Abstract
Topological properties of DNA influence its structure and biochemical interactions. Within the cell, DNA topology is constantly in flux. Transcription and other essential processes, including DNA replication and repair, not only alter the topology of the genome but also introduce additional complications associated with DNA knotting and catenation. These topological perturbations are counteracted by the action of topoisomerases, a specialized class of highly conserved and essential enzymes that actively regulate the topological state of the genome. This dynamic interplay among DNA topology, DNA processing enzymes, and DNA topoisomerases is a pervasive factor that influences DNA metabolism in vivo. Building on the extensive structural and biochemical characterization over the past four decades that has established the fundamental mechanistic basis of topoisomerase activity, scientists have begun to explore the unique roles played by DNA topology in modulating and influencing the activity of topoisomerases. In this review we survey established and emerging DNA topology-dependent protein–DNA interactions with a focus on in vitro measurements of the dynamic interplay between DNA topology and topoisomerase activity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The topological state of DNA and its impact on cellular activities has been recognized since the double helical structure of DNA was first proposed (Watson and Crick 1953). The anti-parallel configuration of two intertwined complementary strands was recognized to provide genomic stability and protection, but it immediately raised concerns regarding the topological complications associated with replication. Genomic DNA that is organized into topologically closed domains, or closed circular DNA such as plasmids (Dixon et al. 2012; Espéli and Boccard 2006), is physically constrained, limiting rotational motion. Therefore, opening of the two DNA strands associated with many DNA metabolic processes results in downstream overwinding, and upstream underwinding, of the DNA. The discovery of DNA topoisomerases, almost 20 years after the discovery of the double helical structure of DNA, provided a resolution to the topological complications inherent in the double helix (Champoux and Dulbecco 1972; Gellert et al. 1976; Wang 1971). By transiently breaking and resealing the DNA backbone via a transesterification reaction (Champoux 2001; Chen et al. 2013; Forterre et al. 2007; Schoeffler and Berger 2008), topoisomerases can modulate the level of supercoiling and maintain the topological homeostasis of genomic DNA (Nitiss 2009a; Vos et al. 2011; Wang 2002). Given the fundamental roles of topoisomerases in nucleic acid metabolism, and their importance as potent targets for anti-bacterial and anti-cancer agents, they have been, and continue to be, extensively studied (Deweese and Osheroff 2009; Kathiravan et al. 2013; Nitiss 2009b; Pommier 2013).
The effects of DNA topology on DNA processing enzymes
Is DNA topology just a nuisance and a side-effect of the DNA duplex conformation? This antagonistic view is changing as an increasing number of studies have revealed that the dynamic interplay between DNA topology and proteins serves regulatory roles in DNA metabolism within the cell (Baranello et al. 2012; Ma and Wang 2014; Schvartzman et al. 2013; Teves and Henikoff 2014; Travers and Muskhelishvili 2005). Supercoiling of DNA generates conformations with physical properties differing from those of linear B-form DNA. The altered topology impacts the geometry and dynamics of DNA bending, as well as the probabilities of DNA melting and distal site juxtaposition (Vologodskii 2009; Vologodskii and Cozzarelli 1993, 1994). These physical changes in turn affect the interaction of DNA-modifying proteins with their DNA substrates (Lomholt et al. 2009; Vologodskii and Cozzarelli 1994). The impact of DNA supercoiling on enzymatic activity is particularly clear for RNA transcription. Recently, several in vivo and in vitro studies have revealed that DNA supercoiling is extensively involved in modulating transcription and gene expression (Baranello et al. 2012; Chong et al. 2014; Ma et al. 2013; Teves and Henikoff 2014; Travers and Muskhelishvili 2005). In order to establish a comprehensive picture of how DNA topology influences DNA metabolism within the cell, it is crucial to understand how DNA topology specifically affects the myriad of proteins that bind and modify DNA.
In this review, we survey recent in vitro studies of the interplay between DNA topology and topoisomerases. Although topoisomerases are the primary focus of the review, we also discuss RNA transcription as a canonical example to illustrate the interplay between DNA topology and enzyme activity. Many of these results have been made possible by the advent of single-molecule methods to control and measure DNA topology, primarily magnetic tweezers (Charvin et al. 2005a; Ma and Wang 2014; Revyakin et al. 2004), complimented by the development of extensive theoretical and computational approaches (Marko 2007; Neukirch and Marko 2011; Ouldridge et al. 2011; Vologodskii and Frank-Kamenetskii 1992; Vologodskii et al. 1992; Vologodskii and Marko 1997; Zhang and Marko 2008). Single-molecule approaches complement ensemble approaches, but the former uniquely provide a means of distinguishing the effects of twist and writhe that are typically convolved in ensemble measurements. (Charvin et al. 2003, 2004, 2005a, b; Neuman et al. 2009; Seol et al. 2013a; Stone et al. 2003). We can therefore distinguish twist (torsion) and writhe of DNA topology as the two main “effectors” on DNA processing enzymes. After a brief overview of DNA topology, we review recent studies of DNA topology-dependent protein activities that are modulated by twist or writhe.
Several techniques have been employed to study the effects of DNA supercoiling on protein binding and activity. The most common ensemble techniques used to investigate topology-modifying enzymes such as DNA topoisomerases are based on gel electrophoresis approaches that can separate DNA topoisomers at the resolution of a single linking number, in addition to resolving different knotted DNA species (Keller 1975). Topology-dependent protein binding can be measured with several approaches, including the electrophoretic mobility shift assay (Osheroff 1986; Palecek et al. 2001), atomic force microscopy (Alonso-Sarduy et al. 2011; López et al. 2012; Vanderlinden et al. 2014), and an equilibrium topology-dependent binding assay (Litwin et al. 2015). Single-molecule techniques that can control as well as measure the topology of individual DNA molecules extend our capacity to directly observe how DNA topology modulates the activities of proteins and the dynamic nature of the interplay between protein activity and topology (reviewed in Charvin et al. 2004, 2005a; Forth et al. 2013; Koster et al. 2010; Lipfert et al. 2015; Ma and Wang 2014; Neuman 2010; Strick et al. 2000).
DNA topology
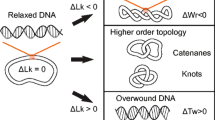
DNA topology describes the tertiary conformations of DNA that include supercoiling, knots, and catenanes (Fig. 1). Most DNA metabolic processes perturb DNA topology such as the simultaneous positive supercoiling due to overwinding ahead of RNA polymerase and negative supercoiling due to underwinding of DNA behind RNA polymerase generated during transcription (Liu and Wang 1987). This example illustrates an important but subtle point when discussing DNA topology. The positive supercoiling ahead of RNA polymerase is precisely balanced by the negative supercoiling behind the polymerase; hence the global topology of the DNA remains constant. In general, enzymatic processes that do not cut the DNA backbone do not alter the global topology of DNA; nonetheless, changes in local topology affect enzymatic processes. For example, replication generates local positive supercoiling ahead of the DNA polymerase due to the separation of the duplex DNA by DNA replication helicases (Lohman and Bjornson 1996) and the formation of links, or pre-catenanes, between the two daughter strands behind the polymerase due to the backward diffusion of accumulating positive supercoils via the rotation of the replication fork (Peter et al. 1998). If the local positive supercoiling and pre-catenanes are not resolved, replication or cell division will fail. This example illustrates the importance of local topological changes in addition to global topology in DNA–enzyme interactions. Random cyclization of DNA, enhanced by entangled DNA conditions in tight spaces, such as bacteriophage DNA in capsid, or site-specific recombination processes, can also lead to the formation of knots in the DNA (reviewed in Darcy and Vazquez 2013).
Graphical overview of DNA topology. Linking number differences (∆Lk) arise due to cellular processes, such as DNA transcription and replication. Differences in the linking number are accommodated by a combination of twist (torsion) (Tw) and writhe (Wr), which change the structure and mechanics of DNA. Knotting within a DNA molecule and links between DNA molecules (catenanes) represent higher order topological conformations of DNA
The topological conformations of a closed circular or rotationally constrained domain of DNA can be described in terms of the linking number (Lk) a topological invariant that is defined as the integer number of links between the two complementary single strands of DNA (Bates and Maxwell 2005; Vologodskii et al. 1992). This topological quantity is physically manifested in two distinct geometric properties of DNA: twist (Tw) and writhe (Wr) (Boles et al. 1990) (Figs. 1, 2). Twist is defined as the number of times the Watson and Crick strands twist around each other along the helical axis of the DNA, whereas writhe is a measure of winding and crossing of the double-helical axis in space. A remarkable mathematical result states that the sum of twist and writhe describing a given DNA molecule is equal to the linking number (Fuller 1978; White 1969). Consequently, changes in the linking number for a given closed-circular DNA, or topologically constrained segment of DNA, are partitioned between changes in twist and writhe, i.e.,
(Fuller 1978). For relaxed B-form DNA, the two strands twist around each other once every ∼10.5 bp; therefore, the relaxed linking number (Lk0) is equal to the relaxed twist (Tw0) ∼ n/10.5 for n base pairs as the writhe is identical to 0 for the relaxed DNA. Supercoiling of DNA refers to changes in the linking number from the relaxed linking number. Negative supercoiling, corresponding to a reduction in linking number (ΔLk <0), is accommodated by a reduction in twist and the formation of negative (right-handed) writhe, primarily through the generation of plectonemes in which the helical axis crosses itself, forming an interwound superhelix. The other possible supercoiled conformation in which the helical axis follows a toroidal superhelical path similar to a coiled telephone cord is not observed in free DNA (Bates and Maxwell 2005). Positive supercoiling, corresponding to an increase in linking number (ΔLk >0), is accommodated by an increase in twist and positive (left-handed) writhe. The fractional change in linking number (also termed specific linking difference or superhelical density: σ) is often used to describe the DNA topological state of a plasmid compared to relaxed DNA:
The mechanical and geometrical effects of DNA twist (Tw) and writhe (Wr). Left half of graph Underwound DNA (ΔTw <0) can result in local DNA melting, facilitating single-strand DNA (ssDNA) binding by proteins such as transcription factor IID (TFIID) and TATA box binding protein (TATA), and wrapping of DNA by nucleosomes. On the other hand, overwound DNA (ΔTw >0) increases DNA torsion, thereby hindering enzymatic activities associated with opening of the DNA duplex, including initiation and elongation by RNA polymerase. Increased torsion also hinders and can reverse DNA wrapping interactions of nucleosomes. Right half of graph The formation of writhe facilitates the interactions among distal sites of DNA and promotes local DNA bending. In addition, geometric differences between positive and negative Wr or DNA juxtapositions within knotted or catenated DNA are subject to chiral- and geometric-dependent activities of some Type II topoisomerases. For example, topoisomerase IV has been shown to be sensitive to the DNA crossing angle α depicted in the figure
The topological state of prokaryotic genomic DNA is strictly regulated to maintain negative supercoiling (Worcel and Burgi 1972). In eukaryotes, the supercoiling density is also effectively negative, but it is largely constrained within nucleosomes (Bates and Maxwell 2005; Travers and Muskhelishvili 2007). In Escherichia coli, σ in general ranges from −0.03 to −0.06 depending on the growth phase (Bliska and Cozzarelli 1987; Hatfield and Benham 2002). Roughly 40 % of the superhelical density in prokaryotes is unconstrained and is largely accommodated by negative writhe through the formation of plectonemes (Bliska and Cozzarelli 1987). When free in solution, the superhelical density in plasmids larger than ~2.5 kbp is partitioned into ~30 % twist and ~70 % writhe (Vologodskii et al. 1992). These two manifestations of linking number differences influence enzyme binding and activity in different manners.
DNA twist (torsion)-dependent protein activity
Many DNA-based processes affect DNA topology primarily by changing the local DNA twist (Lavelle 2014; Teves and Henikoff 2014). Untwisting DNA (negative ΔLk, left-handed rotation) destabilizes the duplex and can generate locally denatured regions of DNA, even under low tension (~0.5 pN) (Strick et al. 1998). Negative supercoiling therefore makes single-stranded DNA (ssDNA) more accessible and can also reduce the DNA bending modulus (Fig. 2). Overwinding DNA (positive ΔLk, right-handed rotation), on the other hand, increases DNA torsion that may hinder proteins from binding, tracking (Ma et al. 2013), or wrapping (Li et al. 2015; Nam and Arya 2014) DNA (Fig. 2). The effects of DNA torsion on enzyme activity is perhaps best understood in the context of supercoiling-dependent RNA transcription (reviewed in Kouzine et al. 2014; Teves and Henikoff 2014). In general, higher transcription efficiency is observed with higher (−) DNA twist (Lim et al. 2003). First, the initial DNA opening and binding of proteins are favored by negative twist, which facilitates melting and opening of the DNA duplex. For example, transcription factor IID (TFIID), one of the proteins involved in the initiation of RNA transcription along with TATA box binding protein, is known to untwist DNA upon binding (Kahn 2000). TFIID binding is generally enhanced on negatively supercoiled DNA (Tabuchi et al. 1993), likely because the unwound DNA conformation lowers both the energy barrier of TFIID binding and the free-energy of the TFIID bound state. In contrast to negative supercoiling, positive supercoiling adversely effects promoter opening by RNA polymerase (Chong et al. 2014; Revyakin et al. 2004). The Revyakin et al. study also revealed that the life-times of the open states of certain promoters are shorter than others, even though the opening-rates at the same degree of supercoiling are comparable, indicating that the stability of the open promoter as well as the rate of opening play roles in supercoiling-dependent transcription initiation. In addition to torsional effects on transcription initiation, Ma et al. (2013) and Chong et al. (2014) independently showed that DNA torsion modulated transcription elongation at the single-molecule level. These authors showed that increasing DNA torsion not only decreased the rate of transcription, but also led to the stalling of RNA polymerase and the inhibition of transcription. Chong et al. (2014) further revealed that the addition of gyrase allows transcription to restart, reflecting the dynamic interplay between transcription DNA torsion and topoisomerase activity in vivo. These two studies underscore the significance of DNA topoisomerases in regulating not only the level of supercoiling but also in facilitating vital cellular processes that can be inhibited by excessive levels of supercoiling or other topological barriers (Wang 2002).
DNA torsion (Tw) has different effects on the catalytic activity of DNA topoisomerases depending on the catalytic mechanism of the specific topoisomerase (Fig. 3). Type IB topoisomerases (Topo IB) generate a transient DNA nick in one strand of duplex DNA by forming a 3′-phosphotyrosine bond which allows a controlled rotation of the free 5′-end around the intact strand inside the protein until the enzyme reseals the nick (Champoux 2001; Pommier 2013). Topo V (topoisomerase Type IC) shares a similar catalytic mechanism with Topo IB despite the lack of structural or sequence similarity (Schoeffler and Berger 2008). DNA torsion lowers the rotational energy barrier (Wereszczynski and Andricioaei 2010) and thus enhances the rate of rotation, i.e., the relaxation rate for both types of topoisomerases (Koster et al. 2005; Seol et al. 2012; Taneja et al. 2007). DNA twist also affects the efficacy of human Topo IB inhibitors, some of which are Federal Drug Administration-approved chemotherapeutic agents that act by preventing religation of the cleaved DNA and trapping the cleavage complex comprised of the topoisomerase, DNA, and inhibitor (Pommier and Cushman 2009). Topo IB inhibitor-mediated DNA cleavage complex formation is enhanced by supercoiling of the DNA substrate (Seol et al. 2015), although the relative enhancement is modulated by the chirality of supercoiling, the specific type of inhibitor, and the inherent chiral-dependence of cleavage (Gentry et al. 2011; McClendon and Osheroff 2006; Seol et al. 2015). This enhancement may arise from the higher affinity of Topo IB for supercoiled DNA (Madden et al. 1995), possibly due to preferential binding at DNA crossovers (Patel et al. 2010; Zechiedrich and Osheroff 1990); however, recent measurements performed at low Topo IB concentrations suggest there is little difference in affinity between relaxed and supercoiled DNA (Litwin et al. 2015). Alternatively, enhanced inhibitor-mediated cleavage with supercoiled DNA may reflect the torque-dependent increase in the occupancy of a cleaved, but not rotating, DNA–protein conformation that promotes inhibitor binding (Seol et al. 2012). Supercoiling-mediated effects have also been demonstrated for the inhibition of human Topo IIα and Topo IIβ, the two isoforms of human Topo IIA, by inhibitors (e.g., etoposide) that act to prevent religation and stabilize enzyme-mediated cleavage complexes in a similar manner as the Topo IB inhibitors (McClendon and Osheroff 2006). Generally speaking, the degree of intrinsic cleavage complex formation is higher for negatively than for positively supercoiled DNA for both human Topo II isoforms. This chiral-dependent difference persists for the formation of the inhibitor-stabilized cleavage complex, resulting in comparable relative changes in the formation of the cleavage complex with increasing concentration of inhibitor for both chiralities of supercoiling (McClendon and Osheroff 2006).
Overview of DNA topoisomerase (Topo) activities and the effects of DNA twist (Tw) and writhe (Wr) on Topo activity. Top 2 rows During a catalytic cycle, Topo IA creates a break in the ssDNA (inset: green circle indicates the enzyme–DNA bond) and passes the intact ssDNA through the gap, resulting in a change of Lk of +1. As the ssDNA region is required for Topo IA binding, underwound DNA facilitates enzymatic activity. Topo IA can only relax negatively supercoiled DNA, whereas reverse gyrases, a combination of a Type IA Topo and a helicase found in hyperthermophiles, can generate positive supercoils with ATP. The rate and degree of positive supercoiling by reverse gyrase are limited by the torque in the DNA. Middle row Topo IB generates a ssDNA nick (inset: green circle indicates the enzyme–DNA bond) and relaxes both positive and negative supercoils via a controlled rotation mechanism, resulting in an a random change in Lk before religation. The rate of relaxation is proportional to the stored torque, i.e., ΔTw. Although not shown, the mechanism of Topo V is similar to that of Topo IB. Bottom 2 rows Topo IIA generates a double-strand break (inset: green circle indicates the enzyme–DNA bond) and passes the other duplex through a DNA gate formed by the enzyme and DNA bridge. Positive DNA torsion decreases the catalytic activity of Topo IIA. Some Topo IIA (Topo IV, gyrase, and human Topo IIα) are affected by the chirality of DNA Wr
Topo IIA employ a strand passage mechanism in which the enzyme passes one segment of duplex DNA through a transient double-strand break generated in a second segment of DNA (Schoeffler and Berger 2008). Topo IA also employ a strand passage mechanism (Schoeffler and Berger 2008). However, these enzymes pass a single-stranded segment of DNA, or in the case of Topo III either ssDNA or double-stranded (ds)DNA, through a transient ssDNA gap (Gubaev and Klostermeier 2014; Lulchev and Klostermeier 2014; Seol et al. 2013b; Terekhova et al. 2012). The binding and activity of Topo IA therefore critically depend on a sufficient level of negative supercoiling to promote the unwinding of the duplex to generate ssDNA binding regions (Kirkegaard and Wang 1985; Lima et al. 1994). Indeed, this dependence on a critical level of negative supercoiling for Topo IA activity is exploited in prokaryotes as a mechanism to regulate the level of negative supercoiling. DNA gyrase (a Type IIA topoisomerase) introduces negative supercoils into genomic DNA, and Topo IA relax negative supercoils to the critical level at which ssDNA is no longer generated. By working in opposition these two topoisomerases establish a dynamic supercoiling equilibrium that maintains the DNA at a specific level of negative supercoiling (Drlica 1992). Reverse gyrase is a unique Type IA topoisomerase combined with a helicase that introduces positive supercoils in an ATP-dependent manner (Ogawa et al. 2015; Schoeffler and Berger 2008). The mechanism of positive supercoiling has not been completely established, but it is known that the rate and extent of positive supercoiling are limited by the positive torque on the DNA (Ogawa et al. 2015, 2016).
In contrast to Topo IB, the catalytic activities of both Topo IA and IIA decrease with increasing positive torsion, suggesting that the rate-limiting step depends on the torque on the DNA (Charvin et al. 2005a; Dekker et al. 2002; Ogawa et al. 2016; Seol et al. 2013a). The different responses of Topo IB and Topo IIA to DNA torsion likely reflects their different physiological roles: Topo IB, faring better with DNA twist, is more involved in removing excess supercoiling associated with transcription, whereas Topo IIA is uniquely able to resolve higher-level DNA structures such as writhe, knots, and catenanes (Teves and Henikoff 2014) due to its duplex strand passage ability. Type IIA topoisomerases are essential for removing links between replicated genomic DNA prior to cell division, since a single unresolved link results in either the failure of mitosis or chromosome breakage, both of which are highly deleterious to the cell (Wang 2002). Twist has also been shown to affect the strand passage rate of Topo II. Single-molecule measurements of Topo IIA reveal a decrease in activity associated with highly supercoiled or highly linked DNA molecules under significant mechanical load (Charvin et al. 2005a; Seol et al. 2013a). These results raise the question of how Topo IIA performs unlinking and supercoil relaxation of highly supercoiled and stressed DNA, particularly when efficient DNA decatenation is critical, such as during mitosis. One potential solution to these problems is the activity of accessory proteins that may aid Type II topoisomerases in processing these stressed topological states. For example, RECQL5, a member of the RecQ helicase family in humans, promotes the decatenation activity of Topo IIα, and its depletion in cells results in a similar phenotype to that observed with the inhibition of Topo IIα catalytic activity (Ramamoorthy et al. 2012). A similar role for Escherichia coli RecQ helicase working in conjunction with Topo III and ssDNA binding protein has been shown to act as a decatenase (Harmon et al. 2003) and to resolve linked replication intermediates arising at the end of convergent replication in bacteria (Suski and Marians 2008). The mechanistic details of these interactions have yet to be established, but DNA helicases frequently confront DNA topology in line with or in collaboration with DNA topoisomerases as they are extensively involved in DNA metabolism. In addition to the DNA repair helicases, such as the RecQ family of helicases that functionally and physically interact with topoisomerases, the replicative helicases that precede DNA polymerase are directly responsible for and must contend with the local positive supercoiling that accompanies opening of the DNA duplex. Detailed studies of the effect of DNA topology on the activity of helicases are currently lacking; understanding the interplay between DNA helicases and DNA topology is an important and interesting direction of future research.
In addition to the specific proteins mentioned above, many other proteins that are involved in DNA metabolic processes are sensitive to DNA torsion, such as nucleosomes (Li et al. 2015; Vlijm et al. 2015), transcription factors, and regulators (reviewed in Baranello et al. 2012; Ding et al. 2014), RecA (Fulconis et al. 2004), and replication protein A (RPA; De Vlaminck et al. 2010). More generally, an argument can be made that most, if not all, protein DNA interactions are likely influenced by the topology of the DNA, but there is a limited subset of proteins for which the effect of DNA topology has been explicitly determined. Nucleosomes represent a canonical example of the dynamic interplay between DNA and proteins. They play essential roles in regulating the cellular level of supercoiling and the accessibility of DNA. In particular, the inherent dynamic behavior of nucleosome reorganization observed at the single-molecule level (Bancaud et al. 2006) suggests that nucleosome dynamics respond to changes in supercoiling and, more specifically, to DNA torsion, which regulates binding of numerous proteins and controls cellular processes, including transcription and replication.
DNA writhe-dependent protein activity
DNA topology also alters the local and global geometry of DNA manifest through writhe. Although knots and catenanes are considered to be unfavorable for most DNA metabolic processes (Ishii et al. 1991; Portugal and Rodriguez-Campos 1996; Sundin and Varshavsky 1981), the formation of writhe, knots, and catenanes can bring distant sites close to one another to facilitate processes that require long-range genomic interactions (Liu et al. 2009). For example, writhe can promote enhancer–promoter communication (Liu et al. 2001) or regulate recombination processes (Crisona et al. 1994; Merickel and Johnson 2004). Writhe is typically accommodated by changes in local DNA geometry that can facilitate the binding of proteins due to either a local distortion, such as bending, of DNA or the higher probability of juxtaposition of two DNA segments (Fogg et al. 2012; Zechiedrich and Osheroff 2010) (Fig. 2).
The DNA local geometry has been shown to play an important role in two distinct aspects of Topo IIA enzymatic activity, namely, chiral discrimination and below-equilibrium topology simplification. Both human Topo IIα and E coli Topo IV preferentially relax positive supercoils (Charvin et al. 2003; Crisona et al. 2000; McClendon et al. 2005, 2008; Neuman et al. 2009; Seol et al. 2013a; Stone et al. 2003). The underlying mechanism for this chiral discrimination is embedded in differential interactions between positive and negative writhe and the C-terminal domains of the topoisomerases (Corbett et al. 2005; McClendon et al. 2008; Neuman et al. 2009; Seol et al. 2013a). However, the writhe-dependent chiral sensing differently affects the catalytic activities of the two enzymes as chiral discrimination by Topo IV results from chirality-dependent differences in processivity, whereas chiral discrimination by Topo IIα results from chirality-dependent differences in the relaxation rate (Neuman et al. 2009; Seol et al. 2013a) (Fig. 3).
Since the publication by Rybenkov et al. (1997) demonstrating that Type IIA topoisomerases (with the exception of DNA gyrase) reduce the level of DNA linking, knotting, and supercoiling to below thermal equilibrium vales, it has remained a puzzle how these enzymes simplify global topology. Energetically, below-equilibrium topology simplification by Topo IIA does not violate thermodynamics since these topoisomerases utilize ATP as an energy source, so they can work against the thermal equilibrium limit. Topo IA and IB, on the other hand, do not utilize external energy, so they can only achieve thermodynamic equilibrium topological distributions. It remains unclear, however, how Topo IIA employ some of the energy of ATP hydrolysis to distinguish DNA juxtapositions in catenanes and knots and writhe of supercoils, from those occurring within or between DNA molecules due to thermal fluctuations (Rybenkov et al. 1997). This implies that Topo IIA must “sense” which juxtapositions simplify global DNA topology when unlinked. Several mechanistic models of topology simplification by Topo IIA have been proposed (Vologodskii 2009). Two of the models, the G-segment DNA-bending model (Vologodskii et al. 2001) and the hooked-juxtaposition model (Buck and Zechiedrich 2004), propose that local geometry, such as bending by the topoisomerase or selection of “hooked” conformations in juxtaposed DNA segments, results in preferential activity on linked DNA segments which, coupled with the ATP-dependent unidirectional strand passage, results in global simplification of DNA topology. Structural studies and direct mechanical measurements have revealed that Topo IIA bend DNA upon binding (Dong and Berger 2007; Hardin et al. 2011; Lee et al. 2012, 2013; Thomson et al. 2014) and thus that this enzyme likely has a higher binding affinity for bent DNA, such as at the apices of plectonemes in supercoiled DNA. To date, however, the proposed models cannot fully explain the below-equilibrium simplification phenomenon, indicating that the models may only partially reflect the mechanistic basis of this intriguing effect (Hardin et al. 2011; Liu et al. 2010; Martínez-García et al. 2014; Seol et al. 2013b; Thomson et al. 2014; Yan et al. 1999). Recently, it was demonstrated that the binding affinity of Topo IV is linearly related to the linking number for negatively supercoiled plasmid DNA (Litwin et al. 2015), which is consistent with previous observations of Topo IIA binding to DNA crossovers (Zechiedrich and Osheroff 1990). Although the exact mechanism for linking number-dependent binding affinity is uncertain, it highlights how DNA supercoiling and protein activity are dynamically coupled to each other. This supercoil-dependent affinity, in conjunction with other existing models, may shed light on the non-equilibrium topology simplification conundrum. Whereas the below-equilibrium removal of knots and plectonemes may contribute to genomic stability, the temporal dynamics of the process and the exceedingly low energy conversion efficiency (Stuchinskaya et al. 2009) suggest that it may reflect some yet to be determined aspect of Type IIA mechanism that may become clear when the fundamental process of below-equilibrium topology simplification is established and subsequently abolished through mutation.
Concluding remarks
In this review, we provide a broad survey illustrating how DNA topology is both controlled by and modulates topoisomerases and other DNA-processing enzymes that together establish feedback loops promoting the dynamic equilibrium of DNA topology. This dynamic interplay between DNA topology and topoisomerases fundamentally influences the majority of, if not all, DNA–protein interactions in the cell, many of which, in turn, disrupt topological homeostasis. We focused primarily on examples of in vitro ensemble and single-molecule studies to highlight the interactions between individual proteins and DNA topology. Moving forward, we expect the complexity of these experiments will increase to include multiple proteins and protein complexes that may begin to more faithfully recapitulate cellular interactions mediated by and regulating DNA topology. Despite the continuing progress in understanding the dynamic interplay between DNA topology and topoisomerase activity in vitro, the equivalent mechanistic insights and a fundamental understanding of how topology influences and is influenced by this dynamic interplay in vivo is progressing. Recent successes in probing DNA topology in vivo will hopefully herald the development of more sensitive and time-resolved readouts of cellular DNA topology (reviewed in Kouzine et al. 2014; Teves and Henikoff 2014). As these tools improve and as the repertoire of mechanistic insights gleaned from in vitro experiments expands, we may achieve the ultimate goal of understanding the interplay of DNA topology and DNA processing in vivo. As a final note, we want to emphasize that the examples listed in this review represent only a fraction of relevant studies and we apologize for not including other outstanding studies.
References
Alonso-Sarduy L, Roduit C, Dietler G, et al (2011) Human topoisomerase II–DNA interaction study by using atomic force microscopy. FEBS Lett 585:3139–3145. doi:10.1016/j.febslet.2011.08.051
Bancaud A, Conde e Silva N, Barbi M, et al (2006) Structural plasticity of single chromatin fibers revealed by torsional manipulation. Nat Struct Mol Biol 13:444–450. http://www.nature.com/nsmb/journal/v13/n5/suppinfo/nsmb1087_S1.html
Baranello L, Levens D, Gupta A, et al (2012) The importance of being supercoiled: how DNA mechanics regulate dynamic processes. Biochim Biophys Acta 1819:632–638. doi:10.1016/j.bbagrm.2011.12.007
Bates AD, Maxwell A (2005) DNA topology, 2nd edn. Oxford University Press, New York
Bliska JB, Cozzarelli NR (1987) Use of site-specific recombination as a probe of DNA structure and metabolism in vivo. J Mol Biol 194:205–218. doi:10.1016/0022-2836(87)90369-X
Boles TC, White JH, Cozzarelli NR (1990) Structure of plectonemically supercoiled DNA. J Mol Biol 213:931–951. doi:10.1016/S0022-2836(05)80272-4
Buck GR, Zechiedrich LE (2004) DNA disentangling by type-2 topoisomerases. J Mol Biol 340:933–939. doi:10.1016/j.jmb.2004.05.034
Champoux JJ (2001) DNA topoisomerases: structure, function, and mechanism. Annu Rev Biochem 70:369–413. doi:10.1146/annurev.biochem.70.1.369
Champoux JJ, Dulbecco R (1972) An activity from mammalian cells that untwists superhelical DNA–a possible swivel for DNA replication (polyoma–ethidium bromide–mouse-embryo cells–dye binding assay). Proc Natl Acad Sci USA 69:143–146
Charvin G, Bensimon D, Croquette V (2003) Single-molecule study of DNA unlinking by eukaryotic and prokaryotic type-II topoisomerases. Proc Natl Acad Sci USA 100:9820–9825. doi:10.1073/pnas.1631550100
Charvin G, Allemand JF, Strick TR, et al (2004) Twisting DNA: single molecule studies. Contemp Phys 45:383–403. doi:10.1080/00107510410001697279
Charvin G, Strick TR, Bensimon D, et al (2005a) Tracking topoisomerase activity at the single-molecule level. Annu Rev Biophys Biomol Struct 34:201–219. doi:10.1146/annurev.biophys.34.040204.144433
Charvin G, Vologodskii A, Bensimon D, et al (2005b) Braiding DNA: experiments, simulations, and models. Biophys J 88:4124–4136. doi:10.1529/biophysj.104.056945
Chen SH, Chan N-L, Hsieh T-s (2013) New mechanistic and functional insights into DNA topoisomerases. Annu Rev Biochem 82:139–170. doi:10.1146/annurev-biochem-061809-100002
Chong S, Chen C, Ge H, et al (2014) Mechanism of transcriptional bursting in bacteria. Cell 158:314–326. doi:10.1016/j.cell.2014.05.038
Corbett KD, Schoeffler AJ, Thomsen ND, et al (2005) The structural basis for substrate specificity in DNA topoisomerase IV. J Mol Biol 351:545–561. doi:10.1016/j.jmb.2005.06.029
Crisona NJ, Kanaar R, Gonzalez TN, et al (1994) Processive recombination by wild-type gin and an enhancer-independent mutant. Insight into the mechanisms of recombination selectivity and strand exchange. J Mol Biol 243. doi:10.1006/jmbi.1994.1671
Crisona NJ, Strick TR, Bensimon D, et al (2000) Preferential relaxation of positively supercoiled DNA by E. coli topoisomerase IV in single-molecule and ensemble measurements. Genes Dev 14:2881–2892. doi:10.1101/gad.838900
Darcy IK, Vazquez M (2013) Determining the topology of stable protein–DNA complexes. Biochem Soc Trans 41:601–605. doi:10.1042/bst20130004
De Vlaminck I, Vidic I, van Loenhout MTJ, et al (2010) Torsional regulation of hRPA-induced unwinding of double-stranded DNA. Nucleic Acids Res 38:4133–4142. doi:10.1093/nar/gkq067
Dekker NH, Rybenkov VV, Duguet M, et al (2002) The mechanism of type IA topoisomerases. Proc Natl Acad Sci USA 99:12126–12131. doi:10.1073/pnas.132378799
Deweese JE, Osheroff N (2009) The DNA cleavage reaction of topoisomerase II: wolf in sheep's clothing. Nucleic Acids Res 37:738–748. doi:10.1093/nar/gkn937
Ding Y, Manzo C, Fulcrand G, et al (2014) DNA supercoiling: a regulatory signal for the λ repressor. Proc Natl Acad Sci USA 111:15402–15407. doi:10.1073/pnas.1324044111
Dixon JR, Selvaraj S, Yue F, et al (2012) Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485:376–380. http://www.nature.com/nature/journal/v485/n7398/abs/nature11082.html#supplementary-information
Dong KC, Berger JM (2007) Structural basis for gate-DNA recognition and bending by type IIA topoisomerases. Nature 450:1201–1205. http://www.nature.com/nature/journal/v450/n7173/suppinfo/nature06396_S1.html
Drlica K (1992) Control of bacterial DNA supercoiling. Mol Microbiol 6:425–433. doi:10.1111/j.1365-2958.1992.tb01486.x
Espéli O, Boccard F (2006) Organization of the Escherichia coli chromosome into macrodomains and its possible functional implications. J Struct Biol 156:304–310. doi:10.1016/j.jsb.2006.07.010
Fogg JM, Randall GL, Pettitt BM, et al (2012) Bullied no more: when and how DNA shoves proteins around. Q Rev Biophys 45:257–299. doi:10.1017/S0033583512000054
Forterre P, Gribaldo S, Gadelle D, et al (2007) Origin and evolution of DNA topoisomerases. Biochimie 89:427–446. doi:10.1016/j.biochi.2006.12.009
Forth S, Sheinin MY, Inman J, et al (2013) Torque measurement at the single-molecule level. Annu Rev Biophys 42:583–604. doi:10.1146/annurev-biophys-083012-130412
Fulconis R, Bancaud A, Allemand J-F, et al (2004) Twisting and untwisting a single DNA molecule covered by RecA protein. Biophys J 87:2552–2563. doi:10.1529/biophysj.104.043059
Fuller FB (1978) Decomposition of the linking number of a closed ribbon: a problem from molecular biology. Proc Natl Acad Sci USA 75:3557–3561
Gellert M, Mizuuchi K, O'Dea MH, et al (1976) DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc Natl Acad Sci USA 73:3872–3876
Gentry AC, Juul S, Veigaard C, et al (2011) The geometry of DNA supercoils modulates the DNA cleavage activity of human topoisomerase I. Nucleic Acids Res 39:1014–1022. doi:10.1093/nar/gkq822
Gubaev A, Klostermeier D (2014) The mechanism of negative DNA supercoiling: a cascade of DNA-induced conformational changes prepares gyrase for strand passage. DNA Repair 16:23–34. doi:10.1016/j.dnarep.2014.01.011
Hardin AH, Sarkar SK, Seol Y, et al (2011) Direct measurement of DNA bending by type IIA topoisomerases: implications for non-equilibrium topology simplification. Nucleic Acids Res 39:5729–5743. doi:10.1093/nar/gkr109
Harmon FG, Brockman JP, Kowalczykowski SC (2003) RecQ helicase stimulates both DNA catenation and changes in DNA topology by topoisomerase III. J Biol Chem 278:42668–42678. doi:10.1074/jbc.M302994200
Hatfield GW, Benham CJ (2002) DNA topology-mediated control of global gene expression in Escherichia coli. Annu Rev Genet 36:175–203. doi:10.1146/annurev.genet.36.032902.111815
Ishii S, Murakami T, Shishido K (1991) Gyrase inhibitors increase the content of knotted DNA species of plasmid pBR322 in Escherichia coli. J Bacteriol 173:5551–5553
Kahn JD (2000) Topological effects of the TATA box binding protein on minicircle DNA and a possible thermodynamic linkage to chromatin remodeling. Biochemistry 39:3520–3524. doi:10.1021/bi992263f
Kathiravan MK, Khilare MM, Nikoomanesh K, et al (2013) Topoisomerase as target for antibacterial and anticancer drug discovery. J Enzyme Inhib Med Chem 28:419–435. doi:10.3109/14756366.2012.658785
Keller W (1975) Determination of the number of superhelical turns in simian virus 40 DNA by gel electrophoresis. Proc Natl Acad Sci USA 72:4876–4880
Kirkegaard K, Wang JC (1985) Bacterial DNA topoisomerase I can relax positively supercoiled DNA containing a single-stranded loop. J Mol Biol 185:625–637. doi:10.1016/0022-2836(85)90075-0
Koster DA, Croquette V, Dekker C, et al (2005) Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase IB. Nature 434:671–674
Koster DA, Crut A, Shuman S, et al (2010) Cellular strategies for regulating DNA supercoiling: a single-molecule perspective. Cell 142:519–530. doi:10.1016/j.cell.2010.08.001
Kouzine F, Levens D, Baranello L (2014) DNA topology and transcription. Nucleus 5:195–202. doi:10.4161/nucl.28909
Lavelle C (2014) Pack, unpack, bend, twist, pull, push: the physical side of gene expression. Curr Opin Genet Dev 25:74–84. doi:10.1016/j.gde.2014.01.001
Lee S, Jung S-R, Heo K, et al (2012) DNA cleavage and opening reactions of human topoisomerase IIα are regulated via Mg2+−mediated dynamic bending of gate-DNA. Proc Natl Acad Sci USA 109:2925–2930. doi:10.1073/pnas.1115704109
Lee I, Dong KC, Berger JM (2013) The role of DNA bending in type IIA topoisomerase function. Nucleic Acids Res 41:5444–5456. doi:10.1093/nar/gkt238
Li W, Wong WJ, Lim CJ, et al (2015) Complex kinetics of DNA condensation revealed through DNA twist tracing. Phys Rev E 92:022707
Lim HM, Lewis DEA, Lee HJ, et al (2003) Effect of varying the supercoiling of DNA on transcription and its regulation. Biochemistry 42:10718–10725. doi:10.1021/bi030110t
Lima CD, Wang JC, Mondragon A (1994) Three-dimensional structure of the 67K N-terminal fragment of E. coli DNA topoisomerase I. Nature 367:138–146
Lipfert J, van Oene MM, Lee M, et al (2015) Torque spectroscopy for the study of rotary motion in biological systems. Chem Rev 115:1449–1474. doi:10.1021/cr500119k
Litwin TR, Solà M, Holt IJ, et al (2015) A robust assay to measure DNA topology-dependent protein binding affinity. Nucleic Acids Res 43, e43. doi:10.1093/nar/gku1381
Liu LF, Wang JC (1987) Supercoiling of the DNA template during transcription. Proc Natl Acad Sci USA 84:7024–7027
Liu Y, Bondarenko V, Ninfa A, et al (2001) DNA supercoiling allows enhancer action over a large distance. Proc Natl Acad Sci USA 98:14883–14888. doi:10.1073/pnas.261477898
Liu Z, Deibler RW, Chan HS, et al (2009) The why and how of DNA unlinking. Nucleic Acids Res 37:661–671. doi:10.1093/nar/gkp041
Liu Z, Zechiedrich L, Chan HS (2010) Action at hooked or twisted–hooked DNA juxtapositions rationalizes unlinking preference of type-2 topoisomerases. J Mol Biol 400:963–982. doi:10.1016/j.jmb.2010.05.007
Lohman TM, Bjornson KP (1996) Mechanisms of helicase-catalyzed DNA unwinding. Annu Rev Biochem 65:169–214. doi:10.1146/annurev.bi.65.070196.001125
Lomholt MA, van den Broek B, Kalisch S-MJ, et al (2009) Facilitated diffusion with DNA coiling. Proc Natl Acad Sci USA 106:8204–8208. doi:10.1073/pnas.0903293106
López V, Martínez-Robles M-L, Hernández P, et al (2012) Topo IV is the topoisomerase that knots and unknots sister duplexes during DNA replication. Nucleic Acids Res 40:3563–3573. doi:10.1093/nar/gkr1237
Lulchev P, Klostermeier D (2014) Reverse gyrase—recent advances and current mechanistic understanding of positive DNA supercoiling. Nucleic Acids Res 42:8200–8213. doi:10.1093/nar/gku589
Ma J, Wang M (2014) Interplay between DNA supercoiling and transcription elongation. Transcription 5: e28636. doi:10.4161/trns.28636
Ma J, Bai L, Wang MD (2013) Transcription under torsion. Science 340:1580–1583. doi:10.1126/science.1235441
Madden KR, Stewart L, Champoux JJ (1995) Preferential binding of human topoisomerase I to superhelical DNA. EMBO J 14:5399–5409
Marko JF (2007) Torque and dynamics of linking number relaxation in stretched supercoiled DNA. Phys Rev E 76:021926
Martínez-García B, Fernández X, Díaz-Ingelmo O, et al (2014) Topoisomerase II minimizes DNA entanglements by proofreading DNA topology after DNA strand passage. Nucleic Acids Res 42:1821–1830. doi:10.1093/nar/gkt1037
McClendon AK, Osheroff N (2006) The geometry of DNA supercoils modulates topoisomerase-mediated DNA cleavage and enzyme response to anticancer drugs. Biochemistry 45:3040–3050. doi:10.1021/bi051987q
McClendon AK, Rodriguez AC, Osheroff N (2005) Human topoisomerase IIα rapidly relaxes positively supercoiled DNA: implications for enzyme action ahead of replication forks. J Biol Chem 280:39337–39345. doi:10.1074/jbc.M503320200
McClendon AK, Gentry AC, Dickey JS, et al (2008) Bimodal recognition of DNA geometry by human topoisomerase IIα: preferential relaxation of positively supercoiled DNA requires elements in the C-terminal domain. Biochemistry 47:13169–13178. doi:10.1021/bi800453h
Merickel SK, Johnson RC (2004) Topological analysis of Hin-catalysed DNA recombination in vivo and in vitro. Mol Microbiol 51:1143–1154. doi:10.1046/j.1365-2958.2003.03890.x
Nam G-M, Arya G (2014) Torsional behavior of chromatin is modulated by rotational phasing of nucleosomes. Nucleic Acids Res 42:9691–9699. doi:10.1093/nar/gku694
Neukirch S, Marko JF (2011) Analytical description of extension, torque, and supercoiling radius of a stretched twisted DNA. Phys Rev Lett 106:138104
Neuman KC (2010) Single-molecule measurements of DNA topology and topoisomerases. J Biol Chem 285:18967–18971. doi:10.1074/jbc.R109.092437
Neuman KC, Charvin G, Bensimon D, et al (2009) Mechanisms of chiral discrimination by topoisomerase IV. Proc Natl Acad Sci USA 106:6986–6991. doi:10.1073/pnas.0900574106
Nitiss JL (2009a) DNA topoisomerase II and its growing repertoire of biological functions. Nat Rev Cancer 9:327–337
Nitiss JL (2009b) Targeting DNA topoisomerase II in cancer chemotherapy. Nat Rev Cancer 9:338–350
Ogawa T, Yogo K, Furuike S, et al (2015) Direct observation of DNA overwinding by reverse gyrase. Proc Natl Acad Sci USA 112:7495–7500. doi:10.1073/pnas.1422203112
Ogawa T, Sutoh K, Kikuchi A, et al (2016) Torsional stress in DNA limits collaboration among reverse gyrase molecules. FEBS J 283:1372–1384. doi:10.1111/febs.13675
Osheroff N (1986) Eukaryotic topoisomerase II. Characterization of enzyme turnover. J Biol Chem 261:9944–9950
Ouldridge TE, Louis AA, Doye JPK (2011) Structural, mechanical, and thermodynamic properties of a coarse-grained DNA model. J Chem Phys 134:085101. doi:10.1063/1.3552946
Palecek E, Brázdová M, Brázda V, et al (2001) Binding of p53 and its core domain to supercoiled DNA. Eur J Biochem 268:573–581. doi:10.1046/j.1432-1327.2001.01898.x
Patel A, Yakovleva L, Shuman S, et al (2010) Crystal structure of a bacterial topoisomerase IB in complex with DNA reveals a secondary DNA binding site. Structure 18:725–733. doi:10.1016/j.str.2010.03.007
Peter BJ, Ullsperger C, Hiasa H, et al (1998) The structure of supercoiled intermediates in DNA Replication. Cell 94:819–827. doi:10.1016/S0092-8674(00)81740-7
Pommier Y (2013) Drugging topoisomerases: lessons and challenges. ACS Chem Biol 8:82–95. doi:10.1021/cb300648v
Pommier Y, Cushman M (2009) The indenoisoquinoline noncamptothecin topoisomerase I inhibitors: update and perspectives. Mol Cancer Ther 8:1008–1014. doi:10.1158/1535-7163.mct-08-0706
Portugal J, Rodriguez-Campos A (1996) T7 RNA polymerase cannot transcribe through a highly knotted DNA template. Nucleic Acids Res 24:4890–4894. doi: 10.1093/nar/24.24.4890
Ramamoorthy M, Tadokoro T, Rybanska I, et al (2012) RECQL5 cooperates with Topoisomerase II alpha in DNA decatenation and cell cycle progression. Nucleic Acids Res 40:1621–1635. doi:10.1093/nar/gkr844
Revyakin A, Ebright RH, Strick TR (2004) Promoter unwinding and promoter clearance by RNA polymerase: detection by single-molecule DNA nanomanipulation. Proc Natl Acad Sci USA 101:4776–4780. doi:10.1073/pnas.0307241101
Rybenkov VV, Ullsperger C, Vologodskii AV, et al (1997) Simplification of DNA topology below equilibrium values by type II topoisomerases. Science 277:690–693. doi:10.1126/science.277.5326.690
Schoeffler AJ, Berger JM (2008) DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q Rev Biophys 41:41–101. doi:10.1017/S003358350800468X
Schvartzman JB, Martínez-Robles M-L, Hernández P, et al (2013) The benefit of DNA supercoiling during replication. Biochem Soc Trans 41:646–651. doi:10.1042/bst20120281
Seol Y, Zhang H, Pommier Y, et al (2012) A kinetic clutch governs religation by type IB topoisomerases and determines camptothecin sensitivity. Proc Natl Acad Sci USA 109:16125–16130. doi:10.1073/pnas.1240480109
Seol Y, Gentry AC, Osheroff N, et al (2013a) Chiral discrimination and writhe-dependent relaxation mechanism of human topoisomerase IIα. J Biol Chem 288:13695–13703. doi:10.1074/jbc.M112.444745
Seol Y, Hardin AH, Strub M-P, et al (2013b) Comparison of DNA decatenation by Escherichia coli topoisomerase IV and topoisomerase III: implications for non-equilibrium topology simplification. Nucleic Acids Res 41:4640–4649. doi:10.1093/nar/gkt136
Seol Y, Zhang H, Agama K, et al (2015) Single-molecule supercoil relaxation assay as a screening tool to determine the mechanism and efficacy of human topoisomerase IB inhibitors. Mol Cancer Ther 14:2552–2559. doi:10.1158/1535-7163.mct-15-0454
Stone MD, Bryant Z, Crisona NJ, et al (2003) Chirality sensing by Escherichia coli topoisomerase IV and the mechanism of type II topoisomerases. Proc Natl Acad Sci USA 100:8654–8659. doi:10.1073/pnas.1133178100
Strick TR, Croquette V, Bensimon D (1998) Homologous pairing in stretched supercoiled DNA. Proc Natl Acad Sci USA 95:10579–10583
Strick TR, Croquette V, Bensimon D (2000) Single-molecule analysis of DNA uncoiling by a type II topoisomerase. Nature 404:901–904
Stuchinskaya T, Mitchenall LA, Schoeffler AJ, et al (2009) How do type II topoisomerases use ATP hydrolysis to simplify DNA topology beyond equilibrium? Investigating the relaxation reaction of nonsupercoiling type II topoisomerases. J Mol Biol 385:1397–1408. doi:10.1016/j.jmb.2008.11.056
Sundin O, Varshavsky A (1981) Arrest of segregation leads to accumulation of highly intertwined catenated dimers: dissection of final stages of SV40 DNA replication. Cell 25:659–669. doi: 10.1016/0092-8674(81)90173-2
Suski C, Marians KJ (2008) Resolution of converging replication forks by RecQ and topoisomerase III molecular. Cell 30:779–789. doi:10.1016/j.molcel.2008.04.020
Tabuchi H, Handa H, Hirose S (1993) Underwinding of DNA on binding of yeast TFIID to the TATA element. Biochem Biophys Res Commun 192:1432–1438. doi:10.1006/bbrc.1993.1576
Taneja B, Schnurr B, Slesarev A, et al (2007) Topoisomerase V relaxes supercoiled DNA by a constrained swiveling mechanism. Proc Natl Acad Sci USA 104:14670–14675. doi:10.1073/pnas.0701989104
Terekhova K, Gunn KH, Marko JF, et al (2012) Bacterial topoisomerase I and topoisomerase III relax supercoiled DNA via distinct pathways. Nucleic Acids Res 40:10432–10440. doi:10.1093/nar/gks780
Teves SS, Henikoff S (2014) DNA torsion as a feedback mediator of transcription and chromatin dynamics. Nucleus 5:211–218. doi:10.4161/nucl.29086
Thomson NH, Santos S, Mitchenall LA, et al (2014) DNA G-segment bending is not the sole determinant of topology simplification by type II DNA topoisomerases. Sci Rep 4:6158. doi:10.1038/srep06158. http://www.nature.com/articles/srep06158#supplementary-information
Travers A, Muskhelishvili G (2005) DNA supercoiling [mdash] a global transcriptional regulator for enterobacterial growth? Nat Rev Microbiol 3:157–169. doi:10.1038/nrmicro1088
Travers A, Muskhelishvili G (2007) A common topology for bacterial and eukaryotic transcription initiation? EMBO Rep 8:147–151. doi:10.1038/sj.embor.7400898
Vanderlinden W, Lipfert J, Demeulemeester J, et al (2014) Structure, mechanics, and binding mode heterogeneity of LEDGF/p75-DNA nucleoprotein complexes revealed by scanning force microscopy. Nanoscale 6:4611–4619. doi:10.1039/C4NR00022F
Vlijm R, Lee M, Ordu O, et al (2015) Comparing the assembly and handedness dynamics of (H3.3-H4)2 tetrasomes to canonical tetrasomes. PLoS One 10, e0141267. doi:10.1371/journal.pone.0141267
Vologodskii A (2009) Theoretical models of DNA topology simplification by type IIA DNA topoisomerases. Nucleic Acids Res 37:3125–3133. doi:10.1093/nar/gkp250
Vologodskii AV, Cozzarelli NR (1993) Monte Carlo analysis of the conformation of DNA catenanes. J Mol Biol 232. doi:10.1006/jmbi.1993.1465
Vologodskii AV, Cozzarelli NR (1994) Conformational and thermodynamic properties of supercoiled DNA. Annu Rev Biophys Biomol Struct 23:609–643
Vologodskii AV, Frank-Kamenetskii MD (1992) Modeling supercoiled DNA. Methods Enzymol 211:467–480
Vologodskii AV, Marko JF (1997) Extension of torsionally stressed DNA by external force. Biophys J 73:123–132. doi: 10.1016/s0006-3495(97)78053-6
Vologodskii AV, Levene SD, Klenin KV, et al (1992) Conformational and thermodynamic properties of supercoiled DNA. J Mol Biol 227. doi:10.1016/0022-2836(92)90533-p
Vologodskii AV, Zhang W, Rybenkov VV, et al (2001) Mechanism of topology simplification by type II DNA topoisomerases. Proc Natl Acad Sci 98:3045–3049. doi:10.1073/pnas.061029098
Vos SM, Tretter EM, Schmidt BH, et al (2011) All tangled up: how cells direct, manage and exploit topoisomerase function. Nat Rev Mol Cell Biol 12:827–841. http://www.nature.com/nrm/journal/v12/n12/suppinfo/nrm3228_S1.html
Wang JC (1971) Interaction between DNA and an Escherichia coli protein ω. J Mol Biol 55:523–IN516. doi:10.1016/0022-2836(71)90334-2
Wang JC (2002) Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol 3:430–440
Watson JD, Crick FHC (1953) Molecular structure of nucleic acids: a structure for deoxyribose nucleic acid. Nature 171:737–738
Wereszczynski J, Andricioaei I (2010) Free energy calculations reveal rotating-ratchet mechanism for DNA supercoil relaxation by topoisomerase IB and its inhibition. Biophys J 99:869–878. doi:10.1016/j.bpj.2010.04.077
White JH (1969) Self-linking and the gauss integral in higher dimensions. Am J Math 91:693–728. doi:10.2307/2373348
Worcel A, Burgi E (1972) On the structure of the folded chromosome††The terms folded chromosome, folded DNA, folded genome, DNA-RNA protein complex (or plain complex) are used interchangeably and they all refer to the compact, fast sedimenting, DNA-containing structures released from Escherichia coli after careful lysis of the cells by the Stonington & Pettijohn (1971) procedure of Escherichia coli. J Mol Biol 71:127–147. doi:10.1016/0022-2836(72)90342-7
Yan J, Magnasco MO, Marko JF (1999) A kinetic proofreading mechanism for disentanglement of DNA by topoisomerases. Nature 401:932–935. doi:10.1038/44872
Zechiedrich EL, Osheroff N (1990) Eukaryotic topoisomerases recognize nucleic acid topology by preferentially interacting with DNA crossovers. EMBO J 9:4555–4562
Zechiedrich L, Osheroff N (2010) Topoisomerase IB-DNA interactions: X marks the spot. Structure 18:661–663. doi:10.1016/j.str.2010.05.002
Zhang H, Marko JF (2008) Maxwell relations for single-DNA experiments: monitoring protein binding and double-helix torque with force-extension measurements. Phys Rev E 77:031916
Acknowledgments
This research was supported by the Intramural Research Program of the National Heart, Lung, and Blood Institute, National Institutes of Health (HL001056) and by the Human Frontiers Science Program (RGY0072/2010). We thank Lynda Bradly for comments on the manuscript. We thank the anonymous referees for their suggestions and comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Yeonee Seol declares that she has no conflict of interest.
Keir C. Neuman declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
This article is a contribution to Special Issue “DNA Supercoiling” but has already been published in BREV, September 2016, Volume 8, Issue 3, pp 221–231, DOI 10.1007/s12551-016-0206-x.
Rights and permissions
About this article
Cite this article
Seol, Y., Neuman, K.C. The dynamic interplay between DNA topoisomerases and DNA topology. Biophys Rev 8 (Suppl 1), 101–111 (2016). https://doi.org/10.1007/s12551-016-0240-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12551-016-0240-8