the detachment of motor domains from filamentous actin. In this experiment (Ménétret et al. 1991) flash-photolysis of ATP or AMPPNP was used to add nucleotide just milliseconds before rapid freezing. While ATP leads to a rapid detachment of myosin from F-actin, AMPPNP slows down this process and “intermediates” of the detachment process can be followed. In Fig. 4.10b a succession of filament appearances from left (no free nucleotide is added) to right (freezing 2 s after AMPPNP release) is shown. In itself the detachment process is certainly less exciting to study as we cannot expect further insight into the power stroke mechanism. One aspect of the process is the appearance of “curly” filaments, which could be explained by a local symmetry breaking of the actin helical symmetry when individual motor domains are detaching. This is interesting, as helical symmetry of actin filaments does indeed change when proteins bind to actin; a very large symmetry change was discovered by McGough et al. (1997) for cofilin binding to actin. But on a smaller scale this is also observed for the acto-myosin complex as decorated filamentous actin. Chou and Pollard (2019) report a consistent 27.4-Å axial repeat/ −166.6° helical turn for their different F-actin reconstructions, while actin filaments decorated with myosin 5 (Wulf et al. 2016) showed similar axial repeats at a larger helical turn (−167.1 to −167.4°) as refined by an iterative helical reconstruction process (Egelman 2007). Recently, work by Huehn et al. (2018) on cofilin-decorated filaments demonstrated that the change in symmetry of the filaments occurs quite localized at the boundary between decorated and bare filament areas. In that particular case the change in helical symmetry is quite large resulting in a helical turn of −162.1° (McGough et al. 1997; Galkin et al. 2011).
While information about the mechanism of the motor might not be derived from these observations they might help to better understand the dynamics of actin and how different conformations of the filament might be adopted.
Finally, a more complex approach to the visualization of weakly bound states – and in fact all the states in the myosin crossbridge cycle – should be discussed: cryo electron tomography on sarcomeres of vitrified muscle samples. It is possible to quick-freeze contracting muscle fibers (Taylor et al. 1999), and Figs. 4.12 and 4.13 show different views of 3D tomographic reconstructions of ultrathin sections of freeze-substituted and plastic-embedded muscle (Wu et al. 2010, 2012). While there is no doubt that such a sample preparation comes with quite severe fixation, staining, and embedding artifacts, it is still surprising at what level of detail such tomograms can be interpreted. They can certainly not satisfy the need for high-resolution structures, but they do illustrate a possible roadmap to high-resolution structures. What has to be developed next is a routine workflow to prepare cryo samples of sarcomeres for cryo electron tomography. Focused ion beam milling of cellular samples has been developed in recent years (Schaffer et al. 2017) as well as cryo lamella lift out from larger samples (Schaffer et al. 2019). If a really large number of tomograms from such lamella can be recorded, then classifications similar to those discussed in Taylor et al. (2019) might result in many different states of the myosin motor domains. When bringing these states into the correct successive order such work might provide us with movie-like data of all possible structures of actin and myosin and the acto-myosin complex in the crossbridge cycle.
Conventional electron tomography of ultrathin plastic sections of quick-frozen muscle after freeze substitution. Picture from Wu et al. (2010), copyright by PLOS, US. The 3D rendering illustrates different classes of interaction motives found for the crossbridges between thick and thin filaments (cf. also Fig. 4.2). These motives are found after classification and averaging of corresponding motives in 3D. This averaging procedure reduces noise considerably, permitting an initial attempt of docking myosin motor domains into the reconstructed densities
Distribution of possible orientations of the myosin motor domain and lever arm fitted into crossbridge densities of electron tomograms from muscle fibre sections (prepared as in Fig. 4.12). (a–d) Different distributions of orientation for different interaction motives as seen in Fig. 4.12. (Picture from Wu et al. (2010), copyright by PLOS, US)
References
Behrmann E, Müller M, Penczek PA, Mannherz HG, Manstein DJ, Raunser S (2012) Structure of the rigor actin-tropomyosin-myosin complex. Cell 150:327–338
Bradshaw M, Paul DM (2019) After the revolution: how is Cryo-EM contributing to muscle research? J Muscle Res Cell M 40:93–98
Campbell MG, Cheng A, Brilot AF, Moeller A, Lyumkis D, Veesler D, Pan J, Harrison SC, Potter CS, Carragher B, Grigorieff N (2012) Movies of ice-embedded particles enhance resolution in electron cryo-microscopy. Structure 20:1823–1828. https://doi.org/10.1016/j.str.2012.08.026
Chou SZ, Pollard TD (2019) Mechanism of actin polymerization revealed by cryo-EM structures of actin filaments with three different bound nucleotides. Proc Natl Acad Sci U S A 116:4265–4274
Clough RN, Moldovan G, Kirkland A (2014) Direct detectors for electron microscopy. J Phys Conf Ser 522:012046. https://doi.org/10.1088/1742-6596/522/1/012046
Coureux PD, Sweeney HL, Houdusse A (2004) Three myosin V structures delineate essential features of chemo-mechanical transduction. EMBO J 23:4527–4537
Cressey D, Callaway E (2017) Cryo-electron microscopy wins chemistry Nobel. Nature 550:167. https://doi.org/10.1038/nature.2017.22738
Dawson JF, Sablin EP, Spudich JA, Fletterick RJ (2003) Structure of an F-actin trimer disrupted by gelsolin and implications for the mechanism of severing. J Biol Chem 278:1229–1238
Egelman EH (2007) The iterative helical real space reconstruction method: surmounting the problems posed by real polymers. J Struct Biol 157:83–94
Flicker PF, Milligan RA, Applegate D (1991) Cryo-electron microscopy of S1-decorated actin filaments. Adv Biophys 27:185–196
Fujii T, Namba K (2017) Structure of actomyosin rigour complex at 5.2 Å resolution and insights into the ATPase cycle mechanism. Nat Commun 8:13969. https://doi.org/10.1038/ncomms13969
Fujii T, Iwane AH, Yanagida T, Namba K (2010) Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature 467:724–728. https://doi.org/10.1038/nature09372
Galkin VE, Orlova A, Kudryashov DS, Solodukhin A, Reisler E, Schröder GF, Egelman EH (2011) Remodeling of actin filaments by ADF/cofilin proteins. Proc Natl Acad Sci U S A 108:20568–20572. https://doi.org/10.1073/pnas.1110109108
Gurel PS, Kim LY, Ruijgrok PV, Omabegho T, Bryant Z, Alushin GM (2017) Cryo-EM structures reveal specialization at the myosin VI-actin interface and a mechanism of force sensitivity. Elife 6:e31125. https://doi.org/10.7554/eLife.31125
Holmes KC, Angert I, Kull FJ, Jahn W, Schröder RR (2003) Electron cryo-microscopy shows how strong binding of myosin to actin releases nucleotide. Nature 425:423–427
Houdusse A, Szent-Gyorgyi AG, Cohen C (2000) Three conformational states of scallop myosin S1. Proc Natl Acad Sci U S A 97:11238–11243
Huehn A, Cao W, Elam WA, Liu X, De La Cruz EM, Sindelar CV (2018) The actin filament twist changes abruptly at boundaries between bare and cofilin-decorated segments. J Biol Chem 293:5377–5383. https://doi.org/10.1074/jbc.AC118.001843
Huxley HE (1957) The double array of filaments in cross-striated muscle. J Biophys Biochem Cytol 3:631–648
Jontes JD, Wilson-Kubalek EM, Milligan RA (1995) A 32 degree tail swing in brush border myosin I on ADP release. Nature 378:751–753
Kühlbrandt W (2014) The resolution revolution. Science 343:1443–1444
McGough A, Pope B, Chiu W, Weeds A (1997) Cofilin changes the twist of F-actin: implications for actin filament dynamics and cellular function. J Cell Biol 138:771–781
McMullan G, Faruqi AR, Henderson R (2016) Direct electron detectors. Methods Enzymol 579:1–17. https://doi.org/10.1016/bs.mie.2016.05.056
Ménétret JF, Hofmann W, Schröder RR, Rapp G, Goody RS (1991) Time-resolved cryo-electron microscopic study of the dissociation of actomyosin induced by photolysis of photolabile nucleotides. J Mol Biol 219:139–144
Mentes A, Huehn A, Liu X, Zwolak A, Dominguez R, Shuman H, Ostap EM, Sindelaar CV (2018) High-resolution cryo-EM structures of actin-bound myosin states reveal the mechanism of myosin force sensing. Proc Natl Acad Sci U S A 15:1292–1297
Milligan RA, Flicker EE (1987) Structural relationships of actin, myosin, and tropomyosin revealed by cryo-electron microscopy. J Cell Biol 105:29–39
Moore PB, Huxley HE, DeRosier DJ (1970) Three-dimensional reconstruction of F-actin, thin filaments and decorated thin filaments. J Mol Biol 50:279–295
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera – a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612
Prochniewicz E, Zhang Q, Janmey PA, Thomas DD (1996) Cooperativity in F-actin: binding of gelsolin at the barbed end affects structure and dynamics of the whole filament. J Mol Biol 260:756–766
Qu Z, Fujita-Becker S, Ballweber E, Ince S, Herrmann C, Schroeder RR, Mannherz HG (2018) Interaction of isolated cross-linked short actin oligomers with the skeletal muscle myosin motor domain. FEBS J 285:1715–1729. https://doi.org/10.1111/febs.14442
Rayment I, Rypniewski WR, Schmidt-Bäse K, Smith R, Tomchick DR, Benning MM, Winkelmann DA, Wesenberg G, Holden HM (1993a) Three-dimensional structure of myosin subfragment-1: a molecular motor. Science 261:50–58
Rayment I, Holden HM, Whittaker M, Yohn CB, Lorenz M, Holmes KC, Milligan RA (1993b) Structure of the actin-myosin complex and its implications for muscle contraction. Science 261:58–65
Robert-Paganin J, Pylypenko O, Kikuti C, Sweeney HL, Houdusse A (2019) Force generation by myosin motors: a structural perspective. Chem Rev. https://doi.org/10.1021/acs.chemrev.9b00264. [Epub ahead of print]
Schaffer M, Mahamid J, Engel BD, Laugks T, Baumeister W, Plitzko JM (2017) Optimized cryo-focused ion beam sample preparation aimed at in situ structural studies of membrane proteins. J Struct Biol 197:73–82. https://doi.org/10.1016/j.jsb.2016.07.010
Schaffer M, Pfeffer S, Mahamid J, Kleindiek S, Laugks T, Albert S, Engel BD, Rummel A, Smith AJ, Baumeister W, Plitzko JM (2019) A cryo-FIB lift-out technique enables molecular-resolution cryo-ET within native Caenorhabditis elegans tissue. Nat Methods 16:757–762. https://doi.org/10.1038/s41592-019-0497-5
Scheres SHW (2012) RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180:519–530
Schröder RR (2015) Advances in electron microscopy: a qualitative view of instrumentation development for macromolecular imaging and tomography. Arch Biochem Biophys 581:25–38. https://doi.org/10.1016/j.abb.2015.05.010
Schröder RR, Manstein DJ, Jahn W, Holden H, Rayment I, Holmes KC, Spudich JA (1993) Three-dimensional atomic model of F-actin decorated with Dictyostelium myosin S1. Nature 364:171–174
Sellers JR, Veigel C (2006) Walking with myosin V. Curr Opin Cell Biol 18:68–73
Taylor KA, Schmitz H, Reedy MC, Goldman YE, Franzini-Armstrong C, Sasaki H, Tregear RT, Poole K, Lucaveche C, Edwards RJ, Chen LF, Winkler H, Reedy MK (1999) Tomographic 3D reconstruction of quick-frozen, Ca2+-activated contracting insect flight muscle. Cell 99:421–431
Taylor KA, Rahmani H, Edwards RJ, Reedy MK (2019) Insights into actin-myosin interactions within muscle from 3D electron microscopy. Int J Mol Sci 20:E1703. https://doi.org/10.3390/ijms20071703
Veigel C, Coluccio LM, Jontes JD, Sparrow JC, Milligan RA, Molloy JE (1999) The motor protein myosin-I produces its working stroke in two steps. Nature 398:530–533
von der Ecken J, Heissler SM, Pathan-Chhatbar S, Manstein DJ, Raunser S (2016) Cryo-EM structure of a human cytoplasmic actomyosin complex at near-atomic resolution. Nature 534:724–728
Walker M, Zhang XZ, Jiang W, Trinick J, White HD (1999) Observation of transient disorder during myosin subfragment-1 binding to actin by stopped-flow fluorescence and millisecond time resolution electron cryomicroscopy: evidence that the start of the crossbridge power stroke in muscle has variable geometry. Proc Natl Acad Sci U S A 96:465–470
Wells AL, Lin AW, Chen LQ, Safer D, Cain SM, Hasson T, Carragher BO, Milligan RA, Sweeney HL (1999) Myosin VI is an actin-based motor that moves backwards. Nature 401:505–508
Whittaker M, Wilson-Kubalek EM, Smith JE, Faust L, Milligan RA, Sweeney HL (1995) A 35-A movement of smooth muscle myosin on ADP release. Nature 378:748–751
Wu S, Liu J, Reedy MC, Tregear RT, Winkler H, Franzini-Armstrong C, Sasaki H, Lucaveche C, Goldman YE, Reedy MK, Taylor KA (2010) Electron tomography of cryofixed, isometrically contracting insect flight muscle reveals novel actin-myosin interactions. PLoS One 5:e12643. https://doi.org/10.1371/journal.pone.0012643
Wu S, Liu J, Reedy MC, Perz-Edwards RJ, Tregear RT, Winkler H, Franzini-Armstrong C, Sasaki H, Lucaveche C, Goldman YE, Reedy MK, Taylor KA (2012) Structural changes in isometrically contracting insect flight muscle trapped following a mechanical perturbation. PLoS One 7:e39422. https://doi.org/10.1371/journal.pone.0039422
Wulf SF, Ropars V, Fujita-Becker S, Oster M, Hofhaus G, Trabuco LG, Pylypenko O, Sweeney HL, Houdusse AM, Schröder RR (2016) Force-producing ADP state of myosin bound to actin. Proc Natl Acad Sci U S A 113:E1844–E1852. https://doi.org/10.1073/pnas.1516598113
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Schröder, R.R. (2020). The Structure of Acto-Myosin. In: Coluccio, L. (eds) Myosins. Advances in Experimental Medicine and Biology, vol 1239. Springer, Cham. https://doi.org/10.1007/978-3-030-38062-5_4
Download citation
DOI: https://doi.org/10.1007/978-3-030-38062-5_4
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-38061-8
Online ISBN: 978-3-030-38062-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)