Abstract
Many important biological phenomena, ranging from cell signaling to viral infection, are accomplished by transportation of biological substances encapsulated in native nano-sized particles. Thermal fluctuation drives nanoparticles through cellular environments; this movement is facilitated by their small size. To understand how a specific cell function can be achieved through random collisions, it is useful to know the interactions between single particles and the local environment, as determined by measuring cell dynamics at high spatial and temporal resolutions . In this chapter, a simple yet powerful wide-field optical technique, coherent brightfield (COBRI) microscopy , is presented. COBRI microscopy detects linearly scattered light from a nanoparticle through imaging-based interferometry, which enables direct observation of small biological nanoparticles in live cells without labels. Proper image post-processing further improves the detection sensitivity of small particles by removing the scattering background of cell structures. COBRI microscopy can easily operate at a high speed due to its wide-field nature and stable, indefinite scattering signal. Using COBRI, the dynamics of single virus particles and cell vesicles in live cells can be successfully captured at a microsecond temporal resolution and nanometer spatial precision in three dimensions. The ultrahigh spatiotemporal resolution and shot-noise-limited sensitivity of COBRI microscopy provide an opportunity to study the biophysics and biochemistry of live cells at the nanoscale.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
3.1 Introduction to Label-Free Imaging in Live Cells Through Linear Scattering
A biological cell is a highly dynamic system. Cell organelles and other subcellular structures constantly evolve and migrate to facilitate cell functions. Active transportation is an effective method for the redistribution of substance in live cells [1, 2]. In neuronal cells, active transport is especially critical for appropriate neuron activities [3, 4]. For active transportation, cargos are enclosed in small lipid vesicles or micelles, and their translocation is accomplished by motor proteins and cytoskeletons. The fundamental interaction between individual molecules underlies the regulatory mechanisms of intracellular transport, determining when and where the cargo is delivered [5, 6]. However, this sophisticated cell machinery developed through evolution is also utilized by viruses, which hijack the cell machinery by encapsulating their genomes in small particles with specific surface functionalities [7]. Thus, examining how viruses infect cells is not only useful in the battle against disease but also in understanding the cell machinery.
Through biochemical and molecular biology approaches, many proteins have been identified that play crucial roles in cell uptake and transportation [8]. However, these studies have tended to provide static results that lack spatial and temporal resolutions . Optical microscopy is a powerful tool that provides high spatiotemporal resolution. Through labeling with fluorescent proteins and dyes, cell dynamics—including intracellular transport and virus uptake—have been visualized in real time [9,10,11]. Using this technique, individual cell vesicles and virus particles can be seen under the microscope and their trajectories can be reconstructed by single-particle tracking (SPT) [11,12,13]. The main challenge in fluorescence-based optical observation is the limited photon budget that strictly restricts the observation time and data acquisition rate. Fluorescence-based SPT typically provides a spatial precision of 1–10 nm and a temporal resolution of a few to tens of milliseconds [14,15,16]. This spatiotemporal resolution has enabled many studies of interactions between particles and their local environment. However, probing molecular interactions at the nanometer scale requires higher spatiotemporal resolutions that fluorescence-based approaches struggle to provide.
For high-precision and high-speed measurements, a robust signal is required. Linear scattering meets these requirements—unlike fluorescence, which suffers from photobleaching, blinking and saturation , linear scattering from small particles is stable and indefinite, making it a promising contrast mechanism. Using scattering-based optical microscopy , including darkfield and brightfield microscopy, many high-speed and high-precision localization measurements have been reported that uncovered important biophysical and biochemical processes [17,18,19,20]. For a biological nanoparticle with a diameter of 100 nm in an aqueous solution, its scattering cross section in visible light is on the order of 10−13 cm2 (assuming a refractive index of 1.45), which is three orders of magnitude larger than the fluorescence cross section of an organic fluorophore. Therefore, intrinsic scattering from a bio-nanoparticle should be sufficient for direct observation and dynamic study. Indeed, endogenous biological nanoparticles (virus particles and extracellular vesicles) have been detected in cell-free in vitro environments by using several scattering-based imaging techniques, including darkfield [21,22,23], holographic, and other interferometric microscopies [24,25,26,27,28]. Among these imaging techniques, interferometric detection has the advantage of achieving shot-noise-limited sensitivity, independent of the detector noise [29].
Label-free imaging of endogenous bio-nanoparticle is complicated in live cells because of the scattering background of cell structures [30,31,32,33,34]. The smallest particle that can be detected and tracked in a live cell is often limited by imaging specificity, not by optical sensitivity. Having a strategy for distinguishing the signal from the background is therefore highly valuable. In addition, a cell is a three-dimensional (3D) object, and the dynamics of small particles are also 3D. Projection of 3D dynamics onto two-dimensional (2D) can cause misinterpretation. Thus, a label-free 3D imaging and tracking technique is highly desirable.
In this chapter, a simple, yet powerful imaging modality, namely coherent brightfield (COBRI) microscopy , is presented for sensitive and high-speed direct imaging of biological nanoparticles [31, 32]. By using a highly coherent light source (i.e., a laser) for illumination, COBRI microscopy facilitates rapid and sensitive imaging beyond what conventional brightfield microscopy can offer. Through interference, COBRI microscopy also enables tracking in 3D. Section 3.2 describes the concepts, technical details and characterization of COBRI microscopy . In Sect. 3.3, strategies of background estimation and correction for COBRI imaging in live cells are discussed. Section 3.4 presents two examples of COBRI imaging and tracking of single bio-nanoparticles in live cells. Specifically, Sect. 3.4.1 describes how the highly diffusive motion of single virus particles on a cell’s surface is captured with nanometer spatial precision and microsecond temporal resolution ; Sect. 3.4.2 describes how active transport of individual native cell vesicles inside a living cell is resolved with ultrahigh clarity. Finally, Sect. 3.5 concludes the chapter by discussing the sensitivity limit and possible applications of ultrasensitive and ultrahigh-speed label-free imaging.
3.2 COBRI Microscopy
COBRI microscopy essentially works by employing a brightfield microscope and a highly coherent laser light source [31]. The high temporal coherence maximizes imaging contrast and thus sensitivity. The high spatial coherence enables versatile beam shaping (e.g., focusing and structured illumination ). Laser also provides sufficient illumination intensity at the sample for high-speed measurements. A schematic of COBRI microscopy is shown in Fig. 3.1. A laser beam is focused onto the sample through a condenser lens. The choice of the condenser lens determines the available spatial frequency bandwidth in illumination. For example, when using a low-numerical-aperture (low-NA) condenser (as plotted in Fig. 3.1), the illumination has a Gaussian-like profile of a few to tens of micrometers [31]. When using a high-NA condenser, a sharp submicrometer focus can be created at the sample [35]. Although stationary illumination is sufficient for the operation of COBRI microscopy , a rapid, 2D scan of the illumination often makes the measurement easier [31]. Such 2D beam scanning is achieved by using a 2-axis acousto-optic defector (AOD) that typically scans at approximately 100 kHz. The beam scanning is synchronized with image acquisition, which ensures identical beam scanning is performed within each image integration time. When choosing a high-NA condenser together with rapid beam scanning, an arbitrary illumination pattern can be created at the sample. Using rapid beam scanning, stable and uniform laser illumination is created at the sample for measurements at various image acquisition rates, from a few frames per second (fps) to 100,000 fps [31]. For measurements at higher acquisition rates (>100,000 fps), stationary illumination is preferred because beam scanning using an AOD is too slow.

Adapted from [36], with permission from Elsevier
Schematics of COBRI microscopy . SM, single-mode fiber; AOD, acousto-optic deflector; L1–L3, lenses; COND, condenser; OBJ, objective.
Upon illumination, the sample (e.g., a nanoparticle or a biological cell) scatters and absorbs light. The transmitted light, containing the spatial information of the sample, is collected by a high-NA oil-immersion microscope objective. The magnified image is projected onto a camera through a tube lens . COBRI microscopy captures extinction images of the sample in transmission. For small particles, forward scattering light is collected, which interferes with the non-scattered transmitted light. Specifically, the recorded COBRI intensity is written as
where t is the transmitted light field, s is the forward scattered field, and \( {\varphi} \) is the phase difference between the two fields. The first term represents the illumination intensity transmitted through the sample; the second term is the forward scattering intensity of the particle, and the third term is the interference between the two fields. For a small particle in a transparent medium (e.g., bio-nanoparticles in cellular environments), the scattering intensity is considerably weaker than the transmitted light, and thus the second term is negligible in the presence of the first term and the third term. COBRI microscopy detects the particle by using the third interference term. In such interferometric detection, the scattering field of a nanoparticle and the non-scattered transmitted field are often referred to as the signal and reference, respectively. Notably, the phase difference \( {\varphi} \) between the scattering signal and the transmitted reference is a function of the axial position of the particle relative to the imaging microscope objective. This is because the phases of these two fields evolve differently throughout the microscope objective due to their distinct spatial modes [37]. In general, \( {\varphi} \) goes from 0 to π over the depth of focus of the imaging microscope objective, which is approximately 1 μm for visible light and a high-NA objective. In other words, \( {\varphi} \) can be manipulated by controlling the axial position of the particle relative to the optical focus. In 2D imaging and tracking, the axial position of the nanoparticle is adjusted by a sample stage to ensure that its absolute contrast is maximized (as a bright spot when \( {\varphi } \cong 0 \) and as a dark spot when \( {\varphi } \cong {\pi } \)).
To quantify interference visibility, and thus COBRI sensitivity, the COBRI contrast of a particle is defined as the normalized intensity difference caused by the particle. It can be written as
In this definition, zero COBRI contrast represents no detectable interference visibility, and thus no contrast is observed. To compare the sensitivity of COBRI and conventional brightfield microscopes, dielectric nanoparticles, metallic nanoparticles, and biological cells are imaged with increasing illumination spectral bandwidth (Δλ), from Δλ = 0.05 nm of a laser to Δλ > 350 nm of a halogen lamp. As illustrated in Fig. 3.2, COBRI with laser illumination provides the highest contrast, typically two- to three-fold stronger than a conventional brightfield microscope. The stronger contrast of COBRI is mainly due to the higher temporal coherence, not the spatial coherence; the figure shows that narrow spectral filtering of a halogen lamp can provide contrast similar to that provided by laser illumination. However, although the interference visibility of a spectrally filtered low-coherence light source is comparable to that of a laser, the illumination intensity at the sample position of a low-coherence light source is limited and typically insufficient for high-speed measurements.

Reproduced from [31] with permission from the American Chemical Society
Comparison of COBRI imaging and conventional brightfield imaging of nanoparticles and living cells . The laser light source was a solid state laser at 532 nm and the white light source was a halogen lamp. Three bandpass filters were added separately to the white light source: filter 1 (527–537 nm), filter 2 (500–550 nm), and filter 3 (350–700 nm). These filters had similar central wavelengths but increasing bandwidths. Together with white light without a filter, five illumination conditions were prepared for comparison. a 200 nm polystyrene beads, b 60 nm gold particles, and c HeLa cells , all immersed in aqueous solution were imaged under the five illumination conditions. The contrast of the samples increased when the spectral bandwidth decreased, showing that imaging sensitivity is enhanced using light sources of higher temporal coherence.
COBRI microscopy captures extinction image of the sample, and for small particles, extinction is a combined effect of absorption in the particle and scattering in all directions by the particle. It is interesting to note that the measured extinction (i.e., the COBRI contrast) depends only on the scattering amplitude in the forward direction, independent of its absorption [35]. This is because extinction by nanoparticle is nothing more than interference between incident and forward scattered light [38].
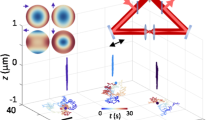
The height-dependent phase difference \( {\varphi} \) enables localization of a particle in the axial direction after appropriate calibration. Figure 3.3 displays how the COBRI contrast of a single vaccinia virus particle changes as a function of the axial position. In this calibration measurement, the virus particle is deposited on a coverglass and its height is adjusted by a sample stage. By calibrating the height-dependent COBRI contrast of the particle, the axial position of the particle can be estimated from the measured contrast. The lateral position of the nanoparticle is determined by finding the center of the spot through image analysis (e.g., 2D Gaussian fitting). Thus, 3D localization from a 2D COBRI image can be accomplished. Taking a vaccinia virus particle as an example, COBRI microscopy can localize it with 1.7–2.3 nm precision in 3D [31].

Reproduced from [31] with permission from the American Chemical Society
Height-dependent COBRI contrast of a vaccinia virus particle deposited on a coverglass. a COBRI contrast changes from dark to bright when the axial position of the virus relative to the microscope objective was varied. b Dependency of COBRI contrast to the axial position was well fitted by a sinusoidal function. c Localization precisions of 2.3, 1.7, and 2.3 nm in the x, y, and z axes were estimated by measuring repeatedly the center position of an immobile virus particle.
COBRI microscopy is a wide-field interferometric transmission microscopy, which shares many common features with holographic microscopy and related techniques [39,40,41]. However, by placing the sample at the focal plane of the microscope objective, COBRI lacks the image reconstruction step which makes COBRI distinct from holographic microscopy. The concept of COBRI microscopy is similar to that of interferometric scattering (iSCAT) microscopy [42,43,44]. The similarities and differences between COBRI and iSCAT are discussed below [35]. The iSCAT microscopy can be considered the reflection counterpart of COBRI microscopy . In COBRI, forward scattering light is collected as signal and transmitted non-scattered light serves as the reference beam, whereas in iSCAT, the backscattering light is the signal and the partial reflection from an interface close to the sample (e.g., the supporting coverglass) serves as the reference beam. Both COBRI and iSCAT provide shot-noise-limited sensitivity through interferometric detection; that is, the sensitivity is determined by the number of detected signal photons. It is demonstrated that COBRI and iSCAT indeed have the same sensitivity given the identical illumination intensity [35]. Different designs of the reference beam, however, lead to several distinctions between COBRI and iSCAT microscopy . First, COBRI and iSCAT have different axial dependencies of particle contrast. In COBRI, when the particle moves in the axial direction, its scattered field has a different phase evolution from the transmitted reference beam, leading to a full contrast inversion over the depth of field of the microscope objective (typically around 1 μm for high NA microscope objective, see Fig. 3.3a). In iSCAT, the reflection geometry makes the particle contrast more sensitive to its axial position relative to the substrate from which the reflected reference beam is created. A full modulation of iSCAT contrast of a particle occurs over a distance of 1/4 optical wavelength, and thus the iSCAT contrast inverts several times over the depth of field [45]. Contrast inversion interrupts continuous imaging and tracking of nanoparticle in the axial direction because the detection of nanoparticle fails when the contrast is zero. As a result, COBRI has a larger working range in the axial direction for continuous imaging and tracking, while iSCAT has a sharper axial dependency of the particle contrast. Second, COBRI imaging is nearly insensitive to the presence of semi-transparent interfaces in the sample because of its transmission geometry. On the contrary, iSCAT detects weak reflections from those interfaces easily, and this property has been exploited to image the flatness of live cell membranes at the nanoscale [34]. However, the sensitive interface detection of iSCAT could complicate the observation of small nanoparticles in live cells due to the highly dynamic and random reflection from the cell membranes. Third, a beamsplitter is inevitably needed in the reflection geometry of iSCAT, which not only increases the complexity of optical alignment but also complicates signal collection at a high efficiency [46]. On the other hand, COBRI does not need a beamsplitter, offering higher signal collection efficiency in principle. Finally, in transmission geometry, COBRI can easily be operated at low light intensity, typically 100-fold lower than the intensity at which iSCAT can be operated under identical image acquisition conditions. For example, COBRI works at an illumination intensity of 0.01 kW/cm2 with an image acquisition rate of 1000 fps, whereas iSCAT requires 1 kW/cm2 correspondingly. The low operational intensity of COBRI makes it useful for live cell imaging where the light dose is of a critical concern [47]. Increasing the illumination intensity in COBRI, and thus its detection sensitivity, is possible through pupil function engineering. By inserting a dot-shaped attenuator at the back focal plane of the microscope objective in the detection path, the illumination intensity of COBRI can reach up to 150 kW/cm2 at 1000 fps where single 10 nm gold nanoparticles are detected [35]. Therefore, COBRI microscopy potentially covers a wider range of illumination intensities, providing a large tunable range of sensitivity for different applications.
3.3 Scattering Background Estimation and Correction
Obtaining a high-quality micrograph where the object of interest (e.g., a nanoparticle) is clearly seen is the goal of microscope imaging. Scattering-based label-free imaging is complicated by the random scattering background because any imperfect optical element along the beam path scatters light and contributes to the background. The general goal of background estimation and correction is to achieve faithful, artifact-free imaging of the signal without the presence of an undesired background. Background correction should not be mistaken for noise reduction or image restoration [48, 49]. Here, the background, in the most general definition, refers to light arriving at the detector that does not carry a signal of interest. The presence of a background complicates or even prevents signal detection. Depending on the application, the signal of interest varies, and so does the background. For example, in some applications, the cell structure and morphology are the signal targets, whereas in other applications, the signal targets are sub-cellular organelles. These different definitions of a signal lead to distinct strategies for estimating and removing the background. In this section, a few methods for background estimation and correction are overviewed, and their performance in live cell imaging is illustrated. In addition, a sophisticated background estimation method that can resolve the nanoscopic motion of a nanoparticle throughout the observation time is discussed [50]. These methods rely on the distinct spatial and temporal characteristics of a signal and background. COBRI and iSCAT share background correction strategies because they both employ scattering-based imaging interferometry.
The simplest measurement in microscopy is perhaps imaging spatially separated particles attached on a clean coverglass. At first glance, such a measurement appears to have no source of an undesired background. However, when the particles are extremely small and have a weak visibility of 0.01 or less, a spatially heterogeneous background due to imperfect, non-uniform illumination begins to complicate any measurement. The illumination background can be measured using several methods. One method is to capture the background before the particle appears on the coverglass—this method is especially useful in the application of “landing assays” [51,52,53]. If the particle is already attached to the coverglass before the measurement, a common method is to modulate the lateral position of the sample, during which a video is recorded. The static illumination background can be extracted from the moving signal in the video. This can be accomplished by calculating the temporal median background of the video. Alternatively, the signal modulated at a specific frequency can be reconstructed through Fourier analysis. This method of spatial modulation works well for estimating and removing the illumination background, but it does not correct the background caused by the sample (e.g., by roughness of the coverglass) because it moves together with the signal. In the most sensitive COBRI and iSCAT imaging, the scattering background caused by the roughness of the coverglass prevents direct visualization of a weak signal [52].
In many applications, a particle is in constant motion, and the task is to measure this motion. This provides an opportunity for convenient background correction. By recording a video containing the dynamic particle of interest, a static background can be extracted that represents the combined effect of illumination, coverglass roughness, and any other background contributions that are relatively static within the observation time. Correcting that background from the raw video gives a background-free video showing only the signal. This method is particularly powerful in applications of SPT where a particle of interest moves over the observation area. In such cases, the temporal median image of the video is often a reasonable estimation of the static background [54].
Single virus particles and intracellular vesicles have been imaged and tracked in live cells using temporal median background correction and COBRI microscopy [31, 32]. Due to their small sizes, virus particles and cell vesicles move and diffuse constantly because of thermal fluctuation. By contrast, cell structures move and evolve slowly. When capturing a COBRI video at high speed (>5,000 fps) for a few seconds, the median background conveniently represents the scattering background of large cell structures and other static background contributions (i.e., the non-uniform illumination). Many endogenous macromolecules in the cell membrane or inside the cell are expected to move rapidly. If these macromolecules provide sufficient optical contrast, they appear in the background-corrected images and make detection of the particle signal more difficult. Thus, for optimal performance, the imaging sensitivity should be sufficient for detecting the particle of interest but not unnecessarily high to ensure that other smaller entities remain indiscernible. The capability of temporal median background correction in removing cell background scattering has been evaluated experimentally [31]. A cell peripheral area was imaged using COBRI microscopy at 5000 fps for 1 s. A median background was calculated from the recorded video and this was used to normalize the raw video frame by frame. In the normalized video, no cell feature remained. Moreover, the temporal fluctuation of each pixel in the background-corrected video corresponded to the photon shot-noise fluctuation, indicating no additional cell background fluctuation. These findings show that temporal median background correction has favorable performance for removing cell scattering background in high-speed COBRI microscopy .
Distinguishing the signal from the background according to their different spatiotemporal characteristics is a powerful strategy in label-free imaging. Temporal median background correction is convenient, but it requires a highly static background and also a dynamic signal over the space. These requirements cannot be met in some applications, for example, when the motion of the signal is highly localized (close to the size of the signal) throughout the observation time. In such cases, some pixels are continuously occupied by the signal, and the median background estimation at those pixels is biased. In principle, superior background estimation can be made by accounting for the signal or background spatiotemporal characteristics, if they are available.
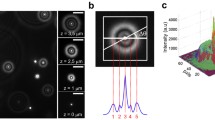
A more general method for background estimation was proposed and demonstrated [50]. Instead of examining the data pixel by pixel independently, as in the case of temporal median filtering, the new method exploits the information encoded in neighboring pixels. Specifically, it takes advantage of a priori knowledge of the shape of the signal. For small particles, their optical image is the point spread function (PSF) of the microscope, typically an airy disk. Specifically, the background estimation is optimized by minimizing the residual error of fitting the background-corrected image with a known PSF through an iterative process. This optimization process repeats itself until the estimation converges. Intuitively, because the signal spot moves over space, every pixel is occupied differently by the signal PSF throughout the video. Even when some pixels are continuously affected by the signal PSF (and thus have no opportunity to reveal their background values directly), their neighboring pixels are likely to be less influenced by the signal because of the moving nature of the signal. For these neighboring pixels, superior estimation of the background values is possible. The estimations of the neighboring pixels eventually help to determine the background value of the pixels that were originally inaccessible, because their intensities are correlated through the signal PSF . Figure 3.4 displays the result of background estimation and correction. The weak signal can barely be seen in the raw image where the signal-to-noise ratio is close to one. By estimating the background with the aforementioned method followed by background removal, the signal appears and its position can be accurately determined in the background-corrected images.

Reproduced from [50] with permission from the American Chemical Society
Background estimation and correction. a Raw image; the yellow arrow points to the position of the signal that is embedded in a heterogeneous background. b Static background image estimated from a series of raw images with a spatially moving signal. c Signal reconstructed by removing the background from the raw image. Note that the range of the color map of (c) is much smaller than that of (a) and (b).
3.4 Label-Free, Ultrahigh-Speed Imaging and Tracking of a Single Bionanoparticle in Live Cells
In this section, two examples of ultrahigh-speed imaging and tracking of native bionanoparticles in live cells by using COBRI microscopy are presented. The first example captured the diffusive motion of a single virus particle on a cell plasma membrane with nanometer spatial precision in 3D at 100,000 fps [31]. Rapid local diffusion and highly transient nanoscopic confinements were disclosed in microsecond timescales. The second example measured the dynamics of single cellular vesicles in the cytoplasm of live cells [32]. Various types of motion were resolved at high spatiotemporal resolutions, including local diffusive motion, stepwise motion by motor proteins, as well as bidirectional and correlated motions. These two examples demonstrated the sensitivity of COBRI microscopy that enables it to conduct nanometer-precise tracking of native biological nanoparticles at ultrahigh speed . Moreover, they illustrated successful live-cell background removal for single particle dynamics on the cell surface and inside the cell.
3.4.1 Single Virus Dynamics on Cell Membrane
Using the 3D tracking capability of COBRI (Sect. 3.2) and cell background correction (Sect. 3.3), highly diffusive motion of a single vaccinia virus particle was revealed on a live HeLa cell membrane captured at an ultrahigh speed of 100,000 fps. The virus particles were locally delivered by a micropipette placed close to the cell under observation. The virus landing process was recorded continuously for a few seconds. The goal of the experiment was to resolve the early interaction between the virus particle and the cell plasma membrane receptors immediately after attachment.
Figure 3.5 displays the 1-s trajectory of the virus exploring the cell membrane right after attachment. The high spatial precision and high temporal resolution unveil the nanoscopic motion of the virus over microsecond timescales. The first surprise is that the virus particle was locally confined to an area of hundreds of nanometers close to the landing site. Moreover, within this confined area, the virus diffused laterally on the plasma membrane with a very high diffusion coefficient (~1 μm2/s), which is as high as its free diffusion in water. This observation indicates that the virus weakly adhered to the membrane and was free to explore the cell surface. Strikingly, from the trajectory, numerous zones of 10–20 nm were present where the virus was transiently confined for sub-milliseconds. The observed viral dynamics are hypothesized to reflect the interaction between the virus and the cell membrane molecules (e.g., glycosaminoglycan). The transient nano-confinements may be evidence of virus interplay with immobile membrane receptors. Notably, the trajectory was 3D, which indicates the 3D cell membrane morphology.

Reproduced from [31] with permission from the American Chemistry Society
Ultrahigh -speed single virus tracking on the plasma membrane of live cells. a Diffusion trajectory of a vaccinia virus on the cell surface captured at 100,000 fps. b Sub-millisecond transient confinements of the virus particle at nanoscopic zones (highlighted in red). c 3D reconstruction of the virus diffusion trajectory. d 3D displacements as a function of time. The displacement in the z axis is much smaller than it is in x and y axes, showing that the virus particle adhered to the cell surface and explored the plasma membrane laterally.
3.4.2 Nanoscopic Dynamics of a Cell Vesicle During Active Transportation
COBRI microscopy also makes it possible to visualize the intracellular dynamics of individual cell vesicles and organelles. With improved sensitivity and speed, a native fibroblast cell appears highly dynamic under COBRI microscopy. Many cell vesicles are locally confined and restricted to diffuse within small volumes created by cytoskeleton networks inside the cytosol. Rapid and directional transportations of cell vesicles are also occasionally observed. Through lipophilic dye labeling, most vesicles were found to be lipid-rich, presumably lipid droplets. Estimated from their COBRI contrast, most vesicles under observation were approximately 300 nm in diameter. The sensitivity of COBRI microscopy is sufficient for tracking single vesicles with nanometer spatial precision in 3D. By capturing vesicle motions at 30,000 fps, their nanoscopic motion is unveiled. Figure 3.6 shows typical transportation of a vesicle with clearly resolved stepwise motion by motor proteins. The bidirectional motion of the vesicle resulting from a tug-of-war between oppositely directed motor proteins attached to the same cargo can clearly be seen. Under ultrahigh-resolution observation, the transient pausing at the moment the vesicle switches directions is clearly resolved where an oscillating behavior over 10-nm is disclosed. In addition to the ultrahigh spatiotemporal resolution, label-free COBRI microscopy offers the opportunity to probe intracellular dynamics in their native condition.

Reproduced from [32] with permission of The Royal Society of Chemistry
Directional intracellular transportation of a native cell vesicle captured using ultrahigh-speed COBRI microscopy . a Reconstructed 3D trajectory of the cell vesicle being translocated inside a live fibroblast cell recorded at 30,000 fps. b Close-up view of the trajectory in (a), revealing the discrete stepping motion of the vesicle. Four clusters of localizations caused by transient pausing between steps are indicated by black arrows. c Analysis of the stepwise motion of the cell vesicle, showing an average step size of 16 nm.
3.5 Conclusions and Future Perspectives
In this chapter, COBRI microscopy was shown to enable ultrahigh-speed imaging with a frame time of microseconds. The high sensitivity of COBRI facilitates nanometer-precise 3D tracking of endogenous biological nanoparticles with diameters as small as 100–200 nm. The cell scattering background can be selectively removed through proper background estimation and correction. The current strategy of background correction is favorable when tracking mobile nanoparticles in a relatively stationary environment. Using COBRI microscopy and background correction, the rapid dynamics of virus particles and vesicles in live cells have been explored at unprecedented resolutions (nanometer spatial precision and microsecond temporal resolution ). In these two experiments described in this chapter, simultaneous high speed and high localization precision made it possible to probe interactions of a nanoparticle with its local environment at the molecular (nanometer) scale. This capability cannot be matched easily by any other techniques.
COBRI microscopy is not a replacement for fluorescence microscopy —instead, simultaneous COBRI and fluorescence imaging can be a powerful combination, with COBRI supporting long-term, high-precision, high-speed measurements, and fluorescence imaging providing excellent imaging specificity through labeling. Indeed, COBRI imaging can easily be added to fluorescence microscopy by operating COBRI simultaneously with an appropriate laser wavelength. For tracking bionanoparticles, fluorescence labeling and imaging can identify nanoparticles of interest, and then COBRI can be employed for long-term, high-resolution observation and tracking of the targeted particles. This approach makes optimal use of the fluorescence photon budget and thus prolongs the observation time. When combining COBRI with fluorescence superresolution microscopy [55], the nanoparticle trajectory can be superimposed on a super-resolved molecular map, providing direct visualization of the local interplay between the nanoparticle and molecules.
COBRI microscopy is more sensitive than conventional brightfield microscopy, but what is the ultimate detection limit? As outlined in Sect. 3.2, the sensitivity of COBRI is shot-noise limited, and thus the number of detected signal photons directly determines whether the target can be perceived. For extremely small nanoparticles, the scattering cross section is very small, and therefore it requires high illumination intensity and long signal integration time to detect them. Conceptually, given sufficient illumination intensity or signal integration time, arbitrarily small nanoparticles can be detected. In practice, illumination intensity is restricted by the damage threshold of the sample and optics, whereas the signal integration time is limited by the signal and background stability that is required for proper background correction. It is worth noting that detecting small signal under strong illumination is complicated by the saturation of photodetector—the full well capacity of the camera sets the maximal light intensity it can detect, determining the strongest illumination intensity of the sample in COBRI and other common-path interferometry. To further increase the illumination intensity without saturation of the camera, pupil function engineering has been employed to selectively attenuate the reference beam and leave the signal of nanoparticle nearly untouched [35, 52, 56,57,58]. This approach effectively enhances the contrast of the signal by matching the amplitude of the reference and the signal. Through pupil function engineering and image postprocessing, COBRI microscopy detects very small nanoparticles (as small as 10 nm gold nanoparticles ) at 1000 fps [35]. By similar approaches of contrast enhancement, ultrahigh sensitivity is achieved by iSCAT microscopy where single protein macromolecules are detected without any labels at an effective image acquisition rate of a few hertz [51, 52, 59, 60].
With enhanced sensitivity, live cells are expected to be highly dynamic and heterogeneous under COBRI microscopy because cell molecules are in constant motion. The spatial resolution of COBRI is diffraction limited , which means that it cannot resolve individual molecules densely packed in live cells. Nevertheless, the spatially resolved fluctuation of the COBRI signal contains information regarding local molecule dynamics that may be revealed by correlation spectroscopy [61, 62]. The high temporal resolution of COBRI microscopy enables investigation of cell molecular dynamics in microsecond timescales. Further investigation could examine whether diffusion, self-assembly, transportation, or even new dynamics are resolvable in a label-free manner.
Light doses are a concern for live cell imaging because of photo-toxicity [47, 63]. To minimize the light dose, ultrahigh-speed COBRI image recording at 100,000 fps is only performed for a few seconds at an illumination intensity of 1 kW/cm2. Otherwise, continuous observation and sample exploration are conducted with a separate camera that captures identical COBRI images at a much lower speed (<1000 fps) that only requires an illumination intensity of <0.01 kW/cm2. Under these conditions, no photo-toxicity is observed.
COBRI microscopy is a simple and powerful imaging modality that provides high sensitivity and high temporal resolution . COBRI is useful for exploring new cell dynamics, including the uptake and release of cell vesicles, cell membrane dynamics, and intracellular or intranuclear dynamics. Research employing COBRI microscopy requires not only optical observation but also sophisticated data processing and analyses. This makes it an exciting multidisciplinary area combining optics, biophysics, chemistry, biology, and engineering.
References
R.D. Vale, Intracellular transport using microtubule-based motors. Annu. Rev. Cell Biol. 3(1), 347–378 (1987)
J. Suh, D. Wirtz, J. Hanes, Efficient active transport of gene nanocarriers to the cell nucleus. Proc. Natl. Acad. Sci. 100(7), 3878–3882 (2003)
N. Hirokawa, S. Niwa, Y. Tanaka, Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron 68(4), 610–638 (2010)
B.W. Guzik, L.S.B. Goldstein, Microtubule-dependent transport in neurons: steps towards an understanding of regulation, function and dysfunction. Curr. Opin. Cell Biol. 16(4), 443–450 (2004)
N. Segev, Coordination of intracellular transport steps by GTPases. Semin. Cell Dev. Biol. 22(1), 33–38 (2011)
N. Hirokawa et al., Kinesin superfamily motor proteins and intracellular transport. Nat. Rev. Mol. Cell Biol. 10, 682 (2009)
D. Mudhakir, H. Harashima, Learning from the viral journey: how to enter cells and how to overcome intracellular barriers to reach the nucleus. AAPS J. 11(1), 65 (2009)
R.D. Vale, The molecular motor toolbox for intracellular transport. Cell 112(4), 467–480 (2003)
D.J. Stephens, V.J. Allan, Light microscopy techniques for live cell imaging. Science 300(5616), 82–86 (2003)
P.L. Leopold et al., Fluorescent virions: dynamic tracking of the pathway of adenoviral gene transfer vectors in living cells. Hum. Gene Ther. 9(3), 367–378 (1998)
I. Wacker et al., Microtubule-dependent transport of secretory vesicles visualized in real time with a GFP-tagged secretory protein. J. Cell Sci. 110(13), 1453–1463 (1997)
M. Lakadamyali et al., Visualizing infection of individual influenza viruses. Proc. Natl. Acad. Sci. U.S.A. 100(16), 9280–9285 (2003)
K. Jaqaman et al., Robust single-particle tracking in live-cell time-lapse sequences. Nat. Methods 5, 695 (2008)
N. Ruthardt, D.C. Lamb, C. Bräuchle, Single-particle tracking as a quantitative microscopy-based approach to unravel cell entry mechanisms of viruses and pharmaceutical nanoparticles. Mol. Ther. 19(7), 1199–1211 (2011)
S. Manley, J.M. Gillette, J. Lippincott-Schwartz, Chapter 5 Single-particle tracking photoactivated localization microscopy for mapping single-molecule dynamics, in Methods in Enzymology, ed. by N.G. Walter (Academic Press, 2010), pp. 109–120
Z. Liu, Luke D. Lavis, E. Betzig, Imaging live-cell dynamics and structure at the single-molecule level. Mol. Cell 58(4), 644–659 (2015)
C. Dietrich et al., Relationship of lipid rafts to transient confinement zones detected by single particle tracking. Biophys. J. 82, 274–284 (2002)
H. Isojima et al., Direct observation of intermediate states during the stepping motion of kinesin-1. Nat. Chem. Biol. 12, 290–297 (2016)
H. Ueno et al., Simple dark-field microscopy with nanometer spatial precision and microsecond temporal resolution. Biophys. J. 98(9), 2014–2023 (2010)
J.L. Martin et al., Anatomy of F1-ATPase powered rotation. Proc. Natl. Acad. Sci. 111(10), 3715–3720 (2014)
S. Faez et al., Fast, label-free tracking of single viruses and weakly scattering nanoparticles in a nanofluidic optical fiber. ACS Nano 9(12), 12349–12357 (2015)
S. Enoki et al., Label-free single-particle imaging of the influenza virus by objective-type total internal reflection dark-field microscopy. PLoS ONE 7(11), e49208 (2012)
B. Agnarsson et al., Evanescent light-scattering microscopy for label-free interfacial imaging: from single sub-100 nm vesicles to live cells. ACS Nano 9(12), 11849–11862 (2015)
E. McLeod et al., High-throughput and label-free single nanoparticle sizing based on time-resolved on-chip microscopy. ACS Nano 9(3), 3265–3273 (2015)
P. Kukura et al., High-speed nanoscopic tracking of the position and orientation of a single virus. Nat. Methods 6, 923–927 (2009)
S.M. Scherr et al., Real-time capture and visualization of individual viruses in complex media. ACS Nano 10(2), 2827–2833 (2016)
G.G. Daaboul et al., Digital detection of exosomes by interferometric imaging. Sci. Rep. 6, 37246 (2016)
H.-M. Wu et al., Nanoscopic substructures of raft-mimetic liquid-ordered membrane domains revealed by high-speed single-particle tracking. Sci. Rep. 6, 20542 (2016)
Y.-H. Lin, W.-L. Chang, C.-L. Hsieh, Shot-noise limited localization of single 20 nm gold particles with nanometer spatial precision within microseconds. Opt. Express 22(8), 9159–9170 (2014)
Y. Yang et al., Label-free tracking of single organelle transportation in cells with nanometer precision using a plasmonic imaging technique. Small 11(24), 2878–2884 (2015)
Y.-F. Huang et al., Coherent brightfield microscopy provides the spatiotemporal resolution to study early stage viral infection in live cells. ACS Nano 11(3), 2575–2585 (2017)
Y.-F. Huang et al., Label-free, ultrahigh-speed, 3D observation of bidirectional and correlated intracellular cargo transport by coherent brightfield microscopy. Nanoscale 9, 6567–6574 (2017)
C. Kural et al., Tracking melanosomes inside a cell to study molecular motors and their interaction. Proc. Natl. Acad. Sci. 104(13), 5378–5382 (2007)
J.-S. Park et al., Label-free and live cell imaging by interferometric scattering microscopy. Chem. Sci. 9(10), 2690–2697 (2018)
C.-Y. Cheng, Y.-H. Liao, C.-L. Hsieh, High-speed imaging and tracking of very small single nanoparticles by contrast enhanced microscopy. Nanoscale. 11, 568–577 (2019)
C.-L. Hsieh, Label-free, ultrasensitive, ultrahigh-speed scattering-based interferometric imaging. Opt. Commun. 422, 69–74 (2018)
J. Hwang, W.E. Moerner, Interferometry of a single nanoparticle using the Gouy phase of a focused laser beam. Opt. Commun. 280(2), 487–491 (2007)
C.F. Bohren, D.R. Huffman, Absorption and Scattering of Light by Small Particles (Wiley-VCH, 2004)
M.K. Kim, Digital holographic microscopy, in Digital Holographic Microscopy: Principles, Techniques, and Applications (Springer, New York, 2011), pp. 149–190
V. Mico, Z. Zalevsky, J. García, Common-path phase-shifting digital holographic microscopy: a way to quantitative phase imaging and superresolution. Opt. Commun. 281(17), 4273–4281 (2008)
J. Garcia-Sucerquia et al., Digital in-line holographic microscopy. Appl. Opt. 45(5), 836–850 (2006)
J. Ortega-Arroyo, P. Kukura, Interferometric scattering microscopy (iSCAT): new frontiers in ultrafast and ultrasensitive optical microscopy. Phys. Chem. Chem. Phys. 14(45), 15625–15636 (2012)
K. Lindfors et al., Detection and spectroscopy of gold nanoparticles using supercontinuum white light confocal microscopy. Phys. Rev. Lett. 93(3), 037401 (2004)
V. Jacobsen et al., Interferometric optical detection and tracking of very small gold nanoparticles at a water-glass interface. Opt. Express 14, 405–414 (2006)
M. Krishnan et al., Geometry-induced electrostatic trapping of nanometric objects in a fluid. Nature 467(7316), 692–695 (2010)
J. Ortega Arroyo, D. Cole, P. Kukura, Interferometric scattering microscopy and its combination with single-molecule fluorescence imaging. Nat. Protocols 11(4), 617–633 (2016)
P.P. Laissue et al., Assessing phototoxicity in live fluorescence imaging. Nat. Methods 14, 657 (2017)
X. Huang et al., Fast, long-term, super-resolution imaging with Hessian structured illumination microscopy. Nat. Biotechnol. 36, 451 (2018)
M. Weigert et al., Content-aware image restoration: pushing the limits of fluorescence microscopy. Nat. Methods 15(12), 1090–1097 (2018)
C.-Y. Cheng, C.-L. Hsieh, Background estimation and correction for high-precision localization microscopy. ACS Photonics 4(7), 1730–1739 (2017)
M. Piliarik, V. Sandoghdar, Direct optical sensing of single unlabelled proteins and super-resolution imaging of their binding sites. Nat. Commun. 5, 4495 (2014)
D. Cole et al., Label-free single-molecule imaging with numerical-aperture-shaped interferometric scattering microscopy. ACS Photonics 4(2), 211–216 (2017)
G. Young et al., Quantitative mass imaging of single biological macromolecules. Science 360(6387), 423–427 (2018)
C.L. Hsieh et al., Tracking single particles on supported lipid membranes: multimobility diffusion and nanoscopic confinement. J. Phys. Chem. B 118(6), 1545–1554 (2014)
B. Huang, M. Bates, X. Zhuang, Super-resolution fluorescence microscopy. Annu. Rev. Biochem. 78(1), 993–1016 (2009)
O. Avci et al., Pupil function engineering for enhanced nanoparticle visibility in wide-field interferometric microscopy. Optica 4(2), 247–254 (2017)
Q.D. Pham et al., Digital holographic microscope with low-frequency attenuation filter for position measurement of a nanoparticle. Opt. Lett. 37(19), 4119–4121 (2012)
K. Goto, Y. Hayasaki, Three-dimensional motion detection of a 20-nm gold nanoparticle using twilight-field digital holography with coherence regulation. Opt. Lett. 40(14), 3344–3347 (2015)
J. Ortega Arroyo et al., Label-free, all-optical detection, imaging, and tracking of a single protein. Nano Lett. 14(4), 2065–2070 (2014)
M.P. McDonald et al., Visualizing single-cell secretion dynamics with single-protein sensitivity. Nano Lett. 18(1), 513–519 (2018)
H. Liu, C. Dong, J. Ren, Tempo-spatially resolved scattering correlation spectroscopy under dark-field illumination and its application to investigate dynamic behaviors of gold nanoparticles in live cells. J. Am. Chem. Soc. 136(7), 2775–2785 (2014)
R. Pecora, Dynamic Light Scattering: Applications of Photon Correlation Spectroscopy (Plenum Press, New York, 1985)
J.-Y. Tinevez et al., Chapter 15 A quantitative method for measuring phototoxicity of a live cell imaging microscope, in Methods in Enzymology, ed. by P.M. Conn (Academic Press, 2012), pp. 291–309
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Hsieh, CL. (2019). Label-Free, Ultrahigh-Speed, Direct Imaging and Tracking of Bionanoparticles in Live Cells by Using Coherent Brightfield Microscopy. In: Astratov, V. (eds) Label-Free Super-Resolution Microscopy. Biological and Medical Physics, Biomedical Engineering. Springer, Cham. https://doi.org/10.1007/978-3-030-21722-8_3
Download citation
DOI: https://doi.org/10.1007/978-3-030-21722-8_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-21721-1
Online ISBN: 978-3-030-21722-8
eBook Packages: Physics and AstronomyPhysics and Astronomy (R0)