Abstract
Although the genetic information is encoded in a one-dimensional array of nucleic acid bases, three-dimensional relationships within DNA play a major role in how this information is accessed and utilized by living organisms. Because of the intertwined nature of the DNA two-braid and its extreme length and compaction in the cell, some of the most important three-dimensional relationships in DNA are topological in nature. Topological linkages within the two-braid and between different DNA segments can be described in simple mathematical terms that account for both the twist and the writhe in the double helix. Topoisomerases are ubiquitous enzymes that regulate the topological state of the genetic material by altering either twist or writhe. To do so, these enzymes transiently open the topological system by breaking one or both strands of the two-braid. This article will review the mathematics of DNA topology, describe the different classes of topoisomerases, and discuss the mechanistic basis for their actions in both biological and mathematical terms. Finally, it will discuss how topoisomerases recognize the topological states of their DNA substrates and products and how some of these enzymes distinguish supercoil handedness during catalysis and DNA cleavage. These latter characteristics make topoisomerases well suited for their individual physiological tasks and impact their roles as targets of important anticancer and antibacterial drugs.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 DNA Topology
DNA (deoxyribonucleic acid) encodes all the inheritable genetic information that makes us what we are. Thus, it is arguably the most important biomolecule in the cell. The structure of DNA represents a perfect biological relationship between form and function. The genetic material is contained in a plectonemically coiled two-braid in which the two strands of the double helix are antiparallel and complementary [1]. This structure serves not only as a framework for the organization and expression of the genetic information, but also provides an elegant mechanism for self-replication and repair [1].
The amount of DNA in a human is staggering. The human genome is encoded in ~3 billion base pairs that are contained on 23 individual chromosomes [2]. Because humans are diploid, each of our cells contains ~6 billion base pairs and 46 chromosomes. At actual size, the DNA in a human cell is ~2 m in length and is compressed into a nucleus that is ~5–10 µm in diameter [3]. The human body is comprised of ~30 trillion cells [4, 5] and therefore contains ~180 sextillion base pairs of DNA that would stretch ~60 billion kilometers in length if laid end-to-end.
Although the human genome is linear, the extreme length and cellular compaction of DNA, the high frictional forces associated with a two-braid of that length, and the fact that the DNA is anchored to cellular scaffolds preclude torsional stress from being translated throughout the genetic material by rotation of DNA ends. Therefore, for all practical purposes, human DNA can be considered to be a closed topological system [6,7,8,9,10,11,12,13,14,15,16]. As long as the ends of DNA are “fixed,” topological relationships are defined as those that can be altered only by breaking one or both strands of the double helix. Even though the genetic information contained within DNA is encoded within a linear sequence of bases, the topological structure of the molecule has profound effects on how this information is accessed and used in the cell.
2 Mathematical and Biological Implications of DNA Topology
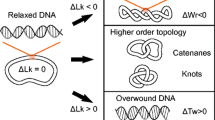
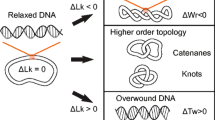
DNA topology can be defined mathematically by three straightforward concepts: twist (Tw), writhe (Wr), and linking number (Lk) [8,9,10,11, 17,18,19,20,21,22]. Twist is the total number of double helical turns in a defined DNA segment. By convention, positive twist is defined as the right-handed twist observed in the normal Watson-Crick DNA structure. Twist represents torsional stress in the double helix. Writhe is defined as the number of times the double helix crosses itself if the DNA segment is projected in two dimensions and represents axial stress in the molecule. Each writhe is assigned an integral value of –1 or +1 based on the handedness of the crossover, which is determined by the direction of rotation that would be required to align the front DNA segment with the back segment without rotating the DNA more than 180° [8, 9]. If the front segment must be rotated clockwise, the writhe is negative (i.e., right-handed); conversely, if it must be rotated counterclockwise, the writhe is positive (i.e., left-handed) (Fig. 1).
Topological relationships in DNA. DNA molecules are drawn as circles for simplicity. Top: DNA that is not under torsional stress is referred to as “relaxed” (middle) and is represented as an unknot (or trivial knot). Underwinding or overwinding the DNA results in negatively supercoiled (left) or positively supercoiled (right) DNA, respectively. Supercoiling is depicted here as writhe (DNA crossovers) for visual clarity, but it should be noted that twist and writhe are interconvertible within these molecules. Bottom: Intramolecular knots (left) and intermolecular tangles (catenanes, shown as a hopf link, right) can also form in DNA. In these cases, twist and writhe are not interconvertible
Mathematically, DNA twist and writhe are related, and the sum of these two values is expressed as the linking number (Lk):
Two key concepts associated with this equation should be emphasized. First, in a topologically closed system, the linking number is invariant [8,9,10,11, 17,18,19,20,21]. The only way to alter this value is to open the system by breaking one or both chains of the DNA double helix. Second, in the absence of knots or tangles, twist and writhe are fluid and interconvertible. The classic means of demonstrating this fluidity (although it is becoming increasingly more dated) is a telephone cord [8,9,10,11, 17,18,19,20,21]. Another example could be a coiled lock of hair such as those seen on a Greek statue. In its unstretched configuration, the cord (or hair) writhes about itself, forming coils without visible twisting. However, when the cord (or hair) is stretched, the writhes are lost and the cord is visibly twisted. Although the coiled and stretched structures are homeomorphic, they contain very different levels of twist and writhe.
When DNA is not under torsional stress, as observed in the canonical Watson-Crick structure, it is said to be “relaxed.” In this form, the double helix makes one helical turn for every 10.5 base pairs (bp) [23]. Therefore, the linking number of a relaxed DNA molecule of 1050 bp would be 100. The magnitude to which topology has the potential to affect the biological function of DNA becomes obvious in the context of the human organism. Considering only links formed between the strands of the DNA two-braid, there are ~600 million links within the ~6 billion bp genome. Every time the genetic information is duplicated, the cell must remove every one of these links. If even one link remains (or if an additional link is generated), two daughter chromosomes will remain intertwined and will not be properly segregated. In total, the ~30 trillion cells of the human body contain ~18 sextillion DNA links!
2.1 DNA Supercoiling
DNA can contain two different kinds of links: those that are formed between the two strands of the DNA two-braid and those that are formed between two separate segments of double helical DNA. This section will address the topological ramifications of links formed between the strands of the two-braid.
The linking number for a right-handed plectonemically coiled double helix is always positive [8, 9, 11, 18]; Lk = 0 would mean that the DNA was completely unwound with no crossings between the two strands of the helix, yielding a paranemic structure. Therefore, DNA topology is often expressed as the change in linking number, ΔLk, which is defined as the difference between the actual Lk of a DNA molecule and the Lk if the molecule were completely relaxed (Lk0).
For example, if the 1050-bp molecule above had an actual Lk of 94, then ΔLk = 94 − 100 = –6, which would mean that the molecule had 6% fewer links than in relaxed DNA.
A DNA molecule with a ΔLk ≠ 0 is under stress, which can be distributed over the molecule as a combination of torsional and axial stress [8, 9, 11, 18]. Axial stress results in the superhelical twists depicted as crossovers in Fig. 1. Consequently, DNA in which ΔLk ≠ 0 is referred to as being “supercoiled.” DNA with a negative ΔLk is referred to as “underwound” or “negatively supercoiled,” and DNA with a positive ΔLk is referred to as “overwound” or “positively supercoiled.”
Because the number of links between the two strands in the DNA two-braid is dependent on the length of a given molecule, ΔLk is also length dependent. Therefore, the term σ (specific linking difference or, more commonly, superhelical density) is utilized to compare levels of supercoiling between DNA molecules of different sizes [8, 9, 11, 18]. The σ value is independent of DNA length and is calculated using the equation
Thus, for the example discussed above, \( \upsigma =\Delta {\text{Lk(}} - 6 )\div {\text{Lk}}_{0} (100) = - 0.06 \). The σ value is always negative for underwound DNA and is always positive for overwound DNA.
Although DNA is typically drawn as a relaxed molecule, this topological form does not usually exist in nature. Organisms generally maintain their genome in an ~6% underwound state [8, 11, 15, 16, 18,19,20], which puts energy into DNA and enhances the opening of the double helix. This negative supercoiling is important because the two-braid is the storage form for the genetic material, and the two strands must be separated in order to express (i.e., transcribe) and duplicate (i.e., replicate) the information encoded in DNA.
While negative supercoiling is beneficial to the cell, DNA overwinding is problematic. Positively supercoiled DNA is generated ahead of replication and transcription machinery, because these tracking systems move through the DNA without rotating [7, 8, 24, 25], thereby pushing extra twists ahead of the replication or transcription bubble (Fig. 2). This overwinding makes it increasingly more difficult to open the double helix and, if unresolved, can block the progression of the tracking system [6,7,8, 11, 14, 15, 24, 25].
Moving DNA tracking machinery creates topological problems. As tracking systems move through the DNA, twists are pushed ahead of the fork, resulting in the accumulation of positive supercoils. In the case of replication, precatenanes (links between newly synthesized daughter chromosomes) form behind the fork.
2.2 DNA Knotting and Tangling
This section will address the topological ramifications of links formed between two separate segments of double helical DNA.
As described above, human cells contain ~2 m of DNA (on 46 chromosomes) that is packed into a nucleus that is only 5–10 µm in diameter [3]. Thus, the DNA two-braid falls prey to the same problems as would be expected if a large number of very long ropes were constrained in a small space. Upon movement or manipulation of the ropes, knots (i.e., links formed within a single rope) and tangles (i.e., links formed between different ropes) are routinely formed. Similarly, biological processes that move or manipulate the double helix often induce the formation of knots and tangles into the DNA (Fig. 1). If unresolved, DNA knots and tangles can have lethal consequences.
Knots are formed as a result of recombination pathways that are used to increase genetic diversity and repair some types of DNA damage [6, 8, 11, 12, 24, 26, 27]. The presence of knots does not allow the two strands of the double helix to be separated and therefore prevents essential DNA processes such as replication and transcription from taking place.
DNA tangles are routinely formed during replication when some of the torsional stress in front of the fork redistributes behind it, resulting in links between the two newly synthesized DNA molecules (Fig. 2) [6, 8, 12, 24, 26, 27]. This tangling prevents the linked chromosomes from being properly segregated into daughter cells during mitosis or meiosis. Because DNA tangles are most easily represented as concatenated circles (shown in Fig. 1 as a hopf link), tangled DNA molecules are often referred to as being “catenated.”
The ΔLk in DNA knots and tangles is caused by the introduction of writhe. It is notable that these writhes are fundamentally different than those present in supercoiled DNA. Whereas writhes generated during supercoiling can be freely converted to twists, the crossovers observed in DNA knots and tangles are constrained as writhes [10, 11].
2.3 Alteration of DNA Topology by Strand Breakage
Assuming a closed topological system, the linking number of DNA can only be changed if one or both strands of the two-braid are cut [6, 8, 11, 12, 15, 24, 26, 27]. Cutting a single strand can alleviate (or, under some circumstances, induce) torsional stress or twist within the molecule. Conversely, cutting both strands can alter the axial stress or writhe in the DNA.
In supercoiled DNA, the ΔLk is caused by a change in the number of links formed between the two strands of the double helix, resulting in a molecule in which twist and writhe are interconvertible. Therefore, supercoils within DNA can be removed by cutting either one or both strands of the two-braid [6, 8, 11, 12, 15, 24, 26,27,28]. In DNA knots and tangles, however, the ΔLk is the result of links between separate segments of DNA two-braids. Thus, the writhes in knots and tangles can only be removed if both strands are broken.
3 Topoisomerases
Cells express multiple enzymes known as topoisomerases that regulate the topological state of the genome [6, 9, 13, 15, 27, 29, 30]. Because DNA topology profoundly affects fundamental cellular processes, topoisomerases are encoded by all species. For simplicity, this article will focus primarily on topoisomerases expressed in humans and bacteria. These enzymes alter the superhelical density of DNA and resolve knots and tangles by creating transient breaks in the DNA backbone, which opens the topological system (Fig. 3) [6, 9, 13, 15, 27,28,29,30,31,32].
Actions of type I and type II topoisomerases. The different cleavage activities of type I and type II topoisomerases allow them to work on different topological structures within double-stranded DNA. Because type I enzymes only cut one strand of the DNA, they are restricted to working on twist. Because type II enzymes cut both strands of the DNA, they are able to work on writhe
Topoisomerases are divided into two major classes based on how many DNA strands they cut to carry out their functions: type I topoisomerases cut one strand, and type II topoisomerases cut both strands of the two-braid [6, 9, 13, 15, 27, 29, 30]. In order to maintain the integrity of the genetic material while the DNA is cut, topoisomerases remain attached to the newly generated termini until they reseal the strand break(s). The stable complexes formed when these enzymes covalently attach to DNA are called “cleavage complexes” and are a hallmark of topoisomerase activity.
3.1 Type I Topoisomerases
There are two subclasses of type I topoisomerases in humans and bacteria: type IA and IB [8, 9, 22, 29, 33,34,35,36,37]. Type I enzymes are denoted by odd numerals and are grouped into the subclasses based on homology and enzymatic mechanism. Type IA topoisomerases use a “single-strand passage” mechanism in which they break one strand of the DNA two-braid, pass the opposite strand through the break, and rejoin the original strand (Fig. 4) [28, 29, 38]. When the enzyme cleaves the DNA, the energy of the broken sugar-phosphate bond is conserved by the formation of a new covalent bond between a tyrosyl residue in the active site of the enzyme and the newly generated 5’-terminal phosphate of the DNA. (DNA strands have a directionality defined by the linkages between the sugar and phosphate groups that make up their backbones. Each phosphate connects the 3′-carbon of one sugar to the 5’ carbon of the following sugar in the chain.) The corresponding 3′-DNA terminus generated by the cleavage event is prevented from rotating by non-covalent interactions with the enzyme. As a result of the single-strand passage mechanism, every catalytic event mediated by type IA topoisomerases changes the linking number by one [13, 28, 29, 38].
Even though type IB topoisomerases also cut one strand of the DNA two-braid, they act by a very different mechanism than the type IA enzymes. Type IB topoisomerases alter DNA topology using a “controlled rotation” mechanism (Fig. 5) [13, 28, 29, 38]. These enzymes covalently attach to the 3’-terminal phosphate during the cleavage event and allow the 5’-DNA terminus of the cleaved strand to rotate about the intact strand. Each rotation changes the linking number by one. The number of strand rotations that occur per catalytic event depends on a number of factors, including the superhelical density of the DNA substrate.
Irrespective of the mechanism used (single-strand passage versus controlled rotation), type I topoisomerases always function by changing the number of links formed between the two strands of the DNA two-braid [13, 28, 29, 38]. Therefore, these enzymes alter DNA topology by changing twist. Consequently, they can alter the superhelical density of DNA, but they cannot resolve DNA knots or tangles formed within double-stranded DNA (Fig. 3).
Humans encode both type IA (topoisomerases IIIα and IIIβ) and type IB (topoisomerase I) enzymes [6, 13, 14, 28, 29, 38]. Human topoisomerase III contributes to genomic stability and prevents inappropriate recombination by relaxing hyper-negatively supercoiled DNA [29, 33,34,35,36,37]. It also works with other proteins to resolve recombination intermediates and stalled replication forks [9, 22, 29, 33,34,35,36,37]. In mice, deletion of the α isoform is lethal, while deletion of the β isoform shortens life span and has deleterious effects on fertility. In humans, deletion of topoisomerase IIIβ is also associated with schizophrenia and neurodevelopmental disorders [39].
The major role of topoisomerase I is likely to remove torsional stress that accumulates ahead of replication forks and other DNA tracking systems [6, 13, 14, 28, 29, 38]. Although the enzyme is dispensable at the cellular level (presumably because of functional redundancies with type II topoisomerases), it appears to be necessary for proper development in multicellular organisms [6, 40,41,42].
Bacteria encode primarily type IA enzymes, topoisomerase I and topoisomerase III. Bacterial topoisomerase I is also known as ω protein and is unrelated to human topoisomerase I. (Unfortunately, a common name was assigned to both proteins before the differences were realized.) Bacterial topoisomerase I works in concert with gyrase (a type II topoisomerase discussed below) to regulate the overall superhelical state of the bacterial chromosome [22, 29, 38]. Bacterial topoisomerase III is a homolog of human topoisomerase IIIα and IIIβ and also plays important roles in maintaining genomic stability [29, 43].
3.2 Type II Topoisomerases
There are two subclasses of type II topoisomerases: type IIA and IIB [9, 12, 29, 33,34,35, 44, 45]. Type II enzymes are denoted by even numerals (with the exception of gyrase, which is discussed below) and are grouped into the subclasses based on homology and reaction mechanism. To date, functional type IIB topoisomerases have been found only in plant and archaeal species. Therefore, only the type IIA enzymes, which are found in humans and bacteria, will be discussed in this article.
Type IIA topoisomerases alter DNA topology by using a “double-strand passage” mechanism in which they cleave both strands of the DNA two-braid, pass a second intact double-helical segment through the break, and rejoin the cleaved strands (Fig. 6) [12, 15, 26, 27, 29,30,31, 46]. The cleaved DNA is known as the “gate-” or “G-segment,” and the intact segment that is transported through the break is known as the “transport-” or “T-segment.” Type IIA enzymes in humans function as homodimers, whereas those in bacteria are A2B2 heterotetramers (the A and B subunits have fused to form the protomer subunit in the human enzyme) [12, 15, 26, 27, 29,30,31, 34, 44,45,46]. The structures of the enzymes have bilateral symmetry that allow for the formation of gated protein annuli at opposite ends. This permits the T-segment to be captured by the protein above the G-segment and exit the protein below it in a controlled fashion (Fig. 6). The double-strand passage reaction involves a series of coordinated protein movements that are coupled to the binding and hydrolysis of the high-energy cofactor ATP.
Due to their bilateral symmetry, type IIA topoisomerases have two active-site tyrosyl residues [29]. When the enzymes cut the double helix, these residues form covalent bonds with the newly generated 5’-terminal phosphates on opposite strands of the DNA. The two scissile bonds are located across the major groove of the DNA, resulting in the formation of 4-base-long single-stranded chains on the 5’-end of each strand [26, 27, 29,30,31, 46,47,48]. As with the type I enzymes, this linkage preserves both genomic integrity and the energy of the broken sugar-phosphate bond.
Because type IIA topoisomerases act on two distinct DNA segments, they modulate DNA topology by altering writhe (Figs. 3 and 6) [9]. As a result of their reaction, they invert the sign of the writhe formed by the DNA crossover (for example, converting a –1 link to a +1 link). Thus, each event catalyzed by type IIA topoisomerases changes the linking number by two. The ability to work on writhe allows type II enzymes to modulate DNA superhelical density. More importantly, it allows them to remove DNA knots and tangles (in which writhe and twist are not interconvertible).
Humans encode two type IIA topoisomerases, topoisomerase IIα and topoisomerase IIβ, which are closely related isoforms [6, 27, 29, 49,50,51,52,53,54,55,56,57]. (It is notable that vertebrates encode two isoforms of topoisomerase II, while invertebrates and lower eukaryotes encode only a single type II enzyme.) Topoisomerase IIα and IIβ share ~70% of their amino acid sequence, but are encoded by separate genes. Although both use the double-strand passage reaction, they differ in their patterns of expression and their cellular functions [12, 27, 35, 44, 45, 54, 58]. Topoisomerase IIα is essential for the survival of proliferating cells (its loss cannot be compensated for by the β isoform), and its expression increases over the cell cycle, peaking in G2/M [59,60,61]. Rapidly growing cells contain ~500,000 copies of topoisomerase IIα, while quiescent cells and differentiated tissues contain virtually none of the α isoform. Topoisomerase IIα is associated with DNA replication complexes and remains bound to chromosomes throughout mitosis, suggesting that it has important functions in growth-related processes such as replication and chromosome segregation [6, 62].
Topoisomerase IIβ is required for neural development in mammals, but is otherwise dispensable at the cellular level [12, 14, 35, 45, 54, 58, 60, 63]. Unlike topoisomerase IIα, topoisomerase IIβ is expressed at high concentrations in most cell types independent of proliferation status. The physiological functions of the β isoform are not yet fully defined. However, it dissociates from chromosomes during mitosis and seems to have an important role in the transcription of developmentally and hormonally regulated genes [35, 63,64,65].
With the exception of a few species, bacteria encode two type IIA topoisomerases, gyrase and topoisomerase IV [31, 46, 66, 67]. Gyrase is the only known topoisomerase that is able to introduce negative supercoils into DNA [29, 33, 35, 58]. To accomplish this task, gyrase wraps the DNA around the C-terminal domain of its A subunit in a right-handed fashion, thereby generating a constrained positive supercoil on the enzyme. (It should be noted that, similar to DNA, proteins have directionality. In this case, directionality is defined by the peptide bonds that link the amino acids in the protein chain and goes from the amino- or N-terminus of the protein to the carboxy- or C-terminus.) In mathematical terms, gyrase performs a type I Reidemeister move on the DNA [68, 69]. Because the DNA wrapping does not change the linking number of the molecule, a compensatory (equal but opposite) type I Reidemeister move is induced elsewhere in the DNA molecule [10, 68, 69], which introduces an unconstrained negative supercoil into the unbound portion of the two-braid. When strand passage occurs, the sign of the induced positive supercoil is inverted, causing a net introduction of two negative supercoils per catalytic cycle [8, 9, 12, 58]. Another important implication of this wrapping mechanism is that the G- and T-segments are on the same DNA molecule and are in close proximity [70]. Consequently, even though gyrase works only on writhe, it is much more efficient at relaxing positive supercoils and introducing negative supercoils than it is at decatenating or unknotting DNA, because these latter reactions require the use of G- and T-segments that are on different DNA molecules or are distal to each other on the same DNA molecule [29, 33, 35, 46, 58].
The major cellular roles of gyrase stem from the ability of the enzyme to carry out intramolecular reactions and to actively underwind DNA. Gyrase functions ahead of replication forks and transcription complexes to alleviate torsional stress induced by DNA overwinding [31, 35, 71]. Additionally, acting in conjunction with bacterial topoisomerase I, gyrase modulates the overall level of DNA supercoiling in the bacterial genome by introducing negative supercoils to maintain the genetic material in an underwound state [31, 35, 71].
Topoisomerase IV uses a “canonical” (i.e., non-wrapping) double-strand passage reaction similar to that utilized by the human type II enzymes [9, 29, 31, 46]. Consequently, topoisomerase IV is able to modulate superhelical density and is also able to carry out the intermolecular strand passage reactions required for decatenation and unknotting. Topoisomerase IV and gyrase display sequence homology. However, due to the differences between the canonical and wrapping mechanisms, they have distinct functions in the bacterial cell [12, 29, 31, 33, 35, 46, 58, 72]. While topoisomerase IV may be involved in regulating DNA over- and underwinding [73,74,75], its primary function is to remove knots and tangles formed by recombination and replication [31, 76,77,78].
4 Recognition of DNA Topology by Topoisomerases
Early studies on the recognition of DNA topology by topoisomerases were concerned with the ability of the enzymes to differentiate between their substrates and products. Consequently, these studies focused primarily on the distinction between negatively supercoiled and relaxed DNA. All of these studies demonstrated that topoisomerases interacted more tightly with their DNA substrates. For example, gyrase (which introduces negative supercoils into relaxed substrates) binds relaxed DNA ~10-fold more tightly than negatively supercoiled molecules [79]. Conversely, human topoisomerase IB [80] and eukaryotic topoisomerase IIA [81] display much higher affinities for supercoiled compared to relaxed DNA. Topoisomerase IIA also hydrolyzes its ATP cofactor more rapidly with negatively supercoiled substrates [81].
The first evidence for the mechanism by which topoisomerases distinguish supercoiled from relaxed DNA came from an electron microscopy study of topoisomerase-DNA complexes, which demonstrated that eukaryotic topoisomerase II binds at DNA crossovers [82]. A later study showed that topoisomerase II simultaneously binds two double-stranded DNA segments [83]. These findings are consistent with the facts that helix-helix juxtapositions are more prevalent in supercoiled molecules and that the type IIA enzyme acts on DNA writhes.
A surprising result of the electron microscopy study was that mammalian topoisomerase IB also binds at DNA crossovers, despite the fact that the enzyme works on twist [82]. The binding of crossovers as a means to differentiate between relaxed and supercoiled molecules was supported by a later study that demonstrated that the type IB enzyme bound equally well to positively and negatively supercoiled DNA, which eliminated topology recognition based on twist [80]. The binding site for the second DNA helix on type IB topoisomerases was later identified by a crystallographic study [84].
In contrast to topoisomerase IB, bacterial topoisomerase I (a type IA enzyme), which recognizes its supercoiled substrate by the single-strand character of the twist associated with negatively supercoiled DNA [85], does not bind at DNA crossovers [82].
5 Recognition of DNA Supercoil Geometry by Topoisomerases
Whereas the studies described above focused on the ability of topoisomerases to discern DNA substrates from reaction products, later studies recognized the fact that topoisomerases work on two very different supercoiled substrates: positively and negatively supercoiled molecules. As discussed above, DNA in organisms ranging from bacteria to humans is globally underwound by ~6% [8, 15, 16, 18,19,20]. However, the torsional stress generated by DNA tracking systems such as replication forks and transcription complexes acutely overwinds the DNA ahead of these molecular machines [6,7,8, 14, 15, 24, 25]. Therefore, these later studies focused on the ability of topoisomerases to discern the geometry (i.e., handedness) of DNA supercoils.
Two different aspects of supercoil geometry recognition by topoisomerases have been examined: the ability to discern supercoil handedness over the entire catalytic event and, more specifically, during the formation of cleavage complexes. Because the mechanisms and ramifications of geometry recognition during these processes differ significantly, they will be discussed separately below.
5.1 Recognition of DNA Supercoil Geometry During Catalysis
This section will discuss the ability of topoisomerases to discern supercoil handedness during catalysis (i.e., the process of changing DNA linking number). For enzymes other than gyrase, studies have examined the removal of positive versus negative supercoils. In the case of gyrase, the removal of positive supercoils has been compared to the introduction of negative supercoils into relaxed DNA.
5.1.1 Type I Topoisomerases
As a result of their mechanisms of action, type IA and type IB topoisomerases recognize DNA supercoil handedness in very different manners. Type IA topoisomerases will not relax positively supercoiled two-braids because they require substantial single-stranded character in their DNA substrate in order to carry out the single-strand passage reaction [85]. The overwinding associated with positive supercoiling impedes this necessary strand separation, whereas negative supercoiling naturally enhances opening of the two strands.
Conversely, type IB topoisomerases, which use a controlled rotation mechanism, can remove both positive and negative supercoils [86]. In fact, human topoisomerase I relaxes positively supercoiled DNA an order of magnitude faster than it does negatively supercoiled substrates [87]. This finding is consistent with simulation studies that suggest mechanistic differences between the removal of positive and negative supercoils, which require the DNA to rotate in opposite directions within the active site of the enzyme [88]. It is also consistent with the primary physiological function of type IB topoisomerases, which is to remove the positive supercoils that accumulate ahead of DNA tracking systems [6, 13, 14, 28, 29, 38].
5.1.2 Type IIA Topoisomerases
All type IIA topoisomerases that use a canonical double-strand passage reaction (as opposed to wrapping) examined to date can remove both positive and negative supercoils. Topoisomerase IV [75, 89,90,91] and human topoisomerase IIα [92, 93] both relax positively supercoiled DNA considerably faster than they do negatively supercoiled molecules. There are two significant differences between positively and negatively supercoiled DNA that could serve as the basis for this chiral recognition. First, the ΔTw (i.e., the difference in twist between the supercoiled and relaxed molecule) is opposite in positively and negatively supercoiled DNA. As discussed above, the differences in ΔTw that accompany over- and underwinding have significant effects on DNA strand separation. Second, the DNA crossings formed in positive and negative writhes occur with different angles (~60° and ~120°, respectively) [94]. The development of single-molecule systems in which two DNA segments can be interwound without altering twist allowed the mechanism of chiral recognition to be addressed. Both topoisomerase IV and human topoisomerase IIα appear to distinguish between positively and negatively supercoiled substrates based on differences in writhe [90, 93]. This finding suggests that these enzymes can discern crossover angles formed at DNA nodes. Elements in the C-terminal domain of both enzymes are required for this recognition [94, 95].
In contrast to topoisomerase IV and human topoisomerase IIα, a number of type IIA topoisomerases cannot discern DNA supercoiling geometry during catalysis and relax positive and negative supercoils at similar rates. Among these are human topoisomerase IIβ and the type IIA enzymes found in yeast, Drosophila, and some viral species [89, 92, 95,96,97]. It is not obvious why these enzymes do not recognize supercoil geometry. However, this once again seems to be related to the C-terminal domains of the type IIA enzymes, which vary widely between species and are lacking in the viral proteins. As further evidence for the role of this protein domain, replacement of the C-terminal domain of topoisomerase IIβ with that of topoisomerase IIα results in a chimeric enzyme that is capable of distinguishing DNA geometry and relaxes positive supercoils an order of magnitude faster than it does negative supercoils [95].
Gyrase differs from other type IIA topoisomerases in that it does not normally relax negatively supercoiled DNA. Therefore, geometry recognition studies with gyrase have compared its abilities to remove positive supercoils and to introduce negative supercoils into relaxed DNA. These processes correspond to the major cellular roles of the enzyme: to remove positive supercoils ahead of DNA tracking systems and to maintain the negative superhelical density of the bacterial chromosome [31, 35, 71]. Gyrase relaxes positive supercoils ~10-fold faster than it negatively supercoils relaxed DNA [91, 98]. The rapid removal of positive supercoils by gyrase reflects its acute function of relaxing overwound DNA ahead of tracking systems and once again requires elements in the C-terminal domain of the enzyme. In this case, it is the specific amino acid residues responsible for DNA wrapping that are necessary [91].
5.2 Recognition of Supercoil Geometry During DNA Cleavage
Although critical to the catalytic function of topoisomerases, the formation of DNA cleavage complexes poses a potential danger to the cell (Fig. 7) [9, 14, 44, 57, 99]. When DNA tracking systems attempt to traverse covalent topoisomerase-cleaved DNA roadblocks in the two-braid, strand breaks can no longer be rejoined by the enzyme and require cellular repair pathways to re-establish the integrity of the double helix. If the strand breaks overwhelm the repair processes, they can induce mutations, chromosomal rearrangements, and cell death pathways. Thus, the enzymes that are necessary for modulating the topological state of DNA also have the potential to fragment the genome.

Figure adapted from Pendleton et al. [57]
Critical balance of DNA cleavage and resealing by topoisomerases. The activity of topoisomerases must be tightly controlled in the cell. When an appropriate level of cleavage complexes is maintained, topological problems are resolved and the cell can grow normally. If the levels of cleavage complexes decrease, slow growth rates and mitotic failure can cause cell death. Conversely, if the levels of cleavage complexes are too high, DNA damage can overwhelm the cell and also lead to cell death.
The inherent danger of cleavage complexes has been exploited for the development of important anticancer and antibacterial drugs that act by stabilizing these complexes. Camptothecin-based drugs that target human topoisomerase I are used to treat ovarian, colorectal, and small-cell lung cancers [100]. Etoposide, doxorubicin, and mitoxantrone, which are used to treat a wide variety of blood-borne and solid tumors, target human topoisomerase IIα and topoisomerase IIβ [44, 57, 99, 101, 102]. Finally, quinolones, such as ciprofloxacin and levofloxacin, which target gyrase and topoisomerase IV, are among the most widely prescribed antibacterial drugs worldwide [46, 67, 103, 104].
Because DNA cleavage complexes formed ahead of tracking systems are the most likely to be converted to permanent strand breaks, topoisomerase-mediated DNA cleavage events that occur on positively supercoiled DNA pose the greatest danger to the cell. Therefore, the ability of topoisomerases to discern supercoil geometry during the cleavage event has been addressed.
5.2.1 Type I Topoisomerases
No data has been reported for the cleavage of positively supercoiled DNA by type IA topoisomerases (presumably because these enzymes do not function on positively supercoiled molecules).
The only type IB topoisomerase for which geometry recognition during cleavage has been investigated is human topoisomerase I [105, 106]. In both the absence and presence of anticancer drugs, this enzyme maintains ~3-fold higher levels of cleavage complexes on positively as compared to negatively supercoiled DNA. Currently, there are no data that address the mechanistic basis for this distinction. However, this result implies that, while type IB topoisomerases work faster on positively supercoiled DNA, they are also inherently more dangerous to the cell while acting on this substrate.
5.2.2 Type IIA Topoisomerases
Even though human topoisomerase IIα is the only eukaryotic type IIA topoisomerase that can recognize supercoil handedness during catalysis, human topoisomerase IIα and IIβ can both discern supercoil geometry during DNA cleavage [105]. A similar result has been found for viral type IIA topoisomerases [96]. All of these enzymes maintain 2–4-fold lower levels of cleavage complexes with positively as compared to negatively supercoiled substrates. Similar results have been found in the absence and presence of anticancer drugs. While this characteristic makes these enzymes safer while operating ahead of DNA tracking systems, it also reduces the potential lethality of chemotherapeutics.
The differential abilities of type IIA enzymes to discern supercoil geometry during catalysis versus cleavage indicates that this recognition must be bimodal in nature. To this point, removal of the C-terminal domain, which is crucial for geometry recognition during catalysis, has no effect on supercoil handedness recognition during cleavage [96, 105]. A later study demonstrated that the ability of human topoisomerase IIα to discern supercoil geometry is embedded within its catalytic core (which contains only the elements needed for the enzyme to cleave and rejoin DNA and does not include either the N-terminal gate or the C-terminal domain) [107]. At the present time, it is not known whether the mechanistic basis for the recognition of supercoil geometry during cleavage is related to the crossover angle of the writhes or the sign of the ΔTw in the DNA.
The effects of supercoil geometry on DNA cleavage mediated by the bacterial type II topoisomerases, gyrase and topoisomerase IV, differ substantially from one another. Gyrase, like the human and viral type IIA enzymes, maintains 2–4-fold lower levels of cleavage complexes on positively supercoiled DNA both in the absence and presence of antibacterial drugs [91, 98, 108]. This recognition is independent of the ability of the enzyme to wrap DNA, which suggests that gyrase also uses a bimodal mechanism to recognize supercoil handedness during catalysis and cleavage. In contrast, topoisomerase IV is the only type IIA enzyme examined to date that does not maintain lower levels of cleavage complexes on positively supercoiled substrates. One study indicated that topoisomerase IV maintains similar levels of cleavage complexes on positively and negatively supercoiled DNA [91], while another suggested that it maintains higher levels of cleavage complexes on overwound substrates [75].
The differential abilities of gyrase and topoisomerase IV to recognize supercoil handedness during cleavage may impact their relative efficacies as targets for quinolone antibacterials. Because gyrase must operate on the overwound DNA formed ahead of replication forks and transcription complexes, it is perfectly positioned to generate cleavage complexes with the potential to be converted to permanent DNA damage. However, the diminished levels of cleavage complexes generated by the enzyme on positively supercoiled DNA may partially abrogate the cytotoxic effects of quinolones. Conversely, topoisomerase IV maintains high levels of cleavage complexes on overwound substrates, but typically acts behind the fork, where cleavage complexes are less likely to be disrupted by tracking systems.
6 Conclusions
DNA topology has a profound effect on how the genetic information is expressed, passed from generation to generation, and recombined in the cell. Topological linkages within the DNA two-braid and between different DNA segments can be described in simple mathematical terms that account for both the twist and the writhe in the double helix. Topoisomerases are enzymes that function as “molecular mathematicians” that regulate the topological state of the genetic material by altering either twist or writhe. To do so, type I and type II topoisomerases transiently open the topological system by breaking one or both strands of the two-braid, respectively.
The importance of topoisomerases is underscored by the fact that they are expressed in all cells and in all species. Topoisomerases can distinguish between the topological states of their substrates and products, which makes them more efficient enzymes. Furthermore, many topoisomerases can distinguish DNA supercoil handedness during catalysis and DNA cleavage, which makes them well suited to their individual physiological tasks and also impacts their roles as targets of important anticancer and antibacterial drugs.
References
J.D. Watson, F.H.C. Crick, Molecular structure of nucleic acids. Nature 171, 737–738 (1953)
International Human Genome Sequencing Consortium, Finishing the euchromatic sequence of the human genome. Nature 431, 931–945 (2004)
B. Alberts, Molecular Biology of the Cell, 6th edn. (Garland Science, Taylor and Francis Group, New York, NY, 2015)
E. Bianconi, A. Piovesan, F. Facchin, A. Beraudi, R. Casadei, F. Frabetti, L. Vitale, M.C. Pelleri, S. Tassani, F. Piva et al., An estimation of the number of cells in the human body. Ann. Hum. Biol. 40, 463–471 (2013)
R. Sender, S. Fuchs, R. Milo, Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14, e1002533 (2016)
J.C. Wang, Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 3, 430–440 (2002)
O. Espeli, K.J. Marians, Untangling intracellular DNA topology. Mol. Microbiol. 52, 925–931 (2004)
A.D. Bates, A. Maxwell, DNA Topology (Oxford University Press, New York, USA, 2005)
J.E. Deweese, M.A. Osheroff, N. Osheroff, DNA topology and topoisomerases: teaching a “knotty” subject. Biochem. Mol. Biol. Educ. 37, 2–10 (2008)
C. Adams, A brief introduction to knot theory from the physical point of view, in Proceedings of Symposia in Applied Mathematics: Applications of Knot Theory, vol. 66, eds. by D. Buck, E. Flapan (American Mathematical Society, Providence, 2009), pp. 1–20
D. Buck DNA topology, in Proceedings of Symposia in Applied Mathematics: Applications of Knot Theory, vol. 66, eds. by D. Buck, E. Flapan (American Mathematical Society, Providence, 2009), pp. 47–80
Z. Liu, R.W. Deibler, H.S. Chan, L. Zechiedrich, The why and how of DNA unlinking. Nucleic Acids Res. 37, 661–671 (2009)
S.H. Chen, N.L. Chan, T.S. Hsieh, New mechanistic and functional insights into DNA topoisomerases. Annu. Rev. Biochem. 82, 139–170 (2013)
Y. Pommier, Y. Sun, S.N. Huang, J.L. Nitiss, Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat. Rev. Mol. Cell Biol. 17, 703–721 (2016)
Y. Seol, K.C. Neuman, The dynamic interplay between DNA topoisomerases and DNA topology. Biophys. Rev. 8, 101–111 (2016)
L. Finzi, W.K. Olson, The emerging role of DNA supercoiling as a dynamic player in genomic structure and function. Biophys. Rev. 8, 1–3 (2016)
F.B. Fuller, The writhing number of a space curve. Proc. Natl. Acad. Sci. USA 68, 815–819 (1971)
W.R. Bauer, F.H. Crick, J.H. White, Supercoiled DNA. Sci. Am. 243, 100–113 (1980)
J.H. White, N.R. Cozzarelli, A simple topological method for describing stereoisomers of DNA catenanes and knots. Proc. Natl. Acad. Sci. USA 81, 3322–3326 (1984)
A.V. Vologodskii, N.R. Cozzarelli, Conformational and thermodynamic properties of supercoiled DNA. Annu. Rev. Biophys. Biomol. Struct. 23, 609–643 (1994)
M.R. Dennis, J.H. Hannay, Geometry of Calugareanu’s theorem. Proc. Roy. Soc. A 461, 3245–3254 (2005)
A.C. Ketron, N. Osheroff, DNA topology and topoisomerases, in Molecular Life Sciences: An Encyclopedic Reference, ed. by E. Bell (Springer, New York, New York, NY, 2014), pp. 1–19
D. Shore, R.L. Baldwin, Energetics of DNA twisting. II. Topoisomer analysis. J. Mol. Biol. 170, 983–1007 (1983)
A. Falaschi, G. Abdurashidova, O. Sandoval, S. Radulescu, G. Biamonti, S. Riva, Molecular and structural transactions at human DNA replication origins. Cell Cycle 6, 1705–1712 (2007)
A. Travers, G. Muskhelishvili, A common topology for bacterial and eukaryotic transcription initiation? EMBO Rep. 8, 147–151 (2007)
J.M. Fortune, N. Osheroff, Topoisomerase II as a target for anticancer drugs: when enzymes stop being nice. Prog. Nucleic Acid Res. Mol. Biol. 64, 221–253 (2000)
A.K. McClendon, N. Osheroff, DNA topoisomerase II, genotoxicity, and cancer. Mutat. Res. 623, 83–97 (2007)
J.B. Leppard, J.J. Champoux, Human DNA topoisomerase I: relaxation, roles, and damage control. Chromosoma 114, 75–85 (2005)
J.J. Champoux, DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 70, 369–413 (2001)
A.J. Schoeffler, J.M. Berger, Recent advances in understanding structure-function relationships in the type II topoisomerase mechanism. Biochem. Soc. Trans. 33, 1465–1470 (2005)
C. Levine, H. Hiasa, K.J. Marians, DNA gyrase and topoisomerase IV: biochemical activities, physiological roles during chromosome replication, and drug sensitivities. Biochim. Biophys. Acta 1400, 29–43 (1998)
Y. Pommier, Topoisomerase I inhibitors: camptothecins and beyond. Nat. Rev. Cancer 6, 789–802 (2006)
K.D. Corbett, J.M. Berger, Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu. Rev. Biophys. Biomol. Struct. 33, 95–118 (2004)
P. Forterre, S. Gribaldo, D. Gadelle, M.C. Serre, Origin and evolution of DNA topoisomerases. Biochimie 89, 427–446 (2007)
S.M. Vos, E.M. Tretter, B.H. Schmidt, J.M. Berger, All tangled up: how cells direct, manage and exploit topoisomerase function. Nat. Rev. Mol. Cell Biol. 12, 827–841 (2011)
T. Viard, C.B. de la Tour, Type IA topoisomerases: a simple puzzle? Biochimie 89, 456–467 (2007)
N.M. Baker, R. Rajan, A. Mondragon, Structural studies of type I topoisomerases. Nucleic Acids Res. 37, 693–701 (2009)
Y.C. Tse-Dinh, Bacterial and archeal type I topoisomerases. Biochim. Biophys. Acta 1400, 19–27 (1998)
G. Stoll, O.P. Pietilainen, B. Linder, J. Suvisaari, C. Brosi, W. Hennah, V. Leppa, M. Torniainen, S. Ripatti, S. Ala-Mello et al., Deletion of TOP3β, a component of FMRP-containing mRNPs, contributes to neurodevelopmental disorders. Nat. Neurosci. 16, 1228–1237 (2013)
J.L. Nitiss, Investigating the biological functions of DNA topoisomerases in eukaryotic cells. Biochim. Biophys. Acta 1400, 63–81 (1998)
M.P. Lee, S.D. Brown, A. Chen, T.-S. Hsieh, DNA topoisomerase I is essential in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 90, 6656–6660 (1993)
S.G. Morham, K.D. Kluckman, N. Voulomanos, O. Smithies, Targeted disruption of the mouse topoisomerase I gene by camptothecin selection. Mol. Cell. Biol. 16, 6804–6809 (1996)
C.R. Lopez, S. Yang, R.W. Deibler, S.A. Ray, J.M. Pennington, R.J. Digate, P.J. Hastings, S.M. Rosenberg, E.L. Zechiedrich, A role for topoisomerase III in a recombination pathway alternative to RuvABC. Mol. Microbiol. 58, 80–101 (2005)
J.E. Deweese, N. Osheroff, The DNA cleavage reaction of topoisomerase II: wolf in sheep’s clothing. Nucleic Acids Res. 37, 738–749 (2009)
J.L. Nitiss, DNA topoisomerase II and its growing repertoire of biological functions. Nat. Rev. Cancer 9, 327–337 (2009)
N.G. Bush, K. Evans-Roberts, A. Maxwell, DNA topoisomerases. EcoSal Plus, 6 (2015)
J.M. Berger, Structure of DNA topoisomerases. Biochim. Biophys. Acta 1400, 3–18 (1998)
J.C. Wang, Moving one DNA double helix through another by a type II DNA topoisomerase: the story of a simple molecular machine. Q. Rev. Biophys. 31, 107–144 (1998)
R. Velez-Cruz, N. Osheroff, DNA topoisomerases: type II, in Encyclopedia of Biological Chemistry, eds. by W.J. Lennarz, M.D. Lane (Elsevier, 2004), pp. 806–811
F.H. Drake, J.P. Zimmerman, F.L. McCabe, H.F. Bartus, S.R. Per, D.M. Sullivan, W.E. Ross, M.R. Mattern, R.K. Johnson, S.T. Crooke, Purification of topoisomerase II from amsacrine-resistant P388 leukemia cells. Evidence for two forms of the enzyme. J. Biol. Chem. 262, 16739–16747 (1987)
F.H. Drake, G.A. Hofmann, H.F. Bartus, M.R. Mattern, S.T. Crooke, C.K. Mirabelli, Biochemical and pharmacological properties of p170 and p180 forms of topoisomerase II. Biochemistry 28, 8154–8160 (1989)
M. Tsai-Pflugfelder, L.F. Liu, A.A. Liu, K.M. Tewey, J. Whang-Peng, T. Knutsen, K. Huebner, C.M. Croce, J.C. Wang, Cloning and sequencing of cDNA encoding human DNA topoisomerase II and localization of the gene to chromosome region 17q21-22. Proc. Natl. Acad. Sci. USA. 85, 7177–7181 (1988)
J.R. Jenkins, P. Ayton, T. Jones, S.L. Davies, D.L. Simmons, A.L. Harris, D. Sheer, I.D. Hickson, Isolation of cDNA clones encoding the beta isozyme of human DNA topoisomerase II and localisation of the gene to chromosome 3p24. Nucleic Acids Res. 20, 5587–5592 (1992)
C.A. Austin, K.L. Marsh, Eukaryotic DNA topoisomerase IIβ. BioEssays 20, 215–226 (1998)
K.B. Tan, T.E. Dorman, K.M. Falls, T.D. Chung, C.K. Mirabelli, S.T. Crooke, J. Mao, Topoisomerase IIα and topoisomerase IIβ genes: characterization and mapping to human chromosomes 17 and 3, respectively. Cancer Res. 52, 231–234 (1992)
A.M. Wilstermann, N. Osheroff, Stabilization of eukaryotic topoisomerase II-DNA cleavage complexes. Curr. Top. Med. Chem. 3, 1349–1364 (2003)
M. Pendleton, R.H. Lindsey Jr., C.A. Felix, D. Grimwade, N. Osheroff, Topoisomerase II and leukemia. Ann. NY Acad. Sci. 1310, 98–110 (2014)
A.C. Gentry, N. Osheroff, DNA topoisomerases: type II, in Encyclopedia of Biological Chemistry, 2nd edn., eds. by W.J. Lennarz, M.D. Lane (Academic Press, Waltham, 2013), pp. 163–168
M.M. Heck, W.N. Hittelman, W.C. Earnshaw, Differential expression of DNA topoisomerases I and II during the eukaryotic cell cycle. Proc. Natl. Acad. Sci. USA 85, 1086–1090 (1988)
R.D. Woessner, M.R. Mattern, C.K. Mirabelli, R.K. Johnson, F.H. Drake, Proliferation- and cell cycle-dependent differences in expression of the 170 kilodalton and 180 kilodalton forms of topoisomerase II in NIH-3T3 cells. Cell Growth Differ. 2, 209–214 (1991)
K. Kimura, M. Saijo, M. Ui, T. Enomoto, Growth state- and cell cycle-dependent fluctuation in the expression of two forms of DNA topoisomerase II and possible specific modification of the higher molecular weight form in the M phase. J. Biol. Chem. 269, 1173–1176 (1994)
P. Grue, A. Grasser, M. Sehested, P.B. Jensen, A. Uhse, T. Straub, W. Ness, F. Boege, Essential mitotic functions of DNA topoisomerase IIα are not adopted by topoisomerase IIβ in human H69 cells. J. Biol. Chem. 273, 33660–33666 (1998)
M.O. Christensen, M.K. Larsen, H.U. Barthelmes, R. Hock, C.L. Andersen, E. Kjeldsen, B.R. Knudsen, O. Westergaard, F. Boege, C. Mielke, Dynamics of human DNA topoisomerases IIα and IIβ in living cells. J. Cell Biol. 157, 31–44 (2002)
B.G. Ju, V.V. Lunyak, V. Perissi, I. Garcia-Bassets, D.W. Rose, C.K. Glass, M.G. Rosenfeld, A topoisomerase IIβ-mediated dsDNA break required for regulated transcription. Science 312, 1798–1802 (2006)
I.G. Cowell, Z. Sondka, K. Smith, K.C. Lee, C.M. Manville, M. Sidorczuk-Lesthuruge, H.A. Rance, K. Padget, G.H. Jackson, N. Adachi et al., Model for MLL translocations in therapy-related leukemia involving topoisomerase IIβ-mediated DNA strand breaks and gene proximity. Proc. Natl. Acad. Sci. USA 109, 8989–8994 (2012)
C. Sissi, M. Palumbo, In front of and behind the replication fork: bacterial type IIA topoisomerases. Cell. Mol. Life Sci. 67, 2001–2024 (2010)
V.E. Anderson, N. Osheroff, Type II topoisomerases as targets for quinolone antibacterials: turning Dr. Jekyll into Mr. Hyde. Curr. Pharm. Des. 7, 337–353 (2001)
J.W. Alexander, G.B. Briggs, On types of knotted curves. Ann. Math. 28, 562–586 (1926)
K. Reidemeister, Elementare begründung der knotentheorie. Abh. Math. Sem. Univ. Hamburg 5, 24–32 (1927)
A. Morrison, N.R. Cozzarelli, Contacts between DNA gyrase and its binding site on DNA: features of symmetry and asymmetry revealed by protection from nucleases. Proc. Natl. Acad. Sci. USA 78, 1416–1420 (1981)
D.A. Koster, A. Crut, S. Shuman, M.A. Bjornsti, N.H. Dekker, Cellular strategies for regulating DNA supercoiling: a single-molecule perspective. Cell 142, 519–530 (2010)
J. Kato, Y. Nishimura, R. Imamura, H. Niki, S. Hiraga, H. Suzuki, New topoisomerase essential for chromosome segregation in E. coli. Cell 63, 393–404 (1990)
H. Hiasa, K.J. Marians, Topoisomerase IV can support oriC DNA replication in vitro. J. Biol. Chem. 269, 16371–16375 (1994)
E.L. Zechiedrich, A.B. Khodursky, S. Bachellier, R. Schneider, D. Chen, D.M. Lilley, N.R. Cozzarelli, Roles of topoisomerases in maintaining steady-state DNA supercoiling in Escherichia coli. J. Biol. Chem. 275, 8103–8113 (2000)
N.J. Crisona, T.R. Strick, D. Bensimon, V. Croquette, N.R. Cozzarelli, Preferential relaxation of positively supercoiled DNA by E. coli topoisomerase IV in single-molecule and ensemble measurements. Genes Dev. 14, 2881–2892 (2000)
X. Wang, R. Reyes-Lamothe, D.J. Sherratt, Modulation of Escherichia coli sister chromosome cohesion by topoisomerase IV. Genes Dev. 22, 2426–2433 (2008)
M.C. Joshi, D. Magnan, T.P. Montminy, M. Lies, N. Stepankiw, D. Bates, Regulation of sister chromosome cohesion by the replication fork tracking protein SeqA. PLoS Genet. 9, e1003673 (2013)
P. Zawadzki, M. Stracy, K. Ginda, K. Zawadzka, C. Lesterlin, A.N. Kapanidis, D.J. Sherratt, The localization and action of topoisomerase IV in Escherichia coli chromosome segregation is coordinated by the SMC complex. MukBEF. Cell Rep. 13, 2587–2596 (2015)
N.P. Higgins, N.R. Cozzarelli, The binding of gyrase to DNA: analysis by retention by nitrocellulose filters. Nucleic Acids Res. 10, 6833–6847 (1982)
K.R. Madden, L. Stewart, J.J. Champoux, Preferential binding of human topoisomerase I to superhelical DNA. EMBO J. 14, 5399–5409 (1995)
N. Osheroff, Eukaryotic topoisomerase II. Characterization of enzyme turnover. J. Biol. Chem. 261, 9944–9950 (1986)
E.L. Zechiedrich, N. Osheroff, Eukaryotic topoisomerases recognize nucleic acid topology by preferentially interacting with DNA crossovers. EMBO J. 9, 4555–4562 (1990)
J. Roca, J.M. Berger, J.C. Wang, On the simultaneous binding of eukaryotic DNA topoisomerase II to a pair of double-stranded DNA helices. J. Biol. Chem. 268, 14250–14255 (1993)
A. Patel, L. Yakovleva, S. Shuman, A. Mondragon, Crystal structure of a bacterial topoisomerase IB in complex with DNA reveals a secondary DNA binding site. Structure 18, 725–733 (2010)
K. Kirkegaard, J.C. Wang, Bacterial DNA topoisomerase I can relax positively supercoiled DNA containing a single-stranded loop. J. Mol. Biol. 185, 625–637 (1985)
J.J. Champoux, R. Dulbecco, An activity from mammalian cells that untwists superhelical DNA–a possible swivel for DNA replication (polyoma-ethidium bromide-mouse-embryo cells-dye binding assay). Proc. Natl. Acad. Sci. USA 69, 143–146 (1972)
R.F. Frohlich, C. Veigaard, F.F. Andersen, A.K. McClendon, A.C. Gentry, A.H. Andersen, N. Osheroff, T. Stevnsner, B.R. Knudsen, Tryptophane-205 of human topoisomerase I is essential for camptothecin inhibition of negative but not positive supercoil removal. Nucleic Acids Res. 35, 6170–6180 (2007)
L. Sari, I. Andricioaei, Rotation of DNA around intact strand in human topoisomerase I implies distinct mechanisms for positive and negative supercoil relaxation. Nucleic Acids Res. 33, 6621–6634 (2005)
G. Charvin, D. Bensimon, V. Croquette, Single-molecule study of DNA unlinking by eukaryotic and prokaryotic type-II topoisomerases. Proc. Natl. Acad. Sci. USA 100, 9820–9825 (2003)
K.C. Neuman, G. Charvin, D. Bensimon, V. Croquette, Mechanisms of chiral discrimination by topoisomerase IV. Proc. Natl. Acad. Sci. USA 106, 6986–6991 (2009)
R.E. Ashley, A. Dittmore, S.A. McPherson, C.L. Turnbough Jr., K.C. Neuman, N. Osheroff, Activities of gyrase and topoisomerase IV on positively supercoiled DNA. Nucleic Acids Res. 45, 9611–9624 (2017)
A.K. McClendon, A.C. Rodriguez, N. Osheroff, Human topoisomerase IIα rapidly relaxes positively supercoiled DNA: implications for enzyme action ahead of replication forks. J. Biol. Chem. 280, 39337–39345 (2005)
Y. Seol, A.C. Gentry, N. Osheroff, K.C. Neuman, Chiral discrimination and writhe-dependent relaxation mechanism of human topoisomerase IIα. J. Biol. Chem. 288, 13695–13703 (2013)
K.D. Corbett, A.J. Schoeffler, N.D. Thomsen, J.M. Berger, The structural basis for substrate specificity in DNA topoisomerase IV. J. Mol. Biol. 351, 545–561 (2005)
A.K. McClendon, A.C. Gentry, J.S. Dickey, M. Brinch, S. Bendsen, A.H. Andersen, N. Osheroff, Bimodal recognition of DNA geometry by human topoisomerase IIα: preferential relaxation of positively supercoiled DNA requires elements in the C-terminal domain. Biochemistry 47, 13169–13178 (2008)
A.K. McClendon, J.S. Dickey, N. Osheroff, Ability of viral topoisomerase II to discern the handedness of supercoiled DNA: bimodal recognition of DNA geometry by type II enzymes. Biochemistry 45, 11674–11680 (2006)
T.R. Strick, V. Croquette, D. Bensimon, Single-molecule analysis of DNA uncoiling by a type II topoisomerase. Nature 404, 901–904 (2000)
R.E. Ashley, T.R. Blower, J.M. Berger, N. Osheroff, Recognition of DNA supercoil geometry by Mycobacterium tuberculosis gyrase. Biochemistry 56, 5440–5448 (2017)
J.L. Nitiss, Targeting DNA topoisomerase II in cancer chemotherapy. Nat. Rev. Cancer 9, 338–350 (2009)
Y. Pommier, DNA topoisomerase I inhibitors: chemistry, biology, and interfacial inhibition. Chem. Rev. 109, 2894–2902 (2009)
Y. Pommier, E. Leo, H. Zhang, C. Marchand, DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 17, 421–433 (2010)
Y. Pommier, C. Marchand, Interfacial inhibitors: targeting macromolecular complexes. Nat. Rev. Drug Discov. 11, 25–36 (2012)
K.J. Aldred, R.J. Kerns, N. Osheroff, Mechanism of quinolone action and resistance. Biochemistry 53, 1565–1574 (2014)
D.C. Hooper, G.A. Jacoby, Mechanisms of drug resistance: quinolone resistance. Ann. N. Y. Acad. Sci. 1354, 12–31 (2015)
A.K. McClendon, N. Osheroff, The geometry of DNA supercoils modulates topoisomerase-mediated DNA cleavage and enzyme response to anticancer drugs. Biochemistry 45, 3040–3050 (2006)
A.C. Gentry, S. Juul, C. Veigaard, B.R. Knudsen, N. Osheroff, The geometry of DNA supercoils modulates the DNA cleavage activity of human topoisomerase I. Nucleic Acids Res. 39, 1014–1022 (2011)
R.H. Lindsey Jr., M. Pendleton, R.E. Ashley, S.L. Mercer, J.E. Deweese, N. Osheroff, Catalytic core of human topoisomerase IIα: insights into enzyme-DNA interactions and drug mechanism. Biochemistry 53, 6595–65602 (2014)
E.G. Gibson, T.R. Blower, M. Cacho, B. Bax, J.M. Berger, N. Osheroff, Mechanism of action of Mycobacterium tuberculosis gyrase inhibitors: a novel class of gyrase poisons. ACS Infect. Dis. 4, 1211–1222 (2018)
Acknowledgements
Work in the senior author’s laboratory was supported by National Institutes of Health grants R01 GM033944 and R01 GM126363 and US Veterans Administration Merit Review award I01 Bx002198. R.E.A. was supported by pre-doctoral fellowship DGE-0909667 from the National Science Foundation. We are grateful to Elizabeth G. Gibson for critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Ashley, R.E., Osheroff, N. (2019). Regulation of DNA Topology by Topoisomerases: Mathematics at the Molecular Level. In: Adams, C., et al. Knots, Low-Dimensional Topology and Applications. KNOTS16 2016. Springer Proceedings in Mathematics & Statistics, vol 284. Springer, Cham. https://doi.org/10.1007/978-3-030-16031-9_20
Download citation
DOI: https://doi.org/10.1007/978-3-030-16031-9_20
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-16030-2
Online ISBN: 978-3-030-16031-9
eBook Packages: Mathematics and StatisticsMathematics and Statistics (R0)