Abstract
Balanced gene expression is a high priority in order to maintain optimal functioning since alterations and variations could result in acute consequences. X chromosome inactivation (X-inactivation) is one such strategy utilized by mammalian species to silence the extra X chromosome in females to uphold a similar level of expression between the two sexes. A functionally versatile class of molecules called long noncoding RNA (lncRNA) has emerged as key regulators of gene expression and plays important roles during development. An lncRNA that is indispensable for X-inactivation is X-inactive specific transcript (Xist), which induces a repressive epigenetic landscape and creates the inactive X chromosome (Xi). With recent advents in the field of X-inactivation, novel positive and negative lncRNA regulators of Xist such as Jpx and Tsix, respectively, have broadened the regulatory network of X-inactivation. Xist expression failure or dysregulation has been implicated in producing developmental anomalies and disease states. Subsequently, reactivation of the Xi at a later stage of development has also been associated with certain tumors. With the recent influx of information about lncRNA biology and advancements in methods to probe lncRNA, we can now attempt to understand this complex network of Xist regulation in development and disease. It has become clear that the presence of an extra set of genes could be fatal for the organism. Only by understanding the precise ways in which lncRNAs function can treatments be developed to bring aberrations under control. This chapter summarizes our current understanding and knowledge with regard to how lncRNAs are orchestrated at the X-inactivation center (Xic), with a special focus on how genetic diseases come about as a consequence of lncRNA dysregulation.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Our understanding of the human genome keeps increasing as new technological advances allow us to unravel the genome’s functional elements. Although mapping of the human genome has been completed, it still remains unclear how certain regions are demarcated as coding regions or regulatory elements and what the vast regions without coding account for. The number of genes annotated dwindled from the previously estimated 35,000–100,000 to 25,000 protein-coding regions, which only occupies 1–2 % of the overall genome (Liang et al. 2000). This finding has prompted searches elsewhere for unidentified functional elements. Recently, it has become evident that new classes of molecules, noncoding RNAs, are emerging as crucial functional molecules in development, disease, and physiology. Surprisingly, noncoding transcripts were available in abundance while surveying the human genome but had been conveniently overlooked (Carninci et al. 2005). These noncoding RNA transcripts are classified primarily based on the length of the RNA transcripts. Noncoding RNAs generally fall under two main categories with any length below 200 nucleotide-long classified as small noncoding RNA and anything larger than 200 nucleotide-long classified as long noncoding RNA (lncRNA). Furthermore, evidence supporting the involvement of noncoding RNA in various biological processes, for example, gene expression regulation, is rapidly increasing (Wapinski and Chang 2011; Wang and Chang 2011). Indeed, both small and IncRNAs are involved in bringing about epigenetic changes at a particular locus (Peschansky and Wahlestedt 2014). The significance of noncoding RNAs is increasing with regard to their physiological function as well as their association with diseases (Wapinski and Chang 2011; Maass et al. 2014).
One such widely studied lncRNA is X-inactive specific transcript (Xist), which is indispensable for X chromosome inactivation (X-inactivation). In this chapter, we describe a wide variety of lncRNAs in the X-inactivation center (Xic), which reside on the X chromosome and are required for the induction of X-inactivation, and discuss how these lncRNAs cooperate in inducing monoallelic Xist expression to establish only one inactive X chromosome (Xi) in mammalian females. Finally, the various diseases that arise due to a dysfunctional or skewed X-inactivation, and possible future studies in humans, are discussed. An elucidation of these novel regulators and their interaction networks would provide important insight with respect to the molecular mechanism of diseases such as cancer and help toward designing better therapeutics.
2 X Chromosome Inactivation (X-inactivation)
X-inactivation is a dosage compensation mechanism in female mammals which maintains the balance of X-linked gene expression and is achieved when one of the two available X chromosomes is inactivated at an early stage of embryonic development. Failure of X-inactivation would result in an increased dosage of genes which can alter various pathways pertaining to different vital processes (Lyon 1961; Heard and Disteche 2006; Payer and Lee 2008). In the case of males, only one functional X chromosome exists and is usually referred to as genetic unisomy, whereas in females, two X chromosomes exist and the event of silencing one of them is referred to as functional unisomy. X-inactivation is critical for cellular differentiation, and dysregulation could result in developmental abnormalities (Marahrens et al. 1997). X-inactivation occurs either as imprinted X-inactivation at early embryonic stages and in extraembryonic tissues, wherein the paternal X chromosome is silenced (Takagi and Sasaki 1975; Huynh and Lee 2003; Okamoto et al. 2004), or as an act of random X-inactivation in which both the paternal and the maternal X chromosome have an equal probability of being inactivated in the epiblast (Monk and Harper 1979; Tan et al. 1993). In imprinted X-inactivation, although the paternal X chromosome undergoes complete inactivation around the blastocyst stage, it is reactivated during the peri-implantation stage in the epiblast lineage which then gives rise to a broad range of tissue types in the fetus (Mak et al. 2004). Subsequently, these cells are prone to another wave of random X-inactivation. Random X-inactivation is thought to occur close to the time of implantation in cells of the epiblast at around embryonic days 4.5–5.5 in mice. Once the Xi is established, it is inherited through all subsequent cell divisions.
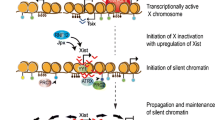
X-inactivation is characterized by Xist lncRNA coating the Xi (Clemson et al. 1996). As the coating masks the chromosome completely, this observable phenomenon is referred to as the “Xist cloud.” Gene silencing is triggered once Xist is upregulated on the future Xi. This designated future Xi goes through the stages of initiation, progression, and maintenance of repressive chromatin states with the aided participation of multiple proteins and machineries (Table 1) (Wutz 2011). Numerous studies have been carried out with respect to dynamic changes occurring on the Xi during X-inactivation. These studies have shown the remarkable changes happening in the epigenetic landscape as well as the chromatin structure during X-inactivation. The epigenetic histone marks accompanying the chromatin state (euchromatin/heterochromatin) are characteristic of its transcriptional activity (active/silent) and influence strongly the chromatin structure. At the onset of X-inactivation, euchromatin markers such as H3K9Ac, H3K4me2, and H3K4me3 are lost, when Xist RNA starts coating the X chromosome, subsequently leading to global H4 hypoacetylation (Keohane et al. 1996; Chaumeil et al. 2002). Meanwhile, loss of RNA polymerase II and nascent transcripts also occurs post-Xist RNA coating (Chaumeil et al. 2006). Interestingly, a whole new array of repressive epigenetic modifications such as H3K27me3, H3K9me2, H4K20me1, and H2AK119ub1 get enriched on the Xi (Wutz 2011). Strikingly, the histone tri-methylations on H3K27 and H3K9 occur at different regions and are recognized by different cofactors in the Xi indicating two different populations of repressed heterochromatins in humans (Chadwick and Willard 2004; Nozawa et al. 2013). The kinetics of X-inactivation is tightly associated with the dynamics of histone epigenetic marks brought about by protein complexes such as polycomb repressive complex, PRC1 and PRC2, which catalyze the repressive histone modifications, H2A119ub and H3K27me3, respectively, in an Xist RNA-dependent manner (Mak et al. 2002; Silva et al. 2003; Plath et al. 2003; de Napoles et al. 2004; Fang et al. 2004).
Additionally, the Xi is marked by a series of epigenetic changes such as histone variant macroH2A along with DNA methylation of the CpG islands and promoters (Norris et al. 1991; Costanzi and Pehrson 1998; Sharp et al. 2011). Smchd1 is also involved in delivering the DNA hypermethylation of the CpG islands associated with the Xi, which is required for long-term maintenance of gene silencing (Blewitt et al. 2008; Gendrel et al. 2012). Another group of proteins associated with the Xi is a member of the trithorax group proteins for transcriptional activation, Ash2L, although its functional role on the Xi remains to be elucidated (Pullirsch et al. 2010).
3 X-inactivation Center (Xic)
Early studies of X chromosome truncations and translocations helped identify the X chromosome locus wherein X-inactivation is induced, called an Xic (Rastan and Brown 1990; Brown 1991). The initial mapping of the Xic was first shown by a series of cytological experiments, which used differential staining of X chromosome material at the metaphase stage of mouse embryos. These showed that chromosomal rearrangements between the X chromosome and autosomes led to random inactivation of the segment hosting the Xic. In experiments examining the T16H reciprocal translocation between the X chromosome and chromosome 16, also referred to as the Searle’s translocation, only the translocated 16X chromosome was inactivated, predicting the presence of the Xic (Takagi 1980; Rastan 1983). Subsequently, truncating one of the X chromosomes at the Xic region in embryonic cells resulted in no X-inactivation, suggesting that two Xic’s are required for X-inactivation (Rastan and Robertson 1985). Furthermore, when one of the X chromosomes was truncated leaving behind a significant chunk of the Xic, termed HD3 translocation, random X-inactivation occurred, indicative of the distal boundary of the Xic (Rastan and Robertson 1985). These results thus suggest that the minimum region required for efficiently triggering X-inactivation lies somewhere between the T16H and the HD3 break points, which was followed by further extensive genetic mapping (Keer et al. 1990). While the majority of experiments were established in mice, a similar strategy was used to determine the XIC in humans using mice/human somatic cell hybrids derived from patient samples with human X chromosome translocations and deletions (Brown et al. 1991b). The human equivalent of the mouse Xic seems to be highly conserved and spans approximately a 1 Mb region in Xq13. Studies increasingly propose that a minimal region is required for X-inactivation to take place in humans. Any abnormalities arising due to rearrangements in this demarcated region could lead to improper functioning and haywire regulations.
X-inactivation is a phenomenon controlled exclusively by the events occurring at the Xic. A series of transgene experiments contributed to delineating the Xic region (Heard et al. 1996; Lee et al. 1996; Lee and Jaenisch 1997). In embryonic stem cells, transgenes containing Xist can induce silencing in surrounding regions at its insertion site. Experiments showed that a mouse transgene was sufficient to induce Xist RNA expression and coating in cis and subsequently silence the LacZ reporter within the transgene (Lee et al. 1996). Copy number and expression levels were also found to play an important role for silencing to take place at the integration site for the genome (Heard et al. 1996). The Xic region, including Xist and its flanking regions, harbors a number of lncRNAs and consists of a complex interplay between each lncRNA to regulate the monoallelic expression of Xist (Fig. 1). Apart from the abundance of both positive and negative lncRNA regulators of Xist at the Xic, a number of proteins are also involved in this tight regulation (Wutz 2011).
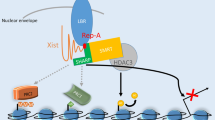
lncRNAs on the Xic. The figure shows the schematic of Xic and lncRNAs originating from the locus at the onset of X-inactivation. Red and blue arrows show the action of lncRNAs as transcriptional activators and repressors, respectively. Orange box with black arrow indicates the actively transcribed gene and direction of transcription. Blue box with black arrows by dashed line indicates repressed gene. While Jpx and Ftx are known as escape genes (Tian et al. 2010; Chureau et al. 2011; Kobayashi et al. 2013), expression of Tsix and Xite persists longer on the Xi at the onset of X-inactivation (Lee et al. 1999a; Ogawa and Lee 2003). Allelic expression of Tsx, DXPas34, and RepA has not yet been reported. Although the Jpx transgene induces Xist expression, it is not clear whether endogenous Jpx acts both in cis and in trans at the initiation of X-inactivation
3.1 Xist
Xist produces a 17 kb transcript in mice and a 19 kb transcript in humans that is processed by polymerase II, polyadenylated, and retained in the nucleus. The Xist lncRNA is exclusively expressed from the Xic on the Xi and a central player of Xic function for both imprinted and random X-inactivation (Brown et al. 1991a; Brockdorff et al. 1991; Penny et al. 1996; Marahrens et al. 1997; Wutz and Jaenisch 2000). Xist lncRNA induces chromosome-wide gene silencing through a cascade of epigenetic modifications on the Xi, which is eventually maintained through multiple rounds of cell division. Xist RNA has multiple functional domains spread across its 8 exons including repeat A-F, which are highly conserved in eutherian mammals, enabling its interaction with transcriptional factors, scaffold proteins, and chromatin-modifying proteins (Sado and Brockdorff 2013).
At an initial phase of X-inactivation, it is proposed that transcriptional factors YY1 (Yin Yang 1) act as an anchoring point to bridge between Xist RNA and Xist gene to serve as a nucleation center for Xist RNA spreading across the Xi (Jeon and Lee 2011). Furthermore, since knockdown of hnRNP U (heterogenous nuclear ribonucleoprotein U, also known as SAF-A and SP120), which has a binding affinity to both DNA and RNA, disrupts Xist RNA localization and X-linked gene silencing, hnRNP U acts as a bridge between matrix/scaffold attached region (MAR/SAR) in the Xi and Xist RNA to facilitate spreading of the silencing machinery such as PRC2 across the Xi (Hasegawa et al. 2010). Recent studies to map Xist RNA and its binding partner, PRC2, on the Xi revealed an orderly fashion of Xist RNA and PRC2 spreading, as well as a strong dependency on Xist RNA for the three-dimensional structure of the Xi (Splinter et al. 2011; Engreitz et al. 2013; Simon et al. 2013). Allele-specific ChIP-seq (chromatin immunoprecipitation with deep sequencing) of a catalytic subunit of PRC2, Ezh2, showed ~50 prominent and ~1500 moderate peaks prior to X-inactivation, suggesting that Ezh2 binds at ~50 strong and ~1500 moderate binding sites (Pinter et al. 2012). The Ezh2 binding sites are frequently associated with canonical H3K4me3/H3K27me3 bivalent domain and CpG islands across the X chromosome. Upon differentiation and induction of X-inactivation, Ezh2 binds to an additional >100 strong and ~4000 moderate binding sites which can then induce spreading of H3K27me3 toward neighboring regions. An Xist RNA binding map produced by combining CHART-seq (capture hybridization analysis of RNA targets with deep sequencing) and RAP (RNA antisense purification) data has also revealed that Xist RNA initially binds to gene-rich regions before spreading to distal, gene-poor regions (Engreitz et al. 2013; Simon et al. 2013). The Xist RNA binding profile overlaps heavily with Ezh2 binding and H3K27me3 density, indicating an Xist RNA-dependent deposition of Ezh2 and H3K27me3. To efficiently induce gene silencing across the entire X chromosome upon differentiation, Xist RNA spreads in a three-dimensional manner away from the Xic toward distal binding sites across the Xi (Lieberman-Aiden et al. 2009; Engreitz et al. 2013; Simon et al. 2013). The repeat A region in exon 1 of Xist RNA is presumed to have an important role in Xist RNA spreading across the gene-rich region of the Xi, since deletion of repeat A has resulted in impairment of this process (Engreitz et al. 2013). It was previously suggested that long interspersed nuclear element 1 (LINE1) repeat elements play a role in assisting Xist RNA to spread along the entire X chromosome, as well as supporting the assembly of heterochromatic nuclear structures and propagation of X-inactivation (Lyon 2000; Chow et al. 2010). However, recent reports show that there is less correlation between the Xist RNA binding site and LINE1 repeats that has been previously speculated (Engreitz et al. 2013; Simon et al. 2013).
4 Long Noncoding RNAs and Elements Controlling Xist Expression
Xist is the central player of X-inactivation: through neighboring lncRNAs, which are involved in the tight regulation of Xist monoallelic expression, Xist induces a repressive chromatin state that leads to X-linked gene silencing along the entire X chromosome. Recent advancements in next-generation sequencing and newer techniques have propelled forward the functional analysis of novel lncRNAs in Xic and increased our understanding of their function and interactions at the molecular level. Here, we describe several lncRNAs in the Xic which regulate Xist expression in both positive and negative ways. Tight regulation in the interplay of these lncRNAs is essential for securing the induction of monoallelic Xist expression and bringing about X-inactivation in only one of the two X chromosomes in females.
4.1 Negative LncRNA Regulators of Xist
4.1.1 Tsix
The noncoding Tsix gene expresses lncRNA antisense to Xist; hence, it is named “Tsix,” which is Xist spelled in reverse order (Lee et al. 1999a). While the Tsix transcript does not coat the X chromosome like its counterpart Xist during X-inactivation, it is detected using RNA fluorescence in situ hybridization as a pinpoint signal at both endogenous loci and is expressed in both male and female undifferentiated cells (Lee et al. 1999a). While monoallelic Tsix expression coincides with Xist silencing on the future active X chromosome (Xa) at the onset of X-inactivation, Tsix extinction and Xist upregulation also occur on the future Xi, thereby suggesting the antagonistic role of Tsix on Xist expression. Tsix expression finally disappears on both X chromosomes at a later stage of differentiation without Xist reactivation on the Xa, suggesting that Tsix represses Xist upregulation at the onset of X-inactivation. Tsix has been reported to play a role for Xist repression both in imprinted and in random X-inactivation (Lee and Lu 1999; Lee 2000; Sado et al. 2001). In Tsix heterozygous mutant female ES cells, a mutation in Tsix always leads to the induction of Xist expression from the Tsix-mutant X chromosome, hence the non-random inactivation of the mutant X chromosome (Lee and Lu 1999). Apart from random X-inactivation, imprinted X-inactivation is controlled by Tsix to prevent the maternal X chromosome from undergoing X-inactivation in the extra embryonic tissues. While female embryos carrying a Tsix mutation on the maternal X chromosome lead to embryonic lethality due to X-inactivation on both X chromosomes, female embryos carrying a mutated Tsix on the paternal X chromosome normally survive to term (Lee 2000; Sado et al. 2001). Since the Tsix-mutant male embryo also carries a Tsix mutant maternal X, this mutation resulted in embryonic lethality due to X-inactivation. These reports demonstrate that Tsix antagonizes Xist expression in cis. Tsix exclusively works to repress Xist by modulating the chromatin structure (Navarro 2005; Sado et al. 2005; Sun et al. 2006). Since the insertion of a polyadenylation signal to produce Tsix truncation at the site close to the 5′ end of Xist effectively abolishes Tsix function, Tsix transcription across the Xist promoter is critical for Xist repression (Ohhata et al. 2008).
4.1.2 Xite (X-inactivation Intergenic Transcription) and DXPas34
Xite resides between minor and major Tsix promoters and is associated with multiple bidirectional long noncoding transcripts (Ogawa and Lee 2003). Since an Xite heterozygous deletion mutation leads to skewed X-inactivation which favors the mutant X chromosome, Xite is involved in choosing which X chromosome will be inactivated. Further analysis revealed that Tsix is downregulated in the Xite deletion mutant in cis; thus, Xite plays a role in the decision of the Xi by positively promoting Tsix expression, which in turn represses Xist (Ogawa and Lee 2003). Many models have been proposed with regard to how Xite could function in X-inactivation. One model suggests that Xite could act as an enhancer for development-specific Tsix regulation at the onset of X-inactivation. Frequent association of multiple bidirectional transcripts and DNaseI hypersensitive sites with enhancer elements supports the Xite enhancer model (Natoli and Andrau 2012; Lam et al. 2014). Indeed, transient enhancer assays revealed that Xite has development-specific enhancer activity in Tsix expression (Stavropoulos et al. 2005).
DXPas34, another region associated with bidirectional promoter activity and DNaseI hypersensitive sites, is also a positive regulator of Tsix expression (Stavropoulos et al. 2005; Vigneau et al. 2006; Cohen et al. 2007). DXPas34 is a 1.2 kb CpG-rich region containing a 34-mer tandem repeat residing 750 bp downstream of the major Tsix promoter. Transient enhancer assays showed that DXPas34, as well as Xite, enhanced Tsix expression (Stavropoulos et al. 2005). Interestingly, deletion of DXPas34 leads to repression of the major Tsix promoter and activation of Xist expression at the onset of X-inactivation, which is followed by Tsix derepression without reversal of X-inactivation. This suggests that DXPas34 functions as both an enhancer and repressor of Tsix in a differentiation-specific manner.
4.1.3 Tsx (Testes-Specific X-Linked Noncoding RNA)
Another noncoding transcript, Tsx, located nearly 40 kb from the 3′-end of Xist, was found to be specifically expressed in the testes and in a very low concentration in both male and female brains (Simmler et al. 1996; Anguera et al. 2011). Although Tsx was initially reported as a potential protein-coding gene, specifically as an encoder of a 144 amino acid protein (Simmler et al. 1996), further investigation has indicated that Tsx is likely to be a noncoding gene (Anguera et al. 2011). Tsx expresses in ES cells as well as in early embryos and is gradually repressed upon differentiation. The homozygous deletion of Tsx in female mice led to a small decrease in fertility, resulting in skewed sex ratios that slightly favored females. Aberrant upregulation of Xist along with Tsix downregulation was observed in a small population of the Tsx mutant cells; thus, Tsx might promote Tsix expression and indirectly upregulate Xist expression (Anguera et al. 2011).
4.2 Positive LncRNA Regulators of Xist
4.2.1 Jpx/Enox
Jpx/Enox is another important lncRNA, which is located 10 kb upstream of Xist and is expressed in the antisense direction of Xist (Chureau et al. 2002; Johnston et al. 2002; Chow et al. 2003). While transgenes including the X-inactivation hub of lncRNAs such as Xist, Tsix, and Xite could only induce Xist activation inefficiently, the additional upstream region of Xist restored the induction of Xist (Lee et al. 1999b). This suggests that the Xic requires an upstream region flanking Xist. Subsequently, further analysis showed that Jpx is required for the proper expression of Xist (Tian et al. 2010; Sun et al. 2013). Jpx escapes from X-inactivation and is upregulated during X-inactivation (Tian et al. 2010). When Jpx was deleted, no X-inactivation was induced in males; however, Jpx heterozygous mutant females exhibited severe phenotypes with massive cell death, significantly impaired Xist upregulation, and compromised X-inactivation induction. These defects and Xist expression levels were restored to normal with overexpression of Jpx using a transgene. These data thereby suggest that Jpx can act in trans to activate Xist, although the Jpx heterozygous mutation shows mildly reduced Xist expression in cis (Tian et al. 2010). As Jpx overexpression with Tsix disruption efficiently induces aberrant X-inactivation even in male differentiating embryonic bodies, both Jpx and Tsix coregulate Xist as an activator and repressor, respectively. That leaves the question of how Jpx RNA promotes Xist expression. A recent study has indicated the unique function of Jpx RNA in Xist expression at the initiation of X-inactivation (Sun et al. 2013). In undifferentiated cells, transcription factor CTCF is loaded on the CTCF binding sites within the Xist P2 promoter by which Xist is repressed. Whereas overexpression of CTCF blocked induction of Xist upregulation, this repression was rescued by overexpression of Jpx. As CTCF binds to both the Xist promoter and Jpx RNA, it is proposed that Jpx RNA replaces CTCF from the Xist promoter, which is followed by induction of Xist expression on the Xi (Sun et al. 2013). Future work will be needed to elucidate the mechanism by which Jpx RNA selectively replaces CTCF from the Xist promoter in the future Xi to induce monoallelic Xist expression.
4.2.2 Ftx (Five Prime to Xist)
Ftx is another gene which encodes a long noncoding transcript located in the upstream of Xist, a potential activator for Xist (Chureau et al. 2002, 2011). Ftx escapes X-inactivation in both imprinted and random X-inactivation and, like Jpx, is upregulated upon induction of random X-inactivation (Chureau et al. 2011; Kobayashi et al. 2013). Deletion of Ftx in male mouse ES cells showed that the expression pattern of X-linked genes in the vicinity of Ftx was altered through a significant drop in expression levels. Furthermore, increased methylation at the 5´ CpG island of Xist was observed, suggesting a positive role of Ftx in Xist expression. However, Ftx has been reported to be dispensable in imprinted X-inactivation in the mouse embryo (Soma et al. 2014). In spite of targeted deletions of Ftx, neither the survival of female embryos nor the expression of Xist was affected during the preimplantation period in the Ftx mutant mice (Soma et al. 2014). Further investigation would be necessary to conclude whether Ftx is essential for random X-inactivation in female mice.
4.2.3 RepA
Apart from the two major isoforms of Xist RNA, known as long and short forms, there is another lncRNA which is derived from Xist. The 1.6 kb RepA RNA, which is transcribed from the repeat A region of Xist, was identified by a PRC2 immunoprecipitation (Zhao et al. 2008). Multiple roles for RepA RNA have been shown. For instance, RepA embedded within Xist is involved in both the recruitment of the PRC2 complex, Ezh2, and the activation of Xist (Zhao et al. 2008). Furthermore, the RepA region in Xist RNA has been shown to be a binding region for the Ezh2 protein, while other works have suggested that RepA plays a role in PRC2 complex spreading and H3K27me3 modification across the Xi (Plath et al. 2003; Kohlmaier et al. 2004; Zhao et al. 2008; Engreitz et al. 2013).
The Xic harbors a variety of lncRNAs such as Tsix and Jpx that are directly or indirectly involved in the regulation of Xist expression. Since lncRNAs are central players for the proper regulation of X-inactivation, further studies need to be extensively performed in order to fully explore the roles played by each lncRNA as well as the cooperative molecular mechanism involved in their interaction. With advancing technologies and novel approaches, it is likely that more and more novel noncoding RNAs will emerge, allowing for a deeper understanding of Xist’s regulation during X-inactivation and its associated epigenetic modifications.
5 X-inactivation and Disease
Maintenance of proper gene dosage in autosome and sex chromosomes is important for ideal development and survival of organisms (Torres et al. 2008). Aneuploidy is referred to as the condition where cells possess an atypical number of chromosomes and is usually detrimental to the organism (Fig. 2a). Although some patients with autosomal aneuploidies can survive, they are at a high risk of congenital malformations. Some examples are the trisomies in chromosomes 13, 18, and 21, usually referred to as Patau, Edwards, and Down syndromes, respectively. Turner syndrome and Klinefelter syndrome are diseases that arise due to an absence of one of the two X chromosomes (XO) in females and an extra X chromosome (XXY) in males, respectively (Sybert and McCauley 2004; Groth et al. 2013). While only one X chromosome remains active per each cell as a result of X-inactivation, 15 % of X-linked genes on the Xi in humans are known to be actively transcribed even in this chromosome-wide silenced environment; these are called escape genes (Carrel and Willard 2005). Absence and excess of escape gene expression on the Xi is proposed to be a cause of Turner and Klinefelter syndromes, respectively.
Genetic and epigenetic failures induce abnormal gene dosage. Deviation of gene dosage leads to detrimental effects on cell survival and development. a Aneuploidy with excess or loss of certain chromosome. b X-autosomal translocation could induce inactivation of the autosome fused to the X chromosome segment containing the Xic. The segment of X chromosome which lacks Xic in turn fails to be inactivated. c Cells with more than two active X chromosome arise by loss of the Xi followed by duplication of the Xa. Reactivation of the Xi could be a potential cause of a supernumerary Xa. Green, orange, and blue bold lines indicate the autosome, X, and Y chromosomes, respectively. Purple zigzag line indicates Xist RNA-induced gene silencing
X-inactivation is a complex enforcement system, which tightly controls the X-linked gene balance between the sexes, as well as secures and maintains a perfect ambience for proper cell differentiation and development. When certain segments from the X chromosome corresponding to the Xic are translocated to an autosomal region, the autosomal genes around the translocation site might be silenced through XIST RNA spreading coupled with accumulation of repressive epigenetic modifications (Fig. 2b) (Brown et al. 1991b), which potentially leads to haploinsufficiency of autosomal genes (White et al. 1998; Giorda et al. 2008; Van Echten-Arends et al. 2013).
While X-linked genes and autosomal genes are present in two copies in females, subsets of genes are expressed only from a single allele in individual cells. X-inactivation and genomic imprinting have allowed us to better understand the mechanism involved in determining whether a particular gene on a single allele will be expressed or repressed, as recently shown by the attribution of lncRNAs as master regulators of monoallelic expression of Xist and imprinted genes (Lee and Bartolomei 2013). The monoallelic expression of female X-linked genes in mammals is crucial for cellular survival and development. In addition, misregulations in proper monoallelic expression of imprinted genes are known to lead to a wide range of diseases such as Beckwith–Weidemann syndrome and Angelman syndrome. Thus, even deviations from normal X-linked gene dosage conditions could potentially give rise to developmental anomalies and disease states (Fig. 2c). In the context of disease conditions induced by dysfunctional X-inactivation, our knowledge remains poor even though the phenomenon is scientifically fascinating.
5.1 X-inactivation and Cancer
While each somatic cell contains a pair of active autosomal chromosomes in mammals, this is not the case in sex chromosomes. When an unfavorable recessive change occurs in an autosomal allele, the secondary chromosome pair can act as a backup to replace the damaged gene; thus, the heterozygous condition might delay or prevent a catastrophic situation. As mentioned previously, mammalian sex chromosomes are characterized as genetic unisomy and functional unisomy in males and females, respectively. Furthermore, the X chromosome is laden with important genes for cellular differentiation and proliferation, as well as those related to cancer. Thus, genetic changes taking place on the delicate sex chromosomes could be immediately detrimental due to the lack of a backup copy and the higher likelihood that the mutations, when carried forward, may be prone to cancer (Spatz et al. 2004).
Aberrant X-inactivation can bring about local as well as chromosome-wide disturbances of X-linked gene silencing to alter the expression of cancer-related and other genes across the Xi which may lead to tumors (Chaligné and Heard 2014). A loss of Xist expression has been reported in many cancer cell lines derived from female breast, cervix, and ovary tumors (Kawakami et al. 2004b; Sirchia et al. 2005; Richardson et al. 2006). In female cancer cells, the frequent disappearance of Xist-expressing cells happens by loss of the Xi followed by a duplication of the Xa instead of reactivation of the Xi. In non-cancerous tissue, reactivation of the Xi by loss of XIST expression could potentially cause a wide range of derepression of X-linked genes, including cancer-related genes, resulting in abnormalities and diseases. X chromosome reactivation in mice is tightly restricted to happen within the inner cell mass at the blastocyst stage followed by random X-inactivation in the epiblast lineage and during the development of the primordial germ cells (Ohhata and Wutz 2013). Thus, reactivation outside of these periods could lead to detrimental effects. Since Xist is constitutively expressed from the Xi in differentiated somatic cells, it is suggestive of Xist’s role in the maintenance of X-inactivation. However, RT-PCR analysis of the mouse/human somatic hybrid cell lines containing human Xi revealed that gene silencing of XIC-lacking human Xi is highly stable, suggesting that no X reactivation takes place on the human Xi without XIC once X-inactivation is established (Brown and Willard 1994) Additionally, when Xist was conditionally deleted in primary mouse embryonic fibroblast cells, the Xi exhibited maintenance of its unique heterochromatic features such as late DNA replication and hypoacetylation on histone H4 even though histone variant macroH2A disappeared due to its Xist RNA-dependent localization (Csankovszki et al. 1999). Based on these observations, it had long been believed that Xist is essential for the initiation of random X-inactivation but dispensable for the maintenance of X-inactivation once the Xi is established. With the recent advantage of high-throughput sequencing and technical improvements, accumulating evidence has indicated otherwise, specifically that depletion of Xist RNA from the Xi can induce partial reactivation of a subset of X-linked genes on the Xi (Csankovszki et al. 2001; Zhang et al. 2007). In conditional Xist-deleted mouse fibroblast, an assessment of individual gene activity revealed that conditional Xist deletion on the Xi led to a slightly increased frequency of reactivation in X-linked GFP and Hprt genes (Csankovszki et al. 2001). This reactivation frequency was largely enhanced by treatment with 5-azadC and trichostatin A, inhibitors for DNA methylation and histone hypoacetylation, respectively, indicating that multiple layers of epigenetic regulation prevent improper reactivation of the Xi. Detailed analysis of conditional Xist knockout in dermal and embryonic fibroblast cells also showed that reactivation of the silenced X-linked genes on the Xi happened in a subset of genes (Zhang et al. 2007). Whereas the repressive histone mark H3K27me3 disappeared on the Xi by conditional Xist knockout, a dearth of active histone marker H3K4me2 remained on the Xi as well, suggesting overall chromosome-wide silencing is somewhat maintained without Xist RNA.
A recent study using human induced pluripotent stem cells (hiPSCs) also supports the role of XIST RNA in the stable repression of X-linked genes. Loss of XIST expression in hiPSCs is significantly correlated with the upregulation of X-linked oncogenes associated with higher growth rate in vitro and poor differentiation in vivo (Anguera et al. 2012). hiPSCs derived from differentiated cells using the Yamanaka factors (OCT-4, SOX2, KLF4, and c-MYC) or its derivatives hold great potential in regenerative medicine (Takahashi and Yamanaka 2006; Takahashi et al. 2007). However, hiPSCs and human embryonic stem cells (hESCs) are known to be genetically and epigenetically unstable (Kim et al. 2010; Bock et al. 2011; Gore et al. 2011); thus, strict validation of hiPSCs would be necessary for therapeutic purposes. Atypical features of female hiPSCs and hESCs are evident in X-inactivation; it is not currently known why these features are not observed in mouse ES cells (Dvash and Fan 2009; Lessing et al. 2013). They are generally classified into three distinct classes based on the status of X-inactivation and XIST expression (Silva et al. 2008; Anguera et al. 2012): Class I lines, which have two Xa’s and can undergo X-inactivation very similarly to mouse ES cells; Class II lines, wherein one X is already inactivated by the XIST cloud and cells may be partially differentiated; and finally Class III lines, wherein X-inactivation is already complete, and however, the expression of XIST is lost. Analysis across different hiPSC cell lines, especially when comparing Class II with Class III lines, showed loss of XIST expression is associated with an upregulation of X-linked genes in the Class III lines. Intriguingly, the upregulated X-linked genes include cancer-related genes such as MAGEA2 and MAGEA6, which are highly expressed in cancers (Rogner et al. 1995). These observations might be a hamper to using hiPSCs as a therapeutic tool. Despite no strong evidence thus far, the changes could be attributed to the loss of XIST expression. Conversely, XIST expression and presence of the Xi could be used as benchmarks to assess hiPSC quality. Furthermore, it should be noted that culture condition to establish and maintain hiPSCs has been improved to create naive Class I hiPSCs with high efficiency (Tomoda et al. 2012; Gafni et al. 2013).
Despite close association between the overexpression of X-linked genes and supernumerary Xa with many human cancers (Liao et al. 2003; Kawakami et al. 2004b; Pageau et al. 2007), it is not clear whether aberrant X-inactivation and reactivation of X-linked genes are a primary cause or just a consequence of cancer transformation and progression. More recently, our understanding of X-inactivation has advanced from its role in dosage compensation to include higher order functions such as tumor suppression. When a conditional knockout of Xist was achieved in the hematopoietic stem cells of mice, highly aggressive forms of hematologic cancer were manifested in a female-specific manner (Yildirim et al. 2013). Histopathological analysis of female mutant mice revealed that Xist deletion in the hematopoietic compartment induced myelodysplastic/myeloproliferative neoplasm (also known as mixed myeloproliferative/myelodysplastic syndrome [MPN/MDS]) (Orazi and Germing 2008). A detailed analysis of Xist deletion mutant mice showed that Xist deletion induced a significant genome-wide gene upregulation, especially in X-linked genes including cancer-related genes, in comparison with autosomal genes. This anomalous gene expression in the Xist mutant mice would potentially promote cancerous effects, suggesting a crucial role of Xist in not only maintenance of X-inactivation but also suppression of cancer. This is the first report that Xist disruption leads to a causal effect on cancer in vivo.
5.2 Skewed X-inactivation and X-linked Diseases
As a consequence of random X-inactivation with an equal probability of either the paternal or maternal originating X chromosome being inactivated, every female’s expression profile is a mosaic with cells having either the paternal or maternal Xi. Such mosaicism is usually advantageous for females since it contributes to physiological diversity and eases the deleterious effects of X-linked mutation. For females who have inherited unfavorable mutations on their X chromosome, there is a chance that approximately half of their somatic cells will express genes from the wild-type allele. This orderly process deviates in a subset of individuals, although the skewing varies by age and cell types (Sharp et al. 2000; Hatakeyama et al. 2004; Minks et al. 2008). It is also known that the clonality of cells varies in different female organs, which could also affect the X-linked phenotype in females (Thomas et al. 1988; Bittel et al. 2008). Since X chromosomes in mammalian female somatic cells are functionally unisomy by random X-inactivation, deviation from random X-inactivation ultimately results in a predominance of either the maternal or paternal X chromosome. This predominance could occur either by chance, by selection after primary choice, or in a predetermined fashion due to the presence of genetic elements or mutations that propel selection bias (Belmont 1996; Minks et al. 2008). The X-linked disorder in female carriers, coupled with skewed X-inactivation in favor of the wild-type X, give rise to greater populations of cells in which the mutated X-linked gene is expressed, therefore manifesting as a severe disease condition (Fig. 3) (Migeon 2006). Less severe variations are more likely to occur in females with smaller populations of mutated X-linked genes.
Skewed X-inactivation and disease phenotype. Female patients carrying a harmful mutation in a single allele show a variety of severity in disease phenotype due to skewed X-inactivation. The skewing of X-inactivation in favor of the wild-type X chromosome specifically leads to a severe phenotype. Orange and blue bold lines indicate the Xa and Xi, respectively. Red X indicates harmful mutation. White and gray circles show healthy and damaged cells, respectively
5.2.1 Rett Syndrome
Rett syndrome, an X-linked neurologic disorder which results in severe intellectual disability, primarily affects females and manifests during early childhood, typically occurring between 6 and 18 months of age; prior to onset, development appears to be normal, but is then followed by developmental regression, reduced brain growth, and severe mental retardation (Weaving et al. 2005). In newborn males, the disease is fatal. Rett syndrome symptoms vary from child to child with severity ranging from subtle abnormalities such as loss in muscle tone, difficulty in feeding, and jerkiness of limbs to more complex mental and physical abnormalities. Rett syndrome arises as a result of a heterozygous mutation in the X-linked gene encoding ubiquitous methyl-CpG binding protein 2 (MECP2) (Amir et al. 1999). MECP2 functions as a transcriptional regulator by binding to the genome in a DNA methylation-dependent manner. Female Rett syndrome patients possess mosaic wild-type/mutant MECP2 expression as a result of the random nature of X-inactivation in somatic cells. Random X-inactivation could be affected by chance, selection, or other genetic factors during early embryonic development. Skewed X-inactivation in patients with a MECP2 mutation progresses into a wide range of clinical presentations and manifests as Rett syndrome (Camus et al. 1996; Krepischi et al. 1998; Weaving et al. 2003). Interestingly, since Rett syndrome is not accompanied by neurodegeneration, attempts to express a Mecp2 transgene in postmitotic neurons have partially rescued the neurologic symptoms in both immature and mature Mecp2 mutant mice (Giacometti et al. 2007; Guy et al. 2007).
5.2.2 X-inactivation in Other Diseases
Similarly, the relation between skewed X-inactivation and phenotype severity in X-linked disease has been shown in a number of X-linked cutaneous genetic diseases such as incontinentia pigmenti, which is associated with characteristic patterns of lines and swirls appearing on the patient’s body called Blaschko’s lines (Happle 2006; Sun and Tsao 2008). Blaschko’s lines are believed to indicate the migration path of ectodermal skin cells during development illuminated by female cells containing a mix of wild-type and mutant Xi (Happle and Frosch 1985). For example, incontinentia pigmenti caused by heterozygous mutation in X-linked NFkB essential modulator (NEMO) (Smahi et al. 2000) commonly develops vesicles that later progress to verrucous and finally to hyperpigmentation in Blaschko’s lines across the trunk. While phenotypic outcome varies in incontinentia pigmenti patients, skewed X-inactivation has been reported in females with heterozygous NEMO mutation (Martinez-Pomar et al. 2005), implicating the correlation between skewed X-inactivation and phenotypic severity of X-linked disease. Skewed X-inactivation is also implicated in X-linked disease conditions such as autoimmune deficiency (Puck et al. 1987), Duchenne muscular dystrophy (Yoshioka et al. 1998; Viggiano et al. 2013), and cancers (Kristiansen et al. 2002, 2005). Clarification of the molecular mechanism that affects probability of skewing toward either X chromosome could help us to develop therapeutic approaches and potential drug targets to improve the condition of these patients.
6 Therapeutic and Diagnostic Applications Using X-inactivation
Characteristic features of Xist RNA-mediated X-inactivation could potentially be harnessed for clinical purposes. Interestingly, the long-range inactivation potential of Xist RNA has already been employed to suppress the extra chromosome 21 in iPS cells established from cells of Down syndrome patients (Jiang et al. 2013). The detrimental condition in Down syndrome arises as a result of three copies of chromosome 21. The manifestations of such an aberration lead to numerous birth defects, stunted growth, reduced intellectual abilities, mental retardation, congenital heart defects, and many physical abnormalities (Mégarbané et al. 2009; Gardiner 2010). To correct gene dosage of chromosome 21, zinc finger nucleases (ZFN) was used to insert an inducible XIST into the DYRK1A locus on one of the three chromosome 21s of iPS cells derived from a Down syndrome patient. Subsequently, the XIST transgene initiated accumulation of the repressive histone markers H3K27me3, H4K20me1, and H2AK119ub1 onto the modified chromosome. This action was associated with hypermethylation of the CpG islands at promoter, and gene repression across the modified chromosome. These results suggest that genes across chromosome 21 undergo chromosome-wide gene silencing by induction of transgenic XIST similar to X-linked gene silencing on the Xi. Most importantly, XIST induction from the transgene on the extra copy of chromosome 21 rescued the defects in proliferation and neural development. A similar application could be used to develop potential therapeutics for other diseases with an extra number of chromosomes such as Edwards syndrome (Trisomy 18) and Patau syndrome (Trisomy 13).
Potential application of XIST as a biomarker has been proposed in testicular germ cell tumors (TGCTs) (Kawakami et al. 2004a). Plasma samples obtained from patients with TGCTs showed hypomethylation of the 5′-end of CpG sites in XIST, while somatic cells showed complete methylation through the CpG sites. The XIST gene is silenced on the Xa in males and usually methylated at its 5′-end. Detection of unmethylated XIST would be due to the extranumerical X with partial inactivation in TGCT patients (Kawakami et al. 2003). Therefore, the methylation profile of XIST might also be used as a potential biomarker for diagnosis of male patients suffering from TGCTs. More recently, advances in the use of Xist expression have led to its potential as a biomarker attributing to its high levels in the urine of patients with membranous nephropathy (MN). MN is an autoimmune-induced glomerular nephritis and the most common cause of nephrotic syndrome in humans (Huang et al. 2014). The primary experiments carried out in MN model mice showed significant upregulation of Xist and long noncoding gene Neat1 in tubular epithelial and glomerular cells. Interestingly, Xist expression levels detected in urine but not serum of MN mice are strongly correlated with the severity of MN. Significantly, this finding could be applied to identify human patients developing different types of glomerular nephritis, particularly as upregulation levels of XIST were detected in the urine, but not in serum samples, of human patients confirmed to have glomerular nephritis. Thus, XIST is a potential noninvasive biomarker to detect this disease.
There is support for examining the validity of underutilized molecular markers for diagnostic purposes. In the most recently released data for the GENCODE project (version October 22, 2014, http://www.gencodegenes.org), the number of long noncoding genes in humans is nearly comparable with the number of conventional protein-coding genes and outnumbers miRNA (Henry and Hayes 2012; Hayes et al. 2014). This offers a new variety of possible diagnostic biomarkers to investigate. Thus, combining the expression profiles of several lncRNAs, including XIST, might allow us to develop better and more reliable diagnostic tools in the future.
7 Conclusions and Future Perspectives
The discovery of novel lncRNAs and their varied functions is emerging at a high rate owing to the advancement of novel techniques used to detect and investigate them. Thus, our understanding of abundant lncRNAs has increased over the decades. In this chapter, we describe lncRNAs residing within the Xic in mice and implicated the role of X-inactivation in the initiation and progression of diverse disease conditions. To further understand the functional mechanism of lncRNAs in a variety of physiological processes, identification of the protein partners in the ribonucleoprotein complex would be inevitable. Recent introduction of techniques such as CHART, RAP, ChIRP (chromatin isolation of RNA purification), RIP-seq (RNA immunoprecipitation-sequencing), and CLIP (crosslinking and immunoprecipitation) help our understanding of the molecular interaction and function between lncRNA and protein partners (Hafner et al. 2010; König et al. 2010; Chu et al. 2011; Simon et al. 2011). Growing evidence linking pathological conditions and developmental anomalies to lncRNAs is emerging, especially in cancer development and progression. Thus, lncRNAs could become valuable therapeutic targets and promote the development of rapid diagnostic tools. Future studies into X-inactivation, a paradigm of lncRNA-mediated gene regulation, will provide additional insight to the molecular mechanisms behind lncRNA function, which will in turn further contribute to lncRNA research and its clinical and diagnostic applications.
Abbreviations
- lncRNA:
-
Long noncoding RNA
- Xist :
-
X-inactive specific transcript
- Xic:
-
X-inactivation center
- ES cells:
-
Embryonic stem cells
- H3K27me3:
-
Histone H3 tri-methylated lysine 27
- LINE:
-
Long interspersed nuclear element
- YY1:
-
Yin Yang 1
- hnRNP U:
-
Heterogenous nuclear ribonucleoprotein U
- Xi:
-
Inactive X chromosome
- Xa:
-
Active X chromosome
- MECP2:
-
Methyl-CpG binding protein 2
- MPN/MDS:
-
Myeloproliferative neoplasm and myelodysplastic syndrome
- CTCF:
-
11 Zinc finger protein/CCCTF binding factor
- hiPSCs:
-
Human induced pluripotent stem cells
References
Amir RE, Van den Veyver IB, Wan M et al (1999) Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet 23:185–188. doi:10.1038/13810
Anguera MC, Ma W, Clift D et al (2011) Tsx produces a long noncoding RNA and has general functions in the germline, stem cells, and brain. PLoS Genet 7:e1002248. doi:10.1371/journal.pgen.1002248
Anguera MC, Sadreyev R, Zhang Z et al (2012) Molecular signatures of human induced pluripotent stem cells highlight sex differences and cancer genes. Cell Stem Cell 11:75–90. doi:10.1016/j.stem.2012.03.008
Belmont JW (1996) Genetic control of X inactivation and processes leading to X-inactivation skewing. Am J Hum Genet 58:1101–1108
Bittel DC, Theodoro MF, Kibiryeva N et al (2008) Comparison of X-chromosome inactivation patterns in multiple tissues from human females. J Med Genet 45:309–313. doi:10.1136/jmg.2007.055244
Blewitt ME, Gendrel A-V, Pang Z et al (2008) SmcHD1, containing a structural-maintenance-of-chromosomes hinge domain, has a critical role in X inactivation. Nat Genet 40:663–669. doi:10.1038/ng.142
Bock C, Kiskinis E, Verstappen G et al (2011) Reference maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell 144:439–452. doi:10.1016/j.cell.2010.12.032
Brockdorff N, Ashworth A, Kay GF et al (1991) Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature 351:329–331. doi:10.1038/351329a0
Brown CJ, Ballabio A, Rupert JL et al (1991a) A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349:38–44. doi:10.1038/349038a0
Brown CJ, Lafreniere RG, Powers VE et al (1991b) Localization of the X inactivation centre on the human X chromosome in Xq13. Nature 349:82–84. doi:10.1038/349082a0
Brown CJ, Willard HF (1994) The human X-inactivation centre is not required for maintenance of X-chromosome inactivation. Nature 368:154–156. doi:10.1038/368154a0
Brown SD (1991) XIST and the mapping of the X chromosome inactivation centre. BioEssays 13:607–612. doi:10.1002/bies.950131112
Camus P, Abbadi N, Perrier MC et al (1996) X chromosome inactivation in 30 girls with Rett syndrome: analysis using the probe. Hum Genet 97:247–250
Carninci P, Kasukawa T, Katayama S et al (2005) The transcriptional landscape of the mammalian genome. Science 309:1559–1563. doi:10.1126/science.1112014
Carrel L, Willard HF (2005) X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 434:400–404. doi:10.1038/nature03479
Chadwick BP, Willard HF (2004) Multiple spatially distinct types of facultative heterochromatin on the human inactive X chromosome. Proc Natl Acad Sci USA 101:17450–17455. doi:10.1073/pnas.0408021101
Chaligné R, Heard E (2014) X-chromosome inactivation in development and cancer. FEBS Lett 588:2514–2522. doi:10.1016/j.febslet.2014.06.023
Chaumeil J, Le Baccon P, Wutz A, Heard E (2006) A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev 20:2223–2237. doi:10.1101/gad.380906
Chaumeil J, Okamoto I, Guggiari M, Heard E (2002) Integrated kinetics of X chromosome inactivation in differentiating embryonic stem cells. Cytogenet Genome Res 99:75–84
Chow JC, Brown CJ, Hall LL et al (2003) Characterization of expression at the human XIST locus in somatic, embryonal carcinoma, and transgenic cell lines. Genomics 82:309–322
Chow JC, Ciaudo C, Fazzari MJ et al (2010) LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell 141:956–969. doi:10.1016/j.cell.2010.04.042
Chow JC, Hall LL, Baldry SEL et al (2007) Inducible XIST-dependent X-chromosome inactivation in human somatic cells is reversible. Proc Natl Acad Sci USA 104:10104–10109. doi:10.1073/pnas.0610946104
Chu C, Chang HY, Qu K et al (2011) Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell 44:667–678. doi:10.1016/j.molcel.2011.08.027
Chureau C, Chantalat S, Romito A et al (2011) Ftx is a non-coding RNA which affects Xist expression and chromatin structure within the X-inactivation center region. Hum Mol Genet 20:705–718. doi:10.1093/hmg/ddq516
Chureau C, Prissette M, Bourdet A et al (2002) Comparative sequence analysis of the X-inactivation center region in mouse, human, and bovine. Genome Res 12:894–908. doi:10.1101/gr.152902
Clemson CM, Willard HF, McNeil JA, Lawrence JB (1996) XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol 132:259–275
Cohen DE, Davidow LS, Erwin JA et al (2007) The DXPas34 repeat regulates random and imprinted X inactivation. Dev Cell 12:57–71. doi:10.1016/j.devcel.2006.11.014
Costanzi C, Pehrson J (1998) Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature 393:599–601
Csankovszki G, Nagy A, Jaenisch R (2001) Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J Cell Biol 153:773–784
Csankovszki G, Panning B, Bates B et al (1999) Conditional deletion of Xist disrupts histone macroH2A localization but not maintenance of X inactivation. Nat Genet 22:323–324. doi:10.1038/11887
de Napoles M, Mermoud JE, Wakao R et al (2004) Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell 7:663–676. doi:10.1016/j.devcel.2004.10.005
Dvash T, Fan G (2009) Epigenetic regulation of X-inactivation in human embryonic stem cells. Epigenetics (official journal of the DNA Methylation Society) 4:19–22
Engreitz JM, Pandya-Jones A, Mcdonel P et al (2013) The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 341:1237973. doi:10.1126/science.1237973
Fang J, Chen T, Chadwick BP et al (2004) Ring1b-mediated H2A ubiquitination associates with inactive X chromosomes and is involved in initiation of X inactivation. J Biol Chem 279:52812–52815. doi:10.1074/jbc.C400493200
Gafni O, Weinberger L, Mansour AA et al (2013) Derivation of novel human ground state naive pluripotent stem cells. Nature 504:282–286. doi:10.1038/nature12745
Gardiner KJ (2010) Molecular basis of pharmacotherapies for cognition in down syndrome. Trends Pharmacol Sci 31:66–73. doi:10.1016/j.tips.2009.10.010
Gendrel A-V, Apedaile A, Coker H et al (2012) Smchd1-dependent and -independent pathways determine developmental dynamics of CpG island methylation on the inactive X chromosome. Dev Cell 23:265–279. doi:10.1016/j.devcel.2012.06.011
Giacometti E, Luikenhuis S, Beard C, Jaenisch R (2007) Partial rescue of MeCP2 deficiency by postnatal activation of MeCP2. Proc Natl Acad Sci USA 104:1931–1936. doi:10.1073/pnas.0610593104
Giorda R, Bonaglia MC, Milani G et al (2008) Molecular and cytogenetic analysis of the spreading of X inactivation in a girl with microcephaly, mild dysmorphic features and t(X;5)(q22.1;q31.1). Eur J Hum Genet 16:897–905. doi:10.1038/ejhg.2008.28
Gore A, Li Z, Fung H-L et al (2011) Somatic coding mutations in human induced pluripotent stem cells. Nature 471:63–67. doi:10.1038/nature09805
Groth KA, Skakkebæk A, Høst C et al (2013) Clinical review: Klinefelter syndrome–a clinical update. J Clin Endocrinol Metab 98:20–30. doi:10.1210/jc.2012-2382
Guy J, Gan J, Selfridge J et al (2007) Reversal of neurological defects in a mouse model of Rett syndrome. Science 315:1143–1147. doi:10.1126/science.1138389
Hafner M, Landthaler M, Burger L et al (2010) PAR-CliP–a method to identify transcriptome-wide the binding sites of RNA binding proteins. J Vis Exp. doi:10.3791/2034
Happle R (2006) X-chromosome inactivation: role in skin disease expression. Acta Paediatr 95:16–23. doi:10.1111/j.1651-2227.2006.tb02384.x
Happle R, Frosch PJ (1985) Manifestation of the lines of Blaschko in women heterozygous for X-linked hypohidrotic ectodermal dysplasia. Clin Genet 27:468–471
Hasegawa Y, Brockdorff N, Kawano S et al (2010) The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev Cell 19:469–476. doi:10.1016/j.devcel.2010.08.006
Hatakeyama C, Anderson CL, Beever CL et al (2004) The dynamics of X-inactivation skewing as women age. Clin Genet 66:327–332. doi:10.1111/j.1399-0004.2004.00310.x
Hayes J, Peruzzi PP, Lawler S (2014) MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med 20:460–469. doi:10.1016/j.molmed.2014.06.005
Heard E, Disteche CM (2006) Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev 20:1848–1867. doi:10.1101/gad.1422906
Heard E, Kress C, Mongelard F et al (1996) Transgenic mice carrying an Xist-containing YAC. Hum Mol Genet 5:441–450
Heard E, Rougeulle C, Arnaud D et al (2001) Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell 107:727–738
Henry NL, Hayes DF (2012) Cancer biomarkers. Molecular Oncology 6:140–146. doi:10.1016/j.molonc.2012.01.010
Huang Y-S, Hsieh H-Y, Shih H-M et al (2014) Urinary Xist is a potential biomarker for membranous nephropathy. Biochem Biophys Res Commun 452:415–421. doi:10.1016/j.bbrc.2014.08.077
Huynh KD, Lee JT (2003) Inheritance of a pre-inactivated paternal X chromosome in early mouse embryos. Nature 426:857–862
Jeon Y, Lee JT (2011) YY1 tethers Xist RNA to the inactive X nucleation center. Cell 146:119–133. doi:10.1016/j.cell.2011.06.026
Jiang J, Brown CJ, Jing Y et al (2013) Translating dosage compensation to trisomy 21. Nature. doi:10.1038/nature12394
Johnston CM, Newall AET, Brockdorff N, Nesterova TB (2002) Enox, a novel gene that maps 10 kb upstream of Xist and partially escapes X inactivation. Genomics 80:236–244
Kawakami T, Okamoto K, Ogawa O, Okada Y (2004a) XIST unmethylated DNA fragments in male-derived plasma as a tumour marker for testicular cancer. Lancet 363:40–42. doi:10.1016/S0140-6736(03)15170-7
Kawakami T, Okamoto K, Sugihara H et al (2003) The roles of supernumerical X chromosomes and XIST expression in testicular germ cell tumors. J Urol 169:1546–1552. doi:10.1097/01.ju.0000044927.23323.5a
Kawakami T, Zhang C, Taniguchi T et al (2004b) Characterization of loss-of-inactive X in Klinefelter syndrome and female-derived cancer cells. Oncogene 23:6163–6169. doi:10.1038/sj.onc.1207808
Keer JT, Hamvas RM, Brockdorff N et al (1990) Genetic mapping in the region of the mouse X-inactivation center. Genomics 7:566–572
Keohane AM, O’neill LP, Belyaev ND et al (1996) X-inactivation and histone H4 acetylation in embryonic stem cells. Dev Biol 180:618–630. doi:10.1006/dbio.1996.0333
Kim K, Doi A, Wen B (2010) Epigenetic memory in induced pluripotent stem cells. Nature 467:285–290. doi:10.1038/nature09342
Kobayashi S, Totoki Y, Soma M et al (2013) Identification of an imprinted gene cluster in the X-inactivation center. PLoS ONE 8:e71222. doi:10.1371/journal.pone.0071222
Kohlmaier A, Savarese F, Lachner M et al (2004) A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol 2:e171. doi:10.1371/journal.pbio.0020171
König J, Zarnack K, Rot G et al (2010) iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol 17:909–915. doi:10.1038/nsmb.1838
Krepischi AC, Kok F, Otto PG (1998) X chromosome-inactivation patterns in patients with Rett syndrome. Hum Genet 102:319–321
Kristiansen M, Knudsen GPS, Maguire P et al (2005) High incidence of skewed X chromosome inactivation in young patients with familial non-BRCA1/BRCA2 breast cancer. J Med Genet 42:877–880. doi:10.1136/jmg.2005.032433
Kristiansen M, Langerød A, Knudsen GP et al (2002) High frequency of skewed X inactivation in young breast cancer patients. J Med Genet 39:30–33. doi:10.1136/jmg.39.1.30
Lam MTY, Li W, Rosenfeld MG, Glass CK (2014) Enhancer RNAs and regulated transcriptional programs. Trends Biochem Sci 39:170–182. doi:10.1016/j.tibs.2014.02.007
Lee JT (2000) Disruption of imprinted X inactivation by parent-of-origin effects at Tsix. Cell 103:17–27
Lee JT, Bartolomei MS (2013) X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell 152:1308–1323. doi:10.1016/j.cell.2013.02.016
Lee JT, Davidow LS, Warshawsky D (1999a) Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet 21:400–404. doi:10.1038/7734
Lee JT, Jaenisch R (1997) Long-range cis effects of ectopic X-inactivation centres on a mouse autosome. Nature 386:275–279. doi:10.1038/386275a0
Lee JT, Lu N (1999) Targeted mutagenesis of Tsix leads to nonrandom X inactivation. Cell 99:47–57
Lee JT, Lu N, Han Y (1999b) Genetic analysis of the mouse X inactivation center defines an 80-kb multifunction domain. Proc Natl Acad Sci U S A 96:3836–3841
Lee JT, Strauss WM, Dausman J, Jaenisch R (1996) A 450 kb transgene displays properties of the mammalian X-inactivation center. Cell 86:83–94
Lessing D, Anguera MC, Lee JT (2013) X chromosome inactivation and epigenetic responses to cellular reprogramming. Annu Rev Genomics Hum Genet. doi:10.1146/annurev-genom-091212-153530
Liang F, Holt I, Pertea G et al (2000) Gene index analysis of the human genome estimates approximately 120,000 genes. Nat Genet 25:239–240. doi:10.1038/76126
Liao DJ, Du Q-Q, Yu BW et al (2003) Novel perspective: focusing on the X chromosome in reproductive cancers. Cancer Invest 21:641–658
Lieberman-Aiden E, van Berkum NL, Williams L et al (2009) Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326:289–293. doi:10.1126/science.1181369
Lyon MF (1961) Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190:372–373
Lyon MF (2000) LINE-1 elements and X chromosome inactivation: a function for “junk” DNA? Proc Natl Acad Sci USA 97:6248–6249
Maass PG, Luft FC, Bahring S (2014) Long non-coding RNA in health and disease. J Mol Med 92:337–346. doi:10.1007/s00109-014-1131-8
Mak W, Nesterova TB, de Napoles M et al (2004) Reactivation of the paternal X chromosome in early mouse embryos. Science 303:666–669. doi:10.1126/science.1092674
Mak W, Silva J, Baxter J et al (2002) Mitotically stable association of polycomb group proteins eed and enx1 with the inactive X chromosome in trophoblast stem cells. Curr Biol 12:1016–1020
Marahrens Y, Panning B, Dausman J et al (1997) Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev 11:156–166
Martinez-Pomar N, Munoz-Saa I, Heine-Suner D et al (2005) A new mutation in exon 7 of NEMO gene: late skewed X-chromosome inactivation in an incontinentia pigmenti female patient with immunodeficiency. Hum Genet 118:458–465. doi:10.1007/s00439-005-0068-y
Mégarbané A, Ravel A, Mircher C et al (2009) The 50th anniversary of the discovery of trisomy 21: the past, present, and future of research and treatment of Down syndrome. Genet Med 11:611–616. doi:10.1097/GIM.0b013e3181b2e34c
Migeon BR (2006) The role of X inactivation and cellular mosaicism in women’s health and sex-specific diseases. JAMA 295:1428–1433. doi:10.1001/jama.295.12.1428
Minks J, Robinson WP, Brown CJ (2008) A skewed view of X chromosome inactivation. J Clin Invest 118:20–23. doi:10.1172/JCI34470
Monk M, Harper MI (1979) Sequential X chromosome inactivation coupled with cellular differentiation in early mouse embryos. Nature 281:311–313
Natoli G, Andrau J-C (2012) Noncoding transcription at enhancers: general principles and functional models. Annu Rev Genet 46:1–19. doi:10.1146/annurev-genet-110711-155459
Navarro P (2005) Tsix transcription across the Xist gene alters chromatin conformation without affecting Xist transcription: implications for X-chromosome inactivation. Genes Dev 19:1474–1484. doi:10.1101/gad.341105
Norris DP, Brockdorff N, Rastan S (1991) Methylation status of CpG-rich islands on active and inactive mouse X chromosomes. Mamm Genome 1:78–83
Nozawa R-S, Nagao K, Igami K-T et al (2013) Human inactive X chromosome is compacted through a PRC2-independent SMCHD1-HBiX1 pathway. Nat Struct Mol Biol. doi:10.1038/nsmb.2532
Ogawa Y, Lee JT (2003) Xite, X-inactivation intergenic transcription elements that regulate the probability of choice. Mol Cell 11:731–743
Ohhata T, Hoki Y, Sasaki H, Sado T (2008) Crucial role of antisense transcription across the Xist promoter in Tsix-mediated Xist chromatin modification. Development 135:227–235. doi:10.1242/dev.008490
Ohhata T, Wutz A (2013) Reactivation of the inactive X chromosome in development and reprogramming. Cell Mol Life Sci 70:2443–2461. doi:10.1007/s00018-012-1174-3
Okamoto I, Otte AP, Allis CD et al (2004) Epigenetic dynamics of imprinted X inactivation during early mouse development. Science 303:644–649. doi:10.1126/science.1092727
Orazi A, Germing U (2008) The myelodysplastic/myeloproliferative neoplasms: myeloproliferative diseases with dysplastic features. Leukemia 22:1308–1319. doi:10.1038/leu.2008.119
Pageau G, Hall LL, Ganesan S et al (2007) The disappearing Barr body in breast and ovarian cancers. Nat Rev Cancer 7:628–633. doi:10.1038/nrc2172
Payer B, Lee JT (2008) X chromosome dosage compensation: how mammals keep the balance. Annu Rev Genet 42:733–772. doi:10.1146/annurev.genet.42.110807.091711
Penny GD, Kay GF, Sheardown SA et al (1996) Requirement for Xist in X chromosome inactivation. Nature 379:131–137. doi:10.1038/379131a0
Peschansky VJ, Wahlestedt C (2014) Non-coding RNAs as direct and indirect modulators of epigenetic regulation. Epigenetics (official journal of the DNA Methylation Society) 9:3–12. doi:10.4161/epi.27473
Pinter SF, Sadreyev R, Yildirim E et al (2012) Spreading of X chromosome inactivation via a hierarchy of defined Polycomb stations. Genome Res 22:1864–1876. doi:10.1101/gr.133751.111
Plath K, Fang J, Mlynarczyk-Evans SK et al (2003) Role of histone H3 lysine 27 methylation in X inactivation. Science 300:131–135. doi:10.1126/science.1084274
Plath K, Talbot D, Hamer KM et al (2004) Developmentally regulated alterations in Polycomb repressive complex 1 proteins on the inactive X chromosome. J Cell Biol 167:1025–1035. doi:10.1083/jcb.200409026
Puck JM, Nussbaum RL, Conley ME (1987) Carrier detection in X-linked severe combined immunodeficiency based on patterns of X chromosome inactivation. J Clin Invest 79:1395–1400. doi:10.1172/JCI112967
Pullirsch D, Härtel R, Kishimoto H et al (2010) The Trithorax group protein Ash2l and Saf-A are recruited to the inactive X chromosome at the onset of stable X inactivation. Development 137:935–943. doi:10.1242/dev.035956
Rastan S (1983) Non-random X-chromosome inactivation in mouse X-autosome translocation embryos–location of the inactivation centre. J Embryol Exp Morphol 78:1–22. doi:10.1016/s0065-2660(08)60074-7
Rastan S, Brown SD (1990) The search for the mouse X-chromosome inactivation centre. Genet Res 56:99–106
Rastan S, Robertson E (1985) X-chromosome deletions in embryo-derived (EK) cell lines associated with lack of X-chromosome inactivation. J Embryol Exp Morphol 90:379–388
Richardson A, Wang Z, de Nicolo A et al (2006) X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell 9:121–132
Rogner UC, Wilke K, Steck E et al (1995) The melanoma antigen gene (MAGE) family is clustered in the chromosomal band Xq28. Genomics 29:725–731. doi:10.1006/geno.1995.9945
Sado T, Brockdorff N (2013) Advances in understanding chromosome silencing by the long non-coding RNA Xist. Philos Trans R Soc Lond B Biol Sci 368:20110325. doi:10.1098/rstb.2011.0325
Sado T, Fenner MH, Tan SS et al (2000) X inactivation in the mouse embryo deficient for Dnmt1: distinct effect of hypomethylation on imprinted and random X inactivation. Dev Biol 225:294–303. doi:10.1006/dbio.2000.9823
Sado T, Hoki Y, Sasaki H (2005) Tsix silences Xist through modification of chromatin structure. Dev Cell 9:159–165. doi:10.1016/j.devcel.2005.05.015
Sado T, Wang Z, Sasaki H, Li E (2001) Regulation of imprinted X-chromosome inactivation in mice by Tsix. Development 128:1275–1286
Sharp A, Robinson D, Jacobs P (2000) Age-and tissue-specific variation of X chromosome inactivation ratios in normal women. Hum Genet 107:343–349. doi:10.1007/s004390000382
Sharp AJ, Stathaki E, Migliavacca E et al (2011) DNA methylation profiles of human active and inactive X chromosomes. Genome Res 21:1592–1600. doi:10.1101/gr.112680.110
Silva J, Mak W, Zvetkova I et al (2003) Establishment of histone H3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev Cell 4:481–495. doi:10.1016/S1534-5807(03)00068-6
Silva SS, Rowntree RK, Mekhoubad S, Lee JT (2008) X-chromosome inactivation and epigenetic fluidity in human embryonic stem cells. Proc Natl Acad Sci USA 105:4820–4825. doi:10.1073/pnas.0712136105
Simmler MC, Cunningham DB, Clerc P et al (1996) A 94 kb genomic sequence 3’ to the murine Xist gene reveals an AT rich region containing a new testis specific gene Tsx. Hum Mol Genet 5:1713–1726
Simon MD, Pinter SF, Fang R et al (2013) High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature 504:465–469. doi:10.1038/nature12719
Simon MD, Wang CI, Kharchenko PV et al (2011) The genomic binding sites of a noncoding RNA. Proc Natl Acad Sci 108:20497–20502. doi:10.1073/pnas.1113536108
Sirchia SM, Ramoscelli L, Grati FR et al (2005) Loss of the inactive X chromosome and replication of the active X in BRCA1-defective and wild-type breast cancer cells. Cancer Res 65:2139–2146. doi:10.1158/0008-5472.CAN-04-3465
Smahi A, Courtois G, Vabres P et al (2000) Genomic rearrangement in NEMO impairs NF-kappaB activation and is a cause of incontinentia pigmenti. The international incontinentia pigmenti (IP) consortium. Nature 405:466–472. doi:10.1038/35013114
Soma M, Fujihara Y, Okabe M et al (2014) Ftx is dispensable for imprinted X-chromosome inactivation in preimplantation mouse embryos. Scientific Reports 4:5181. doi:10.1038/srep05181
Spatz A, Borg C, Feunteun J (2004) X-chromosome genetics and human cancer. Nat Rev Cancer 4:617–629. doi:10.1038/nrc1413
Splinter E, de Wit E, Nora EP et al (2011) The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes Dev 25:1371–1383. doi:10.1101/gad.633311
Stavropoulos N, Rowntree RK, Lee JT (2005) Identification of developmentally specific enhancers for Tsix in the regulation of X chromosome inactivation. Mol Cell Biol 25:2757–2769. doi:10.1128/MCB.25.7.2757-2769.2005
Sun BK, Deaton A, Lee JT (2006) A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol Cell 21:617–628
Sun BK, Tsao H (2008) X-chromosome inactivation and skin disease. J Investig Dermatol 128:2753–2759. doi:10.1038/jid.2008.145
Sun S, Del Rosario BC, Szanto A et al (2013) Jpx RNA activates Xist by evicting CTCF. Cell 153:1537–1551. doi:10.1016/j.cell.2013.05.028
Sybert VP, McCauley E (2004) Turner’s syndrome. N Engl J Med 351:1227–1238. doi:10.1056/NEJMra030360
Takagi N (1980) Primary and secondary nonrandom X chromosome inactivation in early female mouse embryos carrying Searle’s translocation T(X; 16)16H. Chromosoma 81:439–459
Takagi N, Sasaki M (1975) Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature 256:640–642
Takahashi K, Tanabe K, Ohnuki M et al (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131:861–872. doi:10.1016/j.cell.2007.11.019
Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126:663–676
Tan SS, Williams EA, Tam PP (1993) X-chromosome inactivation occurs at different times in different tissues of the post-implantation mouse embryo. Nat Genet 3:170–174. doi:10.1038/ng0293-170
Thomas GA, Williams D, Williams ED (1988) The demonstration of tissue clonality by X-linked enzyme histochemistry. J Pathol 155:101–108. doi:10.1002/path.1711550205
Tian D, Sun S, Lee JT (2010) The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell 143:390–403. doi:10.1016/j.cell.2010.09.049
Tomoda K, Takahashi K, Leung K et al (2012) Derivation conditions impact X-inactivation status in female human induced pluripotent stem cells. Cell Stem Cell 11:91–99. doi:10.1016/j.stem.2012.05.019
Torres EM, Williams BR, Amon A (2008) Aneuploidy: cells losing their balance. Genetics 179:737–746. doi:10.1534/genetics.108.090878
Van Echten-Arends J, Coonen E, Reuters B et al (2013) Preimplantation genetic diagnosis for X;autosome translocations: lessons from a case of misdiagnosis. Hum Reprod 28:3141–3145. doi:10.1093/humrep/det362
Viggiano E, Picillo E, Cirillo A, Politano L (2013) Comparison of X-chromosome inactivation in Duchenne muscle/myocardium-manifesting carriers, non-manifesting carriers and related daughters. Clin Genet 84:265–270. doi:10.1111/cge.12048
Vigneau S, Augui S, Navarro P et al (2006) An essential role for the DXPas34 tandem repeat and Tsix transcription in the counting process of X chromosome inactivation. Proc Natl Acad Sci USA 103:7390–7395. doi:10.1073/pnas.0602381103
Wang KC, Chang HY (2011) Molecular mechanisms of long noncoding RNAs. Mol Cell 43:904–914. doi:10.1016/j.molcel.2011.08.018
Wapinski OL, Chang HY (2011) Long noncoding RNAs and human disease. Trends Cell Biol 21:354–361. doi:10.1016/j.tcb.2011.04.001
Weaving LS, Ellaway CJ, Gécz J, Christodoulou J (2005) Rett syndrome: clinical review and genetic update. J Med Genet 42:1–7. doi:10.1136/jmg.2004.027730
Weaving LS, Williamson SL, Bennetts B et al (2003) Effects ofMECP2 mutation type, location and X-inactivation in modulating Rett syndrome phenotype. Am J Med Genet 118A:103–114. doi:10.1002/ajmg.a.10053
White WM, Willard HF, van Dyke DL, Wolff DJ (1998) The spreading of X inactivation into autosomal material of an x;autosome translocation: evidence for a difference between autosomal and X-chromosomal DNA. Am J Hum Genet 63:20–28. doi:10.1086/301922
Wutz A (2011) Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nat Rev Genet 12:542–553. doi:10.1038/nrg3035
Wutz A, Jaenisch R (2000) A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell 5:695–705
Yildirim E, Kirby JE, Brown DE et al (2013) Xist RNA is a potent suppressor of hematologic cancer in mice. Cell 152:727–742. doi:10.1016/j.cell.2013.01.034
Yoshioka M, Yorifuji T, Mituyoshi I (1998) Skewed X inactivation in manifesting carriers of Duchenne muscular dystrophy. Clin Genet 53:102–107
Zhang L-F, Huynh KD, Lee JT (2007) Perinucleolar targeting of the inactive X during S phase: evidence for a role in the maintenance of silencing. Cell 129:693–706. doi:10.1016/j.cell.2007.03.036
Zhao J, Sun BK, Erwin JA et al (2008) Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322:750–756. doi:10.1126/science.1163045
Acknowledgments
The authors wish to thank members of the Ogawa Lab for helpful comments and Serenity Curtis for editing the manuscript. This work was supported by grant from the NIH (RO1-GM102184) and the March of Dimes Research Foundation (#6-FY12-337) to Y.O.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Charles Richard, J.L., Ogawa, Y. (2015). Understanding the Complex Circuitry of lncRNAs at the X-inactivation Center and Its Implications in Disease Conditions. In: Morris, K. (eds) Long Non-coding RNAs in Human Disease. Current Topics in Microbiology and Immunology, vol 394. Springer, Cham. https://doi.org/10.1007/82_2015_443
Download citation
DOI: https://doi.org/10.1007/82_2015_443
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-23906-4
Online ISBN: 978-3-319-23907-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)