Abstract
Population genomics is advancing our understanding of evolution, ecology, conservation, agriculture, forestry, and human health by allowing new and long-standing questions to be addressed with unprecedented power and accuracy. These advances result from plummeting costs for DNA sequencing, which makes genotyping feasible for hundreds to millions of individuals and loci, and also allows for the study of variation in gene expression, epigenetic variation, and proteins. The increased power also results from the development of innovative software, statistical approaches, and models to extract information from massive next-generation sequencing datasets. Among the most exciting developments are conceptually novel approaches that are advancing understanding about inbreeding and outbreeding depression, adaptive gene flow, population demographic history, and the genomic basis of local adaptation and speciation. Remaining challenges in applying genomics to natural and managed populations include the limited understanding and availability of validated bioinformatics pipelines for genotyping and analyzing genomic data. We also lack knowledge of best practices and general guidelines for filtering and genotyping genomic data including restriction site-associated DNA sequences (RAD), targeted DNA capture, and pooled sequencing. Finally, we emphasize the need for continued rigorous teaching of population genetics theory, so that the next generation of population genomicists can ask well-informed questions and interpret next-generation sequence datasets.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Adaptation
- Community genetics
- Conservation genetics
- Ecological genomics
- Epigenetics
- Evolutionary genomics
- Landscape genomics
- Population genetics
- Selection detection
Molecular markers have totally changed our view of nature (Schlötterer 2004).
Population genomics is a new term for a field of study that is as old as the field of genetics itself, assuming that it means the study of the amount and causes of genome-wide variability in natural populations (Charlesworth 2010).
Population genomic tools have revolutionized many aspects of biology, as detailed throughout the chapters of this volume (Hohenlohe et al. 2018).
1 Introduction
New and long-standing questions in ecology, evolution, conservation biology, and related fields can now be addressed with unprecedented power and accuracy using population genomics approaches. This power results largely from new sequencing and genotyping technologies that produce enormous amounts of data (Schlötterer 2004; Narum et al. 2013; van Dijk et al. 2018; Sedlazeck et al. 2018) but also from new statistical approaches and software (Paradis et al. 2017; Ceballos et al. 2018; Cooke and Nakagome 2018; Faria et al. 2018; Gruber et al. 2018; Hendricks et al. 2018; Knaus and Grünwald 2017; Zhang et al. 2018). These molecular and computational approaches are now within reach of many biologists in terms of costs, ease of data production, and availability of computational tools. This chapter provides an overview of the concepts and primary approaches employed to study genome-wide genetic variation in natural and managed species and populations. Some of these approaches are not yet widely used but are emerging in the literature on population genomics (Hendricks et al. 2018).
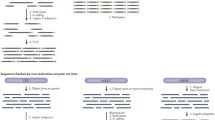
Population genomics has been broadly defined as the simultaneous study of numerous loci and genome regions to better understand the roles of evolutionary processes (such as mutation, genetic drift, gene flow, and natural selection) that influence variation across genomes and populations (Black et al. 2001; Luikart et al. 2003). This definition emphasizes understanding of locus-specific effects like selection against the background of genome-wide effects such as demography and genetic drift in order to improve assessments of adaptive evolution, the effective population size, gene flow, admixture, inbreeding and outbreeding depression, speciation, and the genomic basis of fitness (Fig. 1) (Allendorf et al. 2010; McMahon et al. 2014; Hunter et al. 2018).
Conceptual framework of main steps in a population genomics approach used to identify outlier loci under selection (or genotyping errors) and also to improve estimates of population history and demography using the selectively neutral loci. In Step 1, individuals can be sampled from different phenotypes or environments to help test for adaptive gene marker associations and to dissect the genomic basis of phenotypes, local adaptation, adaptation to captivity, artificial selection, or speciation. Step 2 requires a genetic linkage map or a physical map (Sects. 3.1 and 3.2) to localize genome regions under selection and to ensure high marker density (narrow sense approach). However many unmapped loci can be used in broad sense genomics (Figs. 3 and 4). Step 3 could employ conceptually novel approaches to identify “outlier loci” or chromosomal regions that behave unlike most other loci in the genome and therefore could be under selection or associated with phenotypic traits. Outlier loci under selection can bias estimates of neutral population genetic parameters (Step 4a) such as gene flow, effective population size, and structure. Figure modified from Luikart et al. (2003)
Hohenlohe et al. (2010a) outlined a novel conceptual framework for population genomics that emphasizes the understanding of patterns of genetic variation and evolutionary processes in all genome regions by plotting population genetic statistics across each chromosome using many mapped loci (Fig. 2; Box 1). An example of a population genomics approach is measuring a population genetic summary statistic, e.g., genomic diversity, population differentiation, or gene expression, as a continuous variable along chromosomes to help identify loci under selection, chromosomal islands of adaptive divergence, or alleles associated with a phenotypic trait (see also Fig. 3 in Luikart et al. 2003; Hohenlohe et al. 2010b; Ellegren 2014; Kardos et al. 2015b).
A population genomics perspective and conceptual framework. (A) Traditional population genetics takes data on alleles (colored bars), grouped within individuals (solid boxes) and populations (dashed boxes), and calculates summary statistics to make inferences about evolution, such as nucleotide diversity (π) and population differentiation (F ST). (B) Population genomics takes data on haplotypes within a population and calculates summary statistics as continuous variables along the length of the genome, such as π and the allele frequency spectrum (Tajima’s D). The different types of evolutionary processes leave different signatures in these distributions: (i) hard selective sweep, (ii) region linked to hard sweep, (iii) neutral expectation, (iv) balancing selection, (v) neutral expectation, and (vi) soft sweep. (C) The coalescent structure of ancestral relationships among alleles within a population also reflects these processes along the genome. (D) Given these genomic processes within a population, statistics comparing genetic variation across populations, such as F ST, can also indicate genomic patterns of selection. (E) Collapsing the genomic distribution of a statistic into a frequency distribution provides an estimate of the genome-wide average, allowing identification of statistically significant outliers (shaded regions). Reproduced with permission from Hohenlohe et al. (2010a)
Illustration of how (A) anonymous (unmapped) loci are often detected to be under directional selection (e.g., with high allele frequency differentiation, F ST) among populations and how (B) a genetic linkage map or a physical map (genome assembly) helps to localize the genome regions under selection by positioning loci (SNPs) along a chromosome or entire genome. In panel B, each color represents a different chromosome (linkage group) including the different shades of gray. Knowing the genome position of SNPs allows for multiple, often linked, SNPs to be identified that result from the same selection process and signature (e.g., high F ST), which increases our confidence that the SNPs or genome region are actually under selection and not false positives. Positional information also helps understand the number of loci or genome regions that are under selection. Further, if coding or annotated genes have also been mapped or physically located on a genome sequence, researchers can identify genes in the region of the selection signature, which represent candidate adaptive genes (e.g., Mckinney et al. 2016). Figure (A) represents a broad sense genomics approach, while (B) is narrow sense genomics. Figure modified from Garret McKinney (pers. comm., 2018)
Allendorf (2017) and Hohenlohe et al. (2018) defined population genomics as requiring a sufficient density of DNA markers to detect forces affecting any particular genomic region, e.g., genes under selection, regions of reduced recombination. Here, we provide a narrow sense definition of population genomics as the use of conceptually novel approaches to address questions intractable by traditional genetic methods by using high-density genome-wide markers (e.g., DNA, RNA, epigenetic marks) to provide high power to detect genomic regions associated with traits or evolutionary processes such as fitness, phenotypes, and selection (Box 1). This definition combines the requirement for conceptual novelty aspect from Garner et al. (2016) and Hohenlohe et al. (2010a), with the high-density marker requirement of Allendorf (2017); it also explicitly includes multiple omics approaches (transcriptomics, epigenomics, and proteomics).
Broad sense population genomics can be defined as the use of new genomics technology and numerous loci to address questions in population genetics (e.g., Shafer et al. 2015; Garner et al. 2016; Hohenlohe et al. 2018) (Box 1). We include broad sense approaches here because some are advancing understanding of genomics questions ranging from the discovery of genes underlying adaptive evolution to assessing population parameters and demography using thousands to millions of neutral markers that are often anonymous or not mapped.
Our main goals for this chapter are fourfold. First, we discuss the research topics and questions for which genomics tools are most valuable. We illustrate where genomics methods are improving our ability to address long-standing objectives and also to address previously intractable questions using conceptually novel approaches. Second, we give a brief introduction to new molecular techniques and computational approaches (including bioinformatics workflows and Bayesian methods) to help biologists understand this growing literature and to plan their projects. Third, we provide an overview of the emerging disciplines where population genomics concepts and approaches are being applied. Finally, we discuss future perspectives of applications of population genomics concepts and approaches and conclude the chapter. Throughout, we highlight the opportunities and challenges associated with population genomic analyses in studies of natural and managed populations.
Box 1 How is Narrow Sense Population Genomics Different from Broad Sense Genomics and Traditional Population Genetics?
Defining broad and narrow sense population genomics can be useful because there is often confusion among students and researchers as to what constitutes genomics and also because broad sense population genomics studies include traditional population genetic approaches and the use of more DNA markers (see Charlesworth 2010; Allendorf 2017). An example of a broad sense population genomics study would be using thousands or tens of thousands of anonymous SNPs (Fig. 3) to estimate the inbreeding coefficients of individuals using traditional parameters (e.g., individual heterozygosity; Hoffman et al. 2014; Kardos et al. 2016a), while a narrow sense study would be the mapping of runs of homozygosity (RoH) to infer recent and historical inbreeding (or population bottlenecks) (Bérénos et al. 2015; Howard et al. 2015; Palkopoulou et al. 2015; Pemberton et al. 2017; Kardos et al. 2017; Ceballos et al. 2018). The requirement for narrow sense genomics to include “conceptual novelty” and to address questions not tractable using traditional population genetics addresses the criticism of Charlesworth (2010) and of others saying that population genomics is nothing new.
A narrow sense population genomics study precisely characterizes variation at many specific (mapped) regions of the genome (Allendorf 2017). The density of markers required (see below) varies and depends on phenomena that affect gametic disequilibrium along a chromosome such as mating system (e.g., selfing versus random mating), effective population size, population subdivision, gene flow or admixture, and recombination rates (Slatkin 2008).
2 When Is Population Genomics Most Valuable?
A wide array of fundamental and novel questions can now be reliably addressed thanks to developments in population genomics (Table 1). In this section, we describe several newly invigorated avenues of research in evolutionary biology and conservation genetics. The most exciting developments of population genomics involve using novel approaches to address previously unapproachable questions such as mapping adaptive variation genome wide and resolving the genomic basis of fitness and phenotypes (Hoban et al. 2016; Hendricks et al. 2018; Hunter et al. 2018). Identifying loci underlying adaptive evolution is a long-standing goal in evolutionary biology, and doing so helps to understand the phenotypic traits, biochemical pathways, and nature of the selective forces that have resulted in the bewildering array of biodiversity.
A more common or widespread application of population genomics approaches is improving estimation of population genetic parameters and evolutionary relationships – including assessments of effective population size, population structure, phylogeography, and demography – which are largely broad sense genomics (Luikart et al. 2003). We first discuss these broader sense applications in Sect. 2.1. We then discuss exciting and previously intractable applications including mapping of adaptive genomic variation in Sects. 2.2 through 2.8.
2.1 Estimating Population Genetic Parameters with Genome-Wide Markers: Broad Sense Genomics Approaches
Genomics approaches can be used to address questions that have long been studied using traditional molecular markers such as allozymes or microsatellites (Box 1). In this section, we describe some of those population genetic questions and how genomics can be used to improve them. While traditional molecular markers provide information on a small fraction or subset of the genome, large-scale genomic data (thousands to hundreds of thousands of SNPs) provide a more complete picture of genetic parameters across the entire genome (e.g., Hohenlohe et al. 2010b; Brelsford et al. 2017).
Statistical inference can be used to estimate population genetic parameters, such as genetic diversity, effective population size, population differentiation, or phylogenetic relationships, and these population genetic metrics reflect processes that affect the genome as a whole. However, these metrics can vary tremendously across the genome, which suggests a narrow sense approach (e.g., mapped loci) is advisable. For example, genetic variation and population differentiation often vary tremendously across the genome due to variation in recombination rate, selection intensity (purifying and positive), and the mutation rate (Hohenlohe et al. 2010b).
The primary advantage of broad sense genomics is providing many more genetic markers, often by several orders of magnitude, than previous techniques, and often for similar cost and research effort. This results in the potential for much greater precision of estimates of population genetic parameters. Many more markers can also reduce bias of estimates of population genetic parameters by identifying loci under selection that often should not be used to estimate parameters requiring only neutral loci, such as gene flow, demographic history, and phylogenies. In some cases, recent genomics techniques can also be more cost-effective than traditional techniques, for instance, with the ability to simultaneously detect and genotype loci using RADseq and RAD capture (see Sect. 4) in taxa for which microsatellite or other loci have not previously been developed (Andrews et al. 2016).
In population genomics studies, genome-wide estimates are often considered as the background against which outliers reflect adaptive or functionally important loci (Fig. 1; Luikart et al. 2003), and detection of these loci is central to narrow sense population genomics as described in the sections below (see also Hohenlohe et al. 2018 this volume). The genome-wide background, estimated by either traditional genetic or genomics techniques, is often interpreted to reflect selectively neutral processes. But it is important to remember that the effects of selection and genotype-phenotype relationships are pervasive across the genome due to processes, such as hitchhiking (Maynard Smith and Haigh 1974), background selection (Charlesworth et al. 1993), or isolation by adaptation (Nosil et al. 2008; Corbett-Detig et al. 2015). Whether techniques tend to avoid coding regions (e.g., microsatellites), focus on them (e.g., exon capture, RAD capture with targets in or near genes), or sample randomly across the genome (e.g., RADseq), it can be treacherous to interpret genome-wide patterns as solely reflecting “neutral” processes.
2.1.1 Genetic Variation and Effective Population Size
A central quantity in population genetics is the amount of genetic variation present in a population. This can be quantified in several ways, including expected heterozygosity (H e) or nucleotide diversity (π), which can be estimated from genome-wide SNP data using many analysis programs, such as PLINK (Purcell et al. 2007). Genome-wide genetic variation is the result of multiple interacting processes, including mutation, genetic drift, selection, and population structure, that affect the genome as a whole.
The amount of genetic variation in a population is closely related to the effective population size (N e), which is often a focus of population genomics studies, particularly those relevant to conservation (e.g., Hare et al. 2011; Cammen et al. 2018). While there are several ways to define N e, a common definition derives from the amount of genetic drift in a local population relative to an idealized Wright-Fisher model (Charlesworth 2009; Allendorf et al. 2013). The most direct way to estimate the rate of genetic drift and N e is with temporal genetic samples from a local population, which provide measurements of changes in allele frequencies over time (Wang 2005; Luikart et al. 2010). Often, however, multiple samples over time are not available from natural populations, so other estimation techniques are required.
Random genetic drift due to small population size also leads to nonrandom associations between alleles from different loci, known as gametic disequilibrium (GD). GD provides the basis for methods to estimate N e from a single genetic sample collected at one time point, such as program LDNe in NeEstimator. LDNe requires independent loci such as those on different chromosomes (Do et al. 2014). With the large number of markers available from genomic data, it is likely that physically linked loci (those on the same chromosome) are included. Physically linked loci can downwardly bias estimates of N e by increasing GD (Waples and Do 2010). If markers can be mapped to a reference genome assembly or linkage map, one locus in physically linked pairs of loci can be removed (e.g., as done by Larson et al. (2017)) or a general correction for the number of chromosomes can be applied (Waples et al. 2016). An alternative class of methods uses coalescent-based inference of N e; Nunziata and Weisrock (2018) found that GD methods require more individuals (e.g., n > 30), while coalescent methods require fewer individuals (e.g., n = 15) but more SNP markers (25,000–50,000). Estimates of N e from different methods can vary, and knowledge of population demography or temporal data can improve estimates considerably (Gilbert and Whitlock 2015).
2.1.2 Population Structure and Phylogeography
Populations exist across space, and the spatial distribution of genetic variation is an important focus of population genetics. Quantifying population structure and levels of genetic differentiation among populations (e.g., estimating the parameter F ST) has been tractable with traditional population genetic tools, but again genomic techniques provide greater statistical power and precision for estimating parameters (Hohenlohe et al. 2018 in this volume). Furthermore, the number of markers from genomic data can allow for estimates from fewer individual samples; for instance, Nazareno et al. (2017) report consistent estimates of F ST when using as few as two individuals, genotyped at over 1,500 SNPs.
Many analytical tools are well-suited for assessing and visualizing population structure from large genomic SNP datasets, such as principal components analysis and Bayesian clustering methods, and applying multiple techniques to a single dataset can help reveal important patterns (Fig. 4). When applied to genome-wide data, these approaches illustrate the results of processes that affect the genome as a whole, such as population size and migration rates. In a landscape genetics framework, a combination of genomic and landscape data can identify landscape features associated with variation in dispersal patterns (see Johnson et al. 2018a, b in this volume for a review). Interpolating and mapping genetic similarity across landscapes can reveal areas of high versus low gene flow, e.g., using the estimated effective migration surface (EEMS) approach of Petkova et al. (2016). Recent genomics techniques also provide new power for understanding the relationship between landscape variables and functional genetic variation at specific loci, such as genes; Balkenhol et al. (2017) in this volume review this field of landscape genomics.
Two methods for visualizing patterns of genetic differentiation among populations or closely related taxa: (a) principal components analysis and (b) Bayesian clustering analysis. Here these methods are applied to data from a 48,000 SNP genotyping array from wolves and their relatives. Reproduced with permission from VonHoldt et al. (2011)
2.1.3 Demographic History
A goal of population genomics studies that was considerably less tractable with traditional genetic techniques is a detailed reconstruction of historical demographic patterns, including changes in effective population size and migration rates, using genetic data sampled only from the contemporary populations. A number of techniques have been developed for demographic reconstruction from genetic or genomic data, such as approximate Bayesian computation (ABC; Boitard et al. 2016; Elleouet and Aitken 2018), sequential Markovian coalescent methods (Terhorst et al. 2017), and site frequency spectrum methods (Gutenkunst et al. 2009). See Salmona et al. (2017) in this volume as well as Beichman et al. (2017) for detailed reviews.
As an example, Duranton et al. (2018) estimated the parameters of a demographic model of two populations of European sea bass (Dicentrarchus labrax). Using genomic data mapped to a reference genome, the authors were able to characterize the distribution of lengths of haplotypes and fit model parameters to the observations (Fig. 5). Specifically, they identified tracts of migrant ancestry using the program ChromoPainter (Lawson et al. 2012) and estimated admixture parameters, and they used the method of Harris and Nielsen (2013) to infer demographic history from tracts of identity by state. These results reconstruct the historical details of population isolation and secondary gene flow between Atlantic and Mediterranean populations. This is a narrow sense genomics study because high-density mapped markers are used with a conceptually novel approach (haplotype tracts of immigrant ancestry).
Mapped genomic markers provide information on haplotype lengths, which are informative to assess historic admixture processes. Here the observed distributions of haplotype tract lengths in Atlantic and Mediterranean populations of European sea bass (Dicentrarchus labrax) (red and yellow dots) closely match simulated distributions (dark and light gray dots), allowing estimation of parameters in a model of historic isolation followed by secondary contact and gene flow. The haplotype information and modeling allows estimation of timing, directionality, and amount of gene flow. Reproduced with permission from Duranton et al. (2018)
2.1.4 Phylogenomics
Phylogenetic relationships among taxa can be estimated from a wide range of genetic data types, including genomic data. A complication is that many genetic markers spread across the genome may reflect different evolutionary histories because of recombination, particularly in recently diverged species and where incomplete lineage sorting and admixture play important roles (Edwards et al. 2016). Methods accounting for this, for instance, in estimating phylogeny from large SNP datasets, have been developed (Hohenlohe et al. 2018 this volume; McKain et al. 2018). Ideally, phylogenomic datasets are used not only to estimate a consensus tree among taxa but also to reveal patterns of hybridization and admixture (e.g., using analyses that allow for specific admixture events, such as TreeMix; Pickrell and Pritchard 2012).
2.2 Identifying Adaptive Genetic Variation Underlying Selective Sweeps
Population genomics makes it possible to identify “footprints” of natural selection in genome-wide patterns of genetic variation. The classical genomic signature of positive selection is the hard selective sweep, where fixation of a positively selected de novo mutation dramatically reduces genetic diversity at closely linked loci in a process referred to as genetic hitchhiking (Maynard Smith and Haigh 1974). The size of the region of reduced variation around the positively selected allele depends mainly on the strength of selection (and thus how quickly the sweep progressed) and the recombination rates on either side of the selected site (Jensen et al. 2016).
Hard selective sweeps are characterized by very low nucleotide diversity, and polymorphisms subsequently arising within a swept region display an excess of low-frequency-derived alleles compared to the genome-wide background. Thus, methods used to identify classical selective sweeps generally scan the genome for regions with low diversity (Maynard Smith and Haigh 1974), an excess of rare alleles (Tajima 1989), and a shifted site frequency spectrum (SFS) toward relatively high-frequency-derived alleles (DeGiorgio et al. 2016; Fay and Wu 2000; Huber et al. 2015; Kim and Stephan 2002).
While classical hard selective sweeps strongly reduce genetic variation around the selected site, soft selective sweeps arise from positive selection on standing genetic variation and leave a subtler genomic signature (Hermisson and Pennings 2005). In particular, soft sweeps usually do not strongly reduce genetic variation or result in a large shift in the site frequency spectrum around the selected site because the positively selected allele is present within multiple flanking haplotypes (Pennings and Hermisson 2006; Teshima et al. 2006). Soft sweeps appear to be a dominant mechanism of recent adaptation in humans (McCoy and Akey 2017; Schrider and Kern 2017). Methods based on extended haplotype homozygosity, which look for derived alleles sitting on exceptionally long haplotypes, are thought to have substantially higher power to detect soft selective sweeps than diversity- or site frequency spectrum-based genome scans (Ferrer-Admetlla et al. 2014; Voight et al. 2006). Machine learning appears to also be a powerful method to detect soft sweeps (Schrider and Kern 2017).
Recent studies have detected putative selective sweeps in an array of organisms, ranging from domesticated livestock and humans to natural populations of non-model species. In some cases, these studies have helped to identify the phenotypes and underlying genetic and biochemical pathways involved with the response to positive selection. Recent studies using genome scans based on genome resequencing data have identified putative selective sweeps underlying adaptation to domestication in pigs (Sus scrofa; Rubin et al. 2012), dogs (Canis lupus familiaris; Axelsson et al. 2013), chickens (Gallus gallus; Rubin et al. 2010), and rabbits (Oryctolagus cuniculus; Carneiro et al. 2014).
Schweizer et al. (2016) identified putative selective sweeps in North American gray wolves (Canis lupus) related to coat color and environmental conditions by conducting genome scans via resequencing of exons and intergenic sequences. Kardos et al. (2015b) identified a putative selective sweep in wild bighorn sheep (Ovis canadensis) in the vicinity of the RXFP2 gene associated with horn growth in domestic sheep (RXFP2). Their results suggested that horn morphology (or size) in bighorn sheep evolved at least in part via positive selection on a beneficial variant at RXFP2. See the chapter herein by Hohenlohe et al. (2018) for additional examples of selective sweeps and also Marques et al. (2018), Stetter et al. (2018), and Sugden et al. (2018).
2.3 Genetic Architecture Underlying Adaptive Differentiation
Positive selection acting differently among populations can result in exceptionally strong genetic differentiation in genomic regions containing loci subjected to selection (Lewontin and Krakauer 1973). For example, alleles conferring adaptation to high elevation in humans tend to be at high frequency in high-elevation populations but at low frequency in low-elevation populations in humans (e.g., Lorenzo et al. 2014; Hackinger et al. 2016). Genomic signatures of local adaptation can be detected by scanning a large number of densely mapped loci to detect genes or chromosome regions with exceptionally high genetic differentiation (e.g., F ST outliers) among populations (Hohenlohe et al. 2010b; Paris et al. 2017). Small numbers (100s) of unmapped loci can be tested for adaptive signatures (broad sense genomics), particularly if candidate loci have been identified a priori (e.g., Holliday et al. 2010, 2012), but if adaptation is highly polygenic, some of the causal loci will likely be missed.
Many studies have analyzed large numbers of mapped SNPs to detect F ST outlier chromosomal regions that represent candidate genomic regions for local adaptation (Hohenlohe et al. 2010b; Wang et al. 2016). Gene-environment association (GEA) analyses are also used to identify outlier loci associated with environmental differences (Sect. 2.4; Figs. 3 and 5). Genomic regions displaying exceptionally high genetic differentiation between incipient species can also help to localize loci subjected to divergent selection during speciation (Burri et al. 2015; Ellegren et al. 2012; Harr 2006; Marques et al. 2016; Martin et al. 2013; Poelstra et al. 2014; Renaut et al. 2013; Turner et al. 2005; Wolf and Ellegren 2017).
Problems with F ST outlier tests, and related tests for differentiation, include the use of the wrong null model resulting in false positives. For example, hierarchical population genetic structure can cause higher variance in F ST (e.g., higher F ST’s) than expected assuming a simpler model of population structure. The problem can be assessed and dealt with using simulations to simulate null distributions of F ST (for 1,000s of neutral loci) for a hierarchical population structure (e.g., Lotterhos and Whitlock 2014). False negatives are another problem, which can also be caused by using the wrong or suboptimal spatial model. For example, to avoid many false negatives and increase power to detect selection, Foll et al. (2010) developed a hierarchical Bayesian to improve detection of genes involved in adaptation by humans to living at high altitude and hypoxia.
To avoid false negatives, researchers should use high SNP densities because variation in F ST among SNPs is high even within a strongly selected gene. For example, SNP alleles from the lactose tolerance gene have been under strong positive selection in humans in Northern Europe (Beja-Pereira et al. 2003; Tishkoff et al. 2007). However, only 15 of 61 SNPs across the gene show significantly high F ST (>0.45) between Europeans and other populations (Fig. 6). This suggests that many SNP genotyping strategies (e.g., SNP chips, restriction site-associated DNA sequencing, targeted sequencing) will often have too few SNPs per gene region to reliably detect molecular signatures of adaptive genetic differentiation and perhaps other selection signatures as well (Luikart et al. 2003).
F ST for individual SNPs (dots) randomly sampled from across each of the two genes (CLASP1 and LCT, human chromosome #2) having the highest proportion of SNPs with F ST above 0.45 between the Yorubans in Africa and Utahans representing North Western Europeans. AGFG1 is a typical gene without apparent selection signatures. CLASP1 and LCT are under strong directional selection. An F ST value of 0.45 is approximately the upper 99.9 percentile of empirically observed SNP F ST values across the genome and above which few neutral SNPs are expected. The x-axis represents a randomly chosen SNP (for instance, under random sampling with replacement). Unpublished manuscript by T. Antao and Luikart
2.4 Landscape Genomics
Landscape genomics is an emerging field or approach that strives to identify environmental factors that shape neutral and especially adaptive variation and the genes and their variants that underlie local adaptation (Rellstab et al. 2015; Balkenhol et al. 2017 this book). Environmental conditions vary across time and space, and local conditions can cause fitness differences among individuals that vary for phenotypic traits on which natural selection can act (Blanquart et al. 2013; Hoban et al. 2016). These differences in traits can be associated with underlying genotypic differences and with environmental conditions. Thus landscape genomics methods test for associations among environmental factors, geo-spatial location, or phenotypic traits and genomic variation. Landscape genomics studies focus on local adaptation to environmental conditions within and among different geographic locations (Rellstab et al. 2015; Hoban et al. 2016). The topic of landscape genomics is discussed in detail in the chapter by Balkenhol et al. (2017) in this book.
Genetic differentiation (e.g., F ST) outlier tests alone do not identify the environmental factors or selective pressures driving local adaptation. However, genotype-environment association (GEA) analyses can identify loci associated with specific environmental factors driving local adaptation. Simulation-based studies have found that, in general, GEAs have more power than outlier-based approaches but higher rates (20–50%) of false positives (De Mita et al. 2013; Frichot et al. 2013; Forester et al. 2016). Examples of GEA-based programs are Bayenv2 (Gunther and Coop 2013) that adjusts for population structure using an independent set of markers that are assumed a priori to be neutral and the latent factors mixed model (LFMM, Frichot et al. 2013) approach that uses the covariance structure of all loci being tested to adjust for population history and demographics. There are a large number of tests and software packages available for detecting differentiation outliers and GEAs, and the number of publications using them has grown rapidly, especially for BayeScan, Bayesenv, and LFMM (Ahrens et al. 2018).
Lotterhos and Whitlock (2014) used simulations to show that reliable genetic differentiation test results vary depending on the number of individuals sampled. Their review suggests that F ST outlier tests will detect a higher proportion of outliers as more individuals are sampled. This bias did not occur for GEA where the proportion of associations remained relatively constant as the total number of individuals increased. This finding implies that GEAs are more robust (see also Ahrens et al. 2018).
One recent use of multiple GEA approaches identified a congruent set of candidate genes (among approaches) that are potentially important in the local adaptation of Mediterranean striped red mullet (Mullus surmuletus) populations to their saline environment (Dalongeville et al. 2018). Brauer et al. (2018) used GEA analysis to test for adaptive divergence in the Murray river rainbowfish (Melanotaenia fluviatilis) genome associated with hydroclimate. Brauer et al. (2018) used 17,504 SNPs in a multivariate GEA framework accounting for structure of a river system to identify 146 candidate loci potentially underlying polygenic adaptive responses to seasonal fluctuations in stream flow and periods of extreme temperature and precipitation.
Adjusting or accounting for neutral population structure is necessary to avoid a high rate of false positives with GEA analyses. However, such adjustments can result in false negatives if environmental factors driving local adaptation are correlated with population structure (e.g., from patterns of post-glacial recolonization). Yeaman et al. (2016) addressed this problem using a comparative genomics approach by identifying GEA candidate loci correlated with variation in low temperatures from exome capture and resequencing data based on raw GEA correlations in one conifer species (Pinus contorta). They then looked for significant GEA in those candidate loci in a second species complex (Picea glauca, P. engelmannii, and their hybrids) and vice versa. They also identified shared loci associated with phenotypic variation in cold hardiness. In this way, they identified 47 loci underlying local adaptation to cold in populations of both conifers. For additional examples involving gene expression and epigenetics, see below.
2.4.1 Spatial Signatures of Polygenic Adaptation
Adaptive traits are often polygenic and controlled by a large number of alleles from many loci each having small phenotypic effect (Bourret et al. 2014; Laporte et al. 2016; Stölting et al. 2015; Sork 2016; Yeaman et al. 2016; Boyle et al. 2017). However, methods for detecting adaptive genetic variation often only have the power to detect loci and alleles with large phenotypic effects (Wellenreuther and Hansson 2016). GEA methods can potentially detect weak signatures of adaptation but still might seldom detect alleles with small effect sizes (Coop et al. 2010; Joost et al. 2007).
Many of the early gene-environment association (GEA) methods tested only a single locus at a time, rather than looking at the combined effects of multiple loci simultaneously (Rellstab et al. 2015). More recent work has suggested that multivariate approaches (e.g., redundancy analysis (RDA), canonical correlation analysis (CCA), or using a population graph approach) might help reduce the number of false positives and maintain reasonable power to detect associations under even conditions of weak, multilocus selection (Rajora et al. 2016; Forester et al. 2018). However, multivariate approaches remain seldom used in population genomics literature (Rajora et al. 2016; Wellenreuther and Hansson 2016).
A recent study tested for polygenic signatures of local adaptation using multivariate approaches and 6605 RADseq SNPs in an Australian endemic fish, Murray cod (Maccullochella peelii) (Harrisson et al. 2017). The polygenic multivariate method (redundancy analysis, RDA) supported comparable roles of climate (temperature- and precipitation-related variables) and geography in shaping the distribution of multiple SNP genotypes across the range of Murray cod. Among the candidate SNPs identified by these multivariate and the univariate methods, the top 5% of SNPs contributing to significant RDA axes included 67% of the SNPs identified by univariate methods. The results highlight the value of using a combination of different approaches, including polygenic methods, when looking for signatures of local adaptation in landscape genomics studies.
2.4.2 Landscape Community Genomics: Identifying Loci Underlying Both Species and Landscape Interactions
Genomic variation is influenced by complex interactions between abiotic (e.g., environmental) and biotic (e.g., community) effects. Researchers should consider the effects of both environmental and community factors on evolutionary dynamics simultaneously to avoid potentially incomplete, spurious, or erroneous conclusions about the mechanisms driving patterns of genomic variation among and within populations. Any study of genomic variation and adaptation in nature would ideally begin with a set of predicted abiotic and biotic drivers, including interactions between these two fundamental categories of effects (Hand et al. 2015b). Despite the value of studying concordant patterns of genetic variation in interacting species, there are relatively few empirical examples, in part because of the expense of conducting population genomics on multiple interacting species across heterogeneous landscapes or environmental gradients. Few examples exist but will become more common as it becomes feasible to conduct landscape genomics on multiple interacting species (e.g., see Beja-Pereira et al. 2003).
One recent example of landscape community genomics is a study of the parasitic Alcon blue butterfly (Phengaris alcon) and its two hosts: an ant species (Myrmica scabrinodis) and the marsh gentian (Gentiana pneumonanthe) (De Kort et al. 2018). The female butterfly lays its eggs onto gentian flower buds which develop into caterpillars at the expense of the gentian’s ovules. This has led to coevolutionary shifts in flowering phenology to escape peak times of infestation by the Alcon butterflies (Valdés and Ehrlén 2017). When the caterpillars leave the plant, they are adopted by Myrmica ants as the caterpillar’s chemical signature misleads the ants into accepting and rearing the caterpillar in preference to their own brood. This social parasitism of ants has also lead to coevolutionary changes in the surface chemistry of Myrmica and in the Alcon butterfly larvae (Nash et al. 2008). De Kort et al. (2018) focused on the impact of habitat fragmentation on the Alcon butterfly and subsequently the possible effect on its two obligatory host species (ants and gentians). Some of the among-population genetic variation in the host species could be explained by abiotic variables (e.g., altitude). Additional analyses showed a substantial amount of variation in Alcon butterfly genetic structure could be explained by host genetic structure. De Kort et al. (2018) then suggested that coevolutionary selection has been important in synchronizing genetic structure of this host-parasite system. Habitat fragmentation is impacting the Alcon butterfly (Phengaris rebeli) and will likely impact the genetic structure of its host species as well.
2.5 Genome-Wide Association Studies: Loci Associated with Traits Within Populations
A growing number of population genomics studies have identified loci contributing to phenotypic variation among individuals, including in traits that strongly affect fitness and local adaptation, via genome-wide association studies (GWAS). GWAS typically use a regression model (e.g., a linear mixed-effects [LME] model) to identify loci where genotypes are associated with a trait of interest (Gibson 2018). Population structure is accounted for by fitting a genomic-relatedness matrix (GRM) as a random effect; other potentially informative predictor variables can be included as needed in the random or fixed effects parts of the model. Additional discussions of GWAS and heritability estimation, with emphasis on functional genomics, is provided in the chapter by Pino Del Carpio et al. (2018) in this book (see also Santure and Garant 2018; Armstrong et al. 2018).
The number of studies finding loci associated with variation in fitness-related traits in natural populations is proliferating. Trait-associated loci are often identified in regions that show strong genetic differentiation between individuals with stark differences in morphology. For example, SNPs around the RXFP2 gene included on a 50K SNP array were found to be associated with horn morphology in wild feral Soay sheep (Ovis aries) (Johnston et al. 2011, 2013). Horn morphology strongly affects fitness in Soay sheep (Ovis aries) and in natural populations of wild mountain sheep (e.g., bighorn sheep, Ovis canadensis; Hogg 1984). Thus identifying loci associated with horn size provides an interesting look into the genetic basis of fitness-related variation.
In another recent GWAS example, Brelsford et al. (2017) studied a natural hybrid zone between Audubon’s and myrtle warblers (Setophaga coronata auduboni x S. c. coronata) to identify genomic regions associated with color pigmentation potentially associated with mating success and fitness. RADseq produced 154,683 to 393,755 SNPs, depending on the filtering criteria. For each of five plumage coloration traits studied (eye spot, throat color, eye line, wing bar, and auricular), the authors detected highly significant associations with multiple SNPs genome wide that clustered into chromosomal regions (Fig. 7). The high success in identifying loci associated with these traits likely resulted from the relatively high gametic disequilibrium along chromosomal stretches resulting from hybridization.
Manhattan plots of genomic differentiation (A) and plumage associations (B, C, D). (A) F ST between allopatric myrtle and Audubon’s warblers at 393,755 SNPs across the genome with scaffolds ordered by size. Adjacent scaffolds across the genomes are distinguished by alternating gray or black coloration. Panels B, C, and D are phenotype-genotype associations for three of the five plumage characters studied. The tiny red triangle near the top right of panel (B) shows the cluster of loci that aligns to the zebra finch chromosome 15. This region includes the SCARF2 gene, which is a strong candidate gene for carotenoid pigment transport. Panel (E) shows patterns of divergence and genotype-phenotype associations for eye line (blue points) and eye spot (red points) for a region of chromosome 20. Associations between these two traits are highly correlated with each other as well as patterns of divergence (F ST, small black dots). Coding regions (exons) for genes are shown by the vertical bars, with different adjacent genes colored differently with arbitrarily chosen colors. Modified from Brelsford et al. (2017)
In another study, Husby et al. (2015) identified a locus that was associated with clutch size (a life history trait) in the collared flycatcher (Ficedula albicollis). Similarly, Bérénos et al. (2015) identified two SNPs in Soay sheep (Ovis aries) associated with leg length (a measure of body size), with each of the two SNPs explaining >10% of the additive genetic variance in the trait. One of the SNPs found to be associated with leg length by Bérénos et al. (2015) was also associated with female reproductive success, providing evidence for a link between genotype, phenotype, and fitness in Soay sheep. Lamichhaney et al. (2015) and Küpper et al. (2015) simultaneously identified a large (~4.5 Mb) inversion that controlled mating morphology in the ruff (Philomachus pugnax). Barson et al. (2015) identified a locus with sex-specific dominance and large effects on age at maturation in wild Atlantic salmon.
GWAS methods are also being widely used in conjunction with common garden experiments containing natural or seminatural populations of plants, fish, and other taxa. For example, in black cottonwood (Populus trichocarpa), Mckown et al. (2014) conducted GWAS using 29,355 filtered SNPs using a unified mixed model accounting for population structure effects. They uncovered 410 significant SNPs (from 275 genes) across 19 chromosomes that explained 1–13% of trait variation in trait associations, mostly associations with phenology genes (240 genes) but also biomass (53 genes) and ecophysiology (25 genes).
In the future, association studies will continually find more loci, including loci of small effects associated with adaptive traits, thanks to improved power from sequencing strategies like pool-seq with a reference genome that allow high-density genotyping of populations or lineages (Haussler et al. 2009; Schlötterer et al. 2014; Wessinger et al. 2018; Pruisscher et al. 2018). For example, Narum et al. (2018) used a new genome assembly (2.8 Gb) and pool-seq resequencing for Chinook salmon (Oncorhynchus tshawytscha) to conduct association mapping of important life history traits. The authors pooled individuals from populations of each of three phylogenetic lineages that exhibit different maturation and run-timing phenotypes. Their whole-genome resequencing of pooled (barcoded) individuals suggested that divergent selection was extensive at many loci genome wide within and among phylogenetic lineages. Association mapping with millions of SNPs revealed a genomic region of major effect associated with phenotypes for migration timing. This study illustrates how a genome assembly and high-density markers can help resolve the genetic basis of important phenotypes.
2.6 Quantifying Inbreeding, Inbreeding Depression, and Historical Bottlenecks
The availability of population genomic data is improving our understanding of inbreeding (mating between relatives) and inbreeding depression in the wild (Hedrick and Garcia-Dorado 2016; Kardos et al. 2016a). Inbreeding causes offspring to be homozygous and “identical by descent” (IBD) across large chromosomal segments where the two inherited DNA copies arise from a single DNA copy in a common ancestor of the parents (Kardos et al. 2016a; Speed and Balding 2015; Thompson 2013). The increased homozygosity arising from IBD causes inbreeding depression: reduced fitness of inbred individuals (Charlesworth and Willis 2009).
The pedigree inbreeding coefficient (F P) is a traditional measure of individual inbreeding and predicts the fraction of the genome that is IBD, assuming that pedigree founders are unrelated and noninbred (Keller and Waller 2002; Malécot 1970; Wright 1922). However, F P can be an imprecise measure of the realized fraction of the genome that is IBD (F) due pedigree errors, the stochastic nature of Mendelian segregation and recombination, and the presence of related and inbred pedigree founders (Fisher 1965; Franklin 1977; Stam 1980; Kardos et al. 2016a; Knief et al. 2017; Forstmeier et al. 2012; Goudet et al. 2018). The imprecision of F P and the recent availability of genomic data have led to increased application of genomic estimates of individual inbreeding and inbreeding depression (Hoffman et al. 2014; Huisman et al. 2016; Bérénos et al. 2016).
Genomic measures of individual inbreeding have the advantage that they directly measure patterns of homozygosity across the genome, thus making pedigrees unnecessary to estimate individual inbreeding. Encouragingly, only a few thousand unmapped SNP loci can provide more precise estimates of F (IBD) than a pedigree five to ten generations deep (Kardos et al. 2015a, 2018). Even more powerful, the analysis of many tens of thousands of mapped loci allows the use of runs of homozygosity (ROH) residing within chromosomal segments that are IBD to assess inbreeding (IBD) with very high precision (Kardos et al. 2015a). Genomics studies of inbreeding are greatly advancing our understanding of the extent of inbreeding depression in humans, domestic animals and plants, and natural populations of non-model organisms (Palkopoulou et al. 2015; Xue et al. 2015; Kardos et al. 2018).
ROH can be used to identify and map loci contributing to inbreeding depression by testing for associations between the presence of ROH and individual fitness-related traits (Keller et al. 2012; Kijas 2013; Lander and Botstein 1987; Pryce et al. 2014). Large-scale genomics studies of inbreeding depression (sample sizes >100,000 individuals) based on ROH and other genomic measures of inbreeding are now being done to precisely estimate inbreeding effects on a wide range of human traits (Wessinger et al. 2018; Johnson et al. 2018a, b). Thus, population genomics is beginning to contribute substantially to our understanding of the evolution of fitness-related phenotypes and the genetic basis of inbreeding depression in many species. This understanding has the potential to guide conservation and management of wild population and captive breeding programs, for example, to avoid inbreeding depression and invoke genetic rescue through restoring gene flow (Tallmon et al. 2004; Whiteley et al. 2015).
In another step to identify contributing loci, exons identified by ROH can also be used to bioinformatically identify likely deleterious alleles based on the likely effects of amino acid substitutions and whether such substitutions are common in homologous genes in other organisms using software such as PROVEAN (Choi and Chan 2015). The frequencies of these alleles can be compared among individuals and populations. For example, Conte et al. (2017) found over 13% of all SNP alleles in Picea engelmannii, P. glauca, and hybrid populations had amino acid substitutions predicted to be deleterious, but homozygous genotypes for deleterious alleles were less frequent in hybrid populations due to complementation.
Historical effective population size can be qualitatively inferred from the abundance and length distribution of runs of homozygosity (Fig. 8). For example, analyses of genome-wide runs of homozygosity (ROH) showed inbreeding arising from recent common ancestors of parents (due to small population size) in individuals of recently reintroduced populations of alpine ibex (Capra ibex). The detected ROH were associated with small population size during captive breeding and the founding of small wild populations approximately 20 generations ago. In spite of a rapid population growth in the wild, the ibex carried a genomic signature of their small recent historical population size (Fig. 8). The authors thus suggested that genomic monitoring for ROH could provide an improved indicator for early detection of inbreeding in wild and managed populations (Grossen et al. 2018).
(A) Schematic showing runs of homozygosity (ROH) along a chromosome. (B) Distribution of total genome-wide runs of homozygosity in one representative individual from each of three species including domestic goat (SGB A10), Iberian ibex (Z23), and Alpine ibex (VS0034). The distribution is right-shifted to have longer ROH, >10–20 Mb, in the reintroduced Alpine ibex. (C) Tract length distribution of ROH in wild and reintroduced populations. ROH for individuals from different populations show a range of different tract lengths. Only the reintroduced (captive bred, bottlenecked) individuals have 20 Mb tracts. The wild source population GP (Gran Paradiso) never suffered the captive breeding founder effects, but it did decline to ~100 individuals approximately 100 years ago. Black-outlined circles show the three primary reintroduced populations Albris (orange), Pleureur (light blue), and Brienzer Rothorn (green). Secondary reintroductions established from the primary reintroduced populations share the same color. Populations with mixed ancestry are shown in purple. N, sample size per population. Reproduced with permission from Grossen et al. (2018)
Historical population bottlenecks can also be inferred and approximately dated using ROH and coalescent modeling (Ceballos et al. 2018). Palkopoulou et al. (2015) sequenced genomes from two wooly mammoths from distant populations in terms of both geography (northeastern Siberia versus Wrangel Island, Alaska) and time (~44,800 versus ~4,300 YBP). Intriguingly, both yielded very similar genomic signatures of a nearly identical population decline at the start of the Holocene. One mammoth individual sample was dated to have died just before the species’ went extinct approximately 4,000 years ago. From coalescent modeling, a second genomic signature of a reduced population effective size (and inbreeding) was inferred just before the extinction at the start of the Holocene. The analyses suggested that the wooly mammoth was subject to reduced genetic variation prior to its extinction.
2.7 Delineating Adaptively Differentiated Populations
Population genomics can help identify locally adapted, differentiated populations that are difficult to delineate using selectively neutral markers, especially in high gene flow species, such as forest trees and marine organisms. Prince et al. (2017) used RADseq to assess the evolutionary basis of premature migration among individuals within local populations of Pacific salmonids. Chinook salmon and also steelhead trout exhibit two major migration strategies: premature migrators enter freshwater in the spring with high fat content and stay in freshwater for months until spawning, and mature migrators which enter freshwater sexually mature just prior to the spawn. Gene flow was relatively high between the two very different forms (premature vs normal migration) within a stream (F ST ~ 0.03); F ST between streams was far higher (F ST ~ 0.13). The authors found the same single locus associated with premature migration in multiple populations in each of two different species, Chinook salmon and also steelhead trout.
Results from this study suggest conservation implications. While many traits involved in local adaptation are polygenic, in this case a single locus appears to control migration timing and has significant economic, ecological, and cultural importance (Fig. 9). In particular, extirpation of the premature migration allele and phenotype are unlikely to re-evolve once extirpated from a population in the absence of immigrants carrying the allele from elsewhere. Mutations producing a given important allele are rare evolutionarily, suggesting such alleles will not re-evolve quickly or easily if lost. Furthermore, spatial patterns of adaptive allelic variation can differ from patterns of overall population genetic differentiation. Taken together, these results suggest that conservation units based on genome-wide patterns of genetic differentiation will sometimes fail to protect evolutionarily significant genetic and phenotypic variation.
Genomic basis of premature migration in steelhead. (A) Map of sampling locations of early versus mature (normal) migration types of steelhead trout sampled together in each of many drainages. (B) Association mapping of early vs normal migration of the Eel River steelhead trout with gene annotation, with the (C) gene annotation of a region with strong association; red numbers show genomic locations of the two RAD restriction sites with strongest associated SNPs, and blue asterisks indicate positions of amplicon sequencing, with the candidate gene GREB1L. (D) Phylogenetic tree depicting maximum parsimony of phased amplicon sequences from all individuals; branch lengths, with the exception of terminal tips, reflect nucleotide differences between haplotypes; numbers identify individuals with one haplotype in each migration category clade (i.e., heterozygotes for premature and normal migration haplotypes). Reproduced with permission from Prince et al. (2017)
Adaptively differentiated populations can be identified and prioritized for conservation and breeding (Funk et al. 2012). Population genomics and landscape genomics approaches are often necessary to identify adaptively differentiated populations because common garden or reciprocal transplant experiments are not feasible for many species. Bonin et al. (2007) devised a population adaptive index (PAI), which uses both neutral and adaptive distinctiveness to assess the adaptive value of the population. They suggested that outlier tests could help identify adaptive loci and alleles to then use to identify and prioritize or rank populations for conservation values. In species to which they applied the index (PAI), the neutral and adaptive marker variation among populations were not correlated; Therefore the authors concluded that conservation strategies based on the neutral and adaptive indexes would not protect the same populations.
Other authors have suggested genomics approaches be used to identify adaptively differentiated populations (Funk et al. 2018; Razgour et al. 2018; Hoban 2018). Approaches include genotype-environment associations and gene expression analysis (e.g., Hansen 2010; Chen et al. 2018, see Sects. 2.4 and 4.4). Including environmental variables improves power over differentiation-based methods, helps identify the environmental drivers of adaptation, and facilitates detection of contemporary (and historical) selection (Forester et al. 2018). Ideally, multiple independent data types would be combined to maximize power and reliability of delineating adaptively differentiated populations (geography, environment, behavior, ecology, physiology, transcriptomics, and genomics; Allendorf et al. 2013).
There is enormous risk of prioritizing populations for conservation based on population genomics (or outlier) approaches alone. It can be extremely difficult or impossible to verify whether genes that behave as outliers are genuinely adaptive. The genomic signatures expected from local adaptation (e.g., F ST outliers, GEA) can arise from genetic drift, particularly when small populations and low migration rates are involved. Further, genuine genomic signatures of selection may be due to selective forces in deep history that have since disappeared and thus are irrelevant to adaptation in current or future environments. Third, prioritizing certain populations based on certain particular alleles (even if they are genuinely relevant to adaptation) could actually reduce diversity across the rest of the genome that is necessary for future adaptation (Luikart et al. 2003; Allendorf 2017; Kardos and Shafer 2018).
2.8 Speciation, Hybrid Zones, Admixture, and Adaptive Introgression
Population genomics approaches have opened new avenues to study speciation, admixture events, and hybrid zones in all organisms. A detailed account of this topic is presented by Nadeau and Kawakami (2018) in this book. Here we introduce the topic and provide a few relevant examples.
The European bison (Bison bonasus), Europe’s largest land mammal, was recently shown to be a hybrid of two previously recognized subspecies, by authors using low coverage genome sequence alignments of historical and modern individuals (Wecek et al. 2017). Admixture occurred between subspecies prior to extinction in the wild and also subsequently during recent captive breeding. Admixture with domestic cattle was also significant but was ancient rather than from recent hybridization with domestics. These discoveries would have been difficult or impossible without genome-wide mapped loci and both historical and modern samples.
Kovach et al. (2016) studied genome-wide patterns of admixture and natural selection across recently formed hybrid zones between native cutthroat trout and invasive rainbow trout (Oncorhynchus clarki lewisi and O. mykiss) by genotyping 9,380 species-diagnostic RADtag SNP loci. A significantly greater proportion of the genome appeared to be under selection favoring native cutthroat trout (rather than rainbow trout), in the local native environments. This negative selection against rainbow introgression was found on most chromosomes and was consistent among populations and environments, even in warmer environments where rainbow trout were predicted to have a selective advantage. These data are consistent with previous findings that admixed fish have reduced reproductive success (Muhlfeld et al. 2009). Future studies could use far more loci to precisely map tracts of hybridity and infer timing of introgression of the rainbow haplotype segments into the native cutthroat trout.
Among the most intriguing examples of natural selection favoring “adaptive introgression” of certain alleles following admixture is the introgression of advantageous alleles from Neanderthals (and Denisovans) into modern humans. Genes involved in sugar metabolism, muscle contraction, and oocyte meiosis have been influenced by adaptive introgression from Neanderthals. For example, EPAS1 which influences hemoglobin concentration and response to hypoxia has introgressed from Denisovans into Tibetans, facilitating adaptation to life at high altitude through ancient admixture (Huerta-Sánchez et al. 2014). Other benefits of archaic (Neanderthal) introgression in the past are associated with several neurological and dermatological traits (Kelso and Prüfer 2014; Racimo et al. 2015; Vattathil and Akey 2015).
Evidence for adaptive introgression in nonhuman populations is growing. For example, adaptive introgression was detected in the Tibetan mastiff (Canis domesticus). Alleles for adaptation to high elevation (hypoxia) were identified at several loci, including the EPAS1 and HBB, which were introgression from Tibetan gray wolves (Canis lupus) (Miao et al. 2017). This demonstrates that domestic animals could rapidly become locally adapted by secondary contact with their wild relatives.
Adaptive introgression was also associated with the evolution of seasonal variation in coat color in snowshoe hares (Jones et al. 2018). Snowshoe hare populations molt to white during winter in order to maintain camouflage in environments with consistent winter snow cover. However, snowshoe hares in areas that remain snow-free year round often retain their brown coat color during the winter, thus maintaining effective camouflage in the absence of winter snow. The brown winter coat in snowshoe hares appears to arise from an allele that has introgressed from black-tailed jackrabbits (Jones et al. 2018). Other studies have also shown interesting genome-wide patterns of adaptive introgression (Song et al. 2011; Rieseberg 2011; Pardo-Diaz et al. 2012; Norris et al. 2015; Ozerov et al. 2016; Saint-Pé et al. 2018).
New approaches to analyze mapped loci will advance understanding of hybridization and evolution in hybrid zones. For example, large numbers of mapped loci can be analyzed to infer “local ancestry” across genomes of individuals. This involves mapping the locations of haplotypes arising from different source populations across the genomes of hybrids (Guan 2014; Leitwein et al. 2018). Such ancestry tracts can be used to estimate individual hybridity and population level admixture at both the genome wide and local scale across chromosomes. Additionally, local ancestry information is highly useful for trait mapping in mixed-ancestry populations (Smith and O’Brien 2005). Because the introgressing haplotypes decay in length at a predictable rate with increasing generations since hybridization, analyses of ancestry tract lengths can be informative of the historical timing of admixture events. For example, Leitwein et al. (2018) used 75,684 mapped SNPs obtained from double-digested RAD to identify ancestry tracts and estimate individual admixture proportions along with the timing of admixture in brown trout (Salmo trutta).
3 Benefits of Mapped Loci in Population Genomics
Information on the location of loci in the genome is a defining characteristic of population genomics (narrow sense), as mentioned above (Allendorf 2017). Loci can be mapped in terms of physical and/or genetic (linkage) positions in the genome. Producing both physical and linkage maps is far more tractable with modern genomics methods in non-model organisms than a few years ago. As a result, population genomics research efforts can now feasibly include the construction of a physical or linkage map for most study systems. Below we describe the key features of physical and genetic maps and the relative value of each for population genomic analyses.
Physical and genetic (linkage) mapping are two separate but complementary ways of describing the locations of loci in the genome. A physical map is a genome sequence. Long sequence reads from new (third)-generation sequencers enable high-quality genome assemblies, discovery of novel fitness-affecting structural variation, and the ability to sequence through previously “unsequenceable” repetitive DNA to allow mapping between distant loci along each chromosome. Reference genomes for non-model organisms often, however, are not assembled into chromosomal units, especially when genomes are large and contain a high fraction of highly repetitive content (i.e., retrotransposons) (Ellegren 2014; Epstein et al. 2016).
A linkage map describes the gene order based on the recombination frequency between loci along each chromosome. Linkage maps are constructed by genotyping pedigreed individuals and using linkage analysis, which quantifies how often adjacent loci co-segregate versus segregate independently due to recombination during meiosis. The distance between loci on a linkage map is described in terms of centimorgans (cM), where 1 cM is defined as a 1% recombination frequency between two adjacent loci inherited from a parent. Linkage maps can be developed in some cases where assembly of physical maps remains difficult (e.g., large conifer genomes, De La Torre et al. 2014).
Both physical and linkage maps facilitate population genomics research in at least five ways. First, having large numbers of mapped loci improves the power to identify and localize loci influencing phenotypic variation, fitness, and adaptation (e.g., Burri et al. 2016; Rastas et al. 2016). For example, the availability of densely mapped SNPs along a chromosome allows for localization of the chromosomal region(s) and genes underlying traits or adaptations (Figs. 2, 3, 7, and 10). This helps determine the genetic basis of adaptations or phenotypic variation, including determination of the number, kind, and effect size of genes underlying an adaptation or trait.
(A) Manhattan plot of −log10 (P-value) from a genome-wide association (GWA) analyses of color-patch size based on whole-genome resequencing of 81 male flycatchers. Chromosome identity is shown on the x-axis, and the P-values (open circles) are arranged according to physical SNP positions on each chromosome. Horizontal dashed lines are permutation-based statistical significance thresholds, and the dotted lines are the Bonferroni statistical significance thresholds of statistical significance (no points above the dashed line). (B) The relationship between the strength of linkage disequilibrium (r 2 or nonrandom association between loci) and physical distance in 81 whole-genome resequenced collared flycatcher males. r 2 is shown for each pair of SNPs separated by 50 or fewer kb. The solid line represents a function fitted to the rolling mean of r 2 calculated in nonoverlapping windows of 100 bp. The arrow shows where the mean of r 2 drops below 0.20. The dashed lines represent loess functions fitted to the rolling 5% and 95% quantiles of r 2 in the same nonoverlapping 100 bp windows. (C) Collared flycatcher photo (note forehead patch). (A, B) Reproduced with permission from Kardos et al. (2016b). (C) Copyrighted license and permission to use photo from Jiri Bohdal
Second, physical and linkage maps also help identify independent loci, e.g., loci far apart on the same chromosome or on different chromosomes (although statistical tests for independence can identify independent loci without a map). Independent loci are required for some population genetic inferences, including analyses of effective population size (N e), gene flow, or population relationships (Landry et al. 2002; Storz et al. 2002; Luikart et al. 2003). For example, Larson et al. (2014) estimated N e for wild Chinook salmon using ~10,000 SNPs and the LDNe method (based on gametic disequilibrium) that assumes all loci are independent or not physically linked (Waples and Do 2010). Estimates using only pairs of SNPs from different chromosomes (<1,000 SNPs) consistently gave estimates of N e that were higher than when using all pairs of SNPs; for example, an N e estimate was 1,909 for unlinked SNPs versus only 808 for all SNPs (including linked SNPs), as expected because gametic disequilibrium is stronger for physically linked SNPs, which drives (biases) lower N e estimates.
Third, the combination of a linkage map and a physical genome assembly allows understanding variation in the recombination rate across the genome. This is important because the recombination rate affects the extent of GD (gametic disequilibrium) and genetic diversity across a chromosome. Lower recombination rates result in GD extending over longer physical distances across a chromosome. As described below, the extent of significant GD strongly influences the power to detect footprints of natural selection and the ability to map loci contributing to phenotypic variation.
The recombination rate influences genetic diversity and differentiation among populations or species via its interaction with natural selection. Knowing how recombination rate varies across the genome is, therefore, crucial for interpreting genomic patterns of genetic diversity and differentiation. For example, the recombination rate is known to interact with background selection to generate chromosomal islands of reduced diversity (Charlesworth et al. 1993) and increased differentiation (high F ST, Burri et al. 2015), which might be erroneously interpreted as resulting from positive selection.
Fourth, physical and linkage maps both help researchers determine if they have a sufficient density of loci in the genome to have high power to detect loci subjected to positive selection or genotype-phenotype associations. With a linkage map, researchers can compute how far in centimorgans (cM) significant GD spans across chromosomes or linkage groups. Similarly, with a physical map, researchers can compute how far in base pairs (or kb) GD spans across chromosomes. Knowing the extent of GD is important because detection of phenotype-genotype associations and signatures of selection required GD between genotyped loci and causal loci. In addition, detecting phenotype-genotype associations requires GD between genotyped marker loci and causal loci, and so a relatively high density of markers is needed (Box 2).
Box 2 Importance of Gametic Disequilibrium and Marker Density for Identifying Adaptive Loci
Researchers recently resequenced 81 whole genomes in flycatchers with extreme phenotypes and also genotyped 50K SNP in 415 individuals. Birds were phenotyped for forehead patch size, a sexually selected trait associated with reproductive success. No SNPs were significantly associated with patch size (Fig. 10A). One reason for the failure to detect loci (QTL, quantitative trait loci) using association mapping could be that gametic (linkage) disequilibrium extends only over short chromosome distances (Fig. 10B), which makes the chances of strong associations between a DNA marker and trait loci small even when genotyping many SNP markers (Lowry et al. 2017, but see McKinney et al. 2017a; Catchen et al. 2017).
These results suggest that reliably detecting large-effect trait loci in large natural populations will often require thousands of individuals and the genotyping of hundreds of thousands of loci across the genome. Encouragingly, far fewer individuals and loci will often be sufficient to achieve high power to detect large-effect loci in small populations that typically have widespread strong gametic disequilibrium. This study illustrates the importance of knowing if strong gametic disequilibrium extends over long chromosome distances (e.g., due to low recombination rates, small effective populations size and drift, or perhaps admixture).
We caution that while maps allow quantification of the extent of GD along chromosomes, this quantification must be conducted for each study population of interest because the extent of GD varies among populations with in a species (Table 2) (Whiteley et al. 2011; Gray et al. 2009). GD will be relatively higher (genome wide) in populations with small N e and/or recent admixture (Fig. 11). Quantifying GD along chromosomes also allows researchers to identify hotspots of recombination (low GD) and thus to know which genome regions will require higher densities of markers when screening for loci associated with adaptation or phenotypic variation.
Gametic disequilibrium is stronger and more variable between loci (dots) in small populations of bighorn sheep (National Bison Range, n ~50–75) compared to the moderately larger population (Ram Mountain; n ~100–200). Strong LD (magnitude >0.4, see upper dashed line) stretches over ~30 cM in the National Bison Range population but only to over ~10 cM in Ram Mountain population. Reproduced with permission from Miller et al. (2014)
Directional selection is expected to reduce genetic variation and to alter the site frequency spectrum at the selected site and at closely linked loci (Charlesworth et al. 1993). The expected physical distance over which selection affects genetic variation depends on the local recombination rate. We expect directional selection to affect genetic variation across larger regions when the local recombination rate is low. As described below, accounting for recombination rate variation across the genome is necessary in order to assess differentiation among populations (e.g., F ST) measured across each chromosome. Information on recombination patterns (genome wide) improves interpretation of population genomic tests (GWAS, F ST outliers, etc.) because recombination can influence outlier locus behavior. For example, the rate of recombination is expected to correlate positively with local nucleotide diversity and rates of adaptive evolution, which could influence tests for selective sweeps using heterozygosity or F ST outlier loci (Cutter and Payseur 2013; Campos et al. 2014).
Fifth and finally, GD information from linkage or physical maps can improve theoretical models to advance population genetics beyond bean-bag genetics. Models parameterized with chromosomally explicit GD information can help to understand issues such as the importance of interactions of gene flow, recombination, and selection in adaptation and speciation. Some models stress the importance of recombination and distance among loci in the establishment and maintenance of adaptive alleles in a population (Bürger and Akerman 2011; Yeaman and Whitlock 2011; Feder et al. 2012).
3.1 What Can Physical Maps Provide that Linkage Maps Cannot?
Physical maps (reference genomes) generally provide higher power than linkage maps for detecting selective sweeps or genotype-phenotype associations because millions of SNPs can be mapped (positioned) via sequencing, whereas it is difficult to produce linkage maps with more than approximately 20–30K SNPs. Linkage mapping for tens of thousands of SNPs can require genotyping of many families, which is difficult or impossible in most species due to small family sizes, unavailability of families, or large expense of genotyping tens of thousands of loci in many large families. For example, a map from a single family of Chinook salmon had 5,400 SNP loci while increasing to four families allowed mapping of 13,800 loci (G. McKinney, unpublished data, 2018; see also Mckinney et al. 2016). There are diminishing returns from adding families for mapping because the number of additional loci that can be mapped declines as the number of families increases (unless perhaps genetically divergent families, with different variable loci are mapped).
Physical maps are also useful for improving both the process of discovery of SNP loci and of the subsequent genotyping of SNPs when using next-generation sequencing approaches such as RADseq (Sect. 4.1). For example, physical maps help identify paralogues and duplicated genes to avoid them or genotype them by allowing the alignment of sequencing reads to the physical map. If samples sizes are large, paralogs can be identified in RADseq data (e.g., see HDplot method of McKinney et al. 2017b).
Physical maps can improve genotyping by allowing the alignment of sequencing reads to the entire reference genome during the genotyping process, instead of using only a limited number of putative loci or de novo assembled loci (Hand et al. 2015a; Shafer et al. 2017). A caveat is that reference genomes are never 100% complete, and loci from missing sections of the genome will not be genotyped if doing only reference alignments for genotyping. If a genome is 90% complete, it is possible that 10% of your loci would not be mapped or genotyped when using the reference for genotyping.
Importantly, a physical map (assembly) can be used for genotyping next-generation sequencing reads from a closely related species to help improve genotyping (Cosart et al. 2011; Shafer et al. 2017). In this scenario, reads from one species are aligned to the genome for another for genotyping. This is a benefit of initiatives like Genome 10k that is providing a genome assembly for one species per genus or family of vertebrate, which provides related species a reference genome for mapping and genotyping (Haussler et al. 2009).
3.2 What Can Linkage Maps Provide that Physical Maps Cannot?
A high-density linkage map enables understanding of mechanisms (background/negative selection, positive selection, gene flow, and recombination) that cause heterogeneity along chromosomes in diversity within and differentiation between populations (Burri et al. 2015). A linkage map reveals recombination hot and cold spots which are known to interact with background selection to generate chromosomal islands of divergence (high F ST). Thus, a linkage map can help prevent false positives for local adaptation and improve detection of islands of divergence that are truly indicative of local adaptation (not false positives) (Cruickshank and Hahn 2014). Regional estimates of the recombination rate also help interpret data on runs of homozygosity (RoH) to detect inbreeding and to infer demographic history because recombination hotspots influence the lengths of RoH (Thompson 2013) and the density of SNPs (Charlesworth et al. 1993) and thus the power to detect RoH in genome regions.
Chromosomal level assemblies are often not possible without a linkage map, especially for large genomes with many repetitive sequences (Amores et al. 2011). Assembled chromosomes in turn can be used for identification of chromosomal synteny and structural polymorphisms such as rearrangements (e.g., inversions) within or between species (Amores et al. 2011; O’Quin et al. 2013; Rondeau et al. 2014). Structural changes or polymorphisms can influence fitness and adaptation and thus are important to discover and map (Wellenreuther and Bernatchez 2018). Additionally, assembled chromosomes can improve genome scans for loci associated with adaptation and phenotypic variation, by allowing computation of chromosome-specific distributions of summary statistics (continuously along each chromosome), which can increase power and reliability of outlier tests.
3.3 Combining Linkage and Physical Maps: The Ideal Genomics Approach
Having both a reference genome assembly and linkage map is ideal because they complement each other, and the linkage map improves the accuracy and contiguity of the assembly. Perhaps the most important point is that a linkage map must be combined with a physical map to estimate and map recombination rates across a genome. If researchers must choose between map types when developing genome resources for their species, the physical genome assembly will often be the map of choice because many more SNPs can be mapped physically; It is difficult to build linkage maps including extremely large numbers of SNPs (e.g., because many mapping families are required), as mentioned above.
3.4 Apply Genomics Approaches Without Maps
Many of the methods mentioned above can be applied to sequences from known genes or loci with unmapped locations in the genome. For example, we can conduct tests for loci under selection by testing for different kinds of outlier behavior (F ST, GD, allele frequency skew or heterozygosity excess, excessive locus-specific introgression; Luikart et al. 2003). We can also test for population adaptive differentiation (Bonin et al. 2007) and test for associations between genotypes and the environment or phenotypes (Fig. 1, Step 4a) (Fig. 3a).
4 Genotyping and Sequencing Technologies for Population Genomics
This quote by Schlötterer (2004) at the start of this chapter emphasizes the importance of molecular genetic methods and implies the importance of choosing an appropriate DNA marker or sequencing method for your research question (as did Sunnucks 2000; Benestan et al. 2016). The methods continue to evolve and improve our understanding of nature. SNPs and other markers from a variety of partial genome (and transcriptome) sequencing methods are the mainstay in population genomics studies. Here we provide a short introduction to key marker technologies likely to be most widely useful for non-model species. Low-cost genotyping, including RAD capture, DArT (diversity array technology), and related methods will continue to make population genomics increasingly feasible and widely used. Later in this book, Holliday et al. (2018, Chapter 2) provide more details and merits and demerits of different genotyping and sequencing technologies (see also Andrews et al. 2016; Jones and Good 2016; Holliday et al. 2018). For information on the promising approach of multiplex sequencing of many pooled individuals (pool-seq), see Box 3, Sect. 2.5, Schlötterer et al. (2014), and Narum et al. (2018).
4.1 Reduced Representation and Genotyping-by-Sequencing
Reduced representation sequencing is revolutionizing population genetics, molecular ecology, and conservation biology by making feasible and affordable use of massively parallel sequencing (MPS) on many individuals and loci genome wide (Narum et al. 2013). We can now use MPS to discover and genotype thousands of SNP loci for less cost than genotyping of only ~20 microsatellites. This makes population genomics research feasible for nearly any species. Understanding the strengths and limitations of the many reduced representation approaches is crucial to choose the best method for your research question (Andrews et al. 2016).
Approaches for reduced representation sequencing include general and targeted approaches (Jones and Good 2016). Anonymous approaches include unmapped restriction site-associated DNA sequencing (RADs) and transcriptome sequencing. Targeted approaches allow direct sequencing of loci of interest such as genes or informative RAD loci using capture arrays (below). Informative RAD loci are those in candidate adaptive genes and/or loci that are evenly spaced (mapped) across chromosomes to ensure genome wide coverage and high power for outlier tests, GEA, and association studies (e.g., GWAS) (e.g., Hohenlohe et al. 2010b; Kovach et al. 2016; Simons et al. 2018; Gibson 2018).
4.1.1 RADseq
The development of restriction site-associated DNA sequencing (referred to as RADseq and genotyping-by-sequencing, GBS) was considered among the most important scientific breakthroughs in the first decade of the twenty-first century because it allowed for simultaneous discovery and genotyping of many thousands of SNPs in a single experiment, in non-model species with no genomic resources (Science 2010). It involves the cutting of DNA through digestion with one or more restriction enzymes, labeling fragments from each individual with a unique barcode (short 6–12 bp reads), amplifying fragments using PCR, and high-throughput sequencing of pooled samples from multiple individuals (Andrews et al. 2016).
Another advantage of RADseq is its flexibility in the number of loci that can be genotyped – from hundreds to tens of thousands – by choosing among different restriction enzymes and >15 different RADseq-based techniques (Andrews et al. 2016). A main disadvantage is that there is typically highly uneven coverage of genotypic data among individuals and among loci, with many individuals missing data for many loci unless very stringent filtering is conducted with deep coverage sequencing.
This method has become extremely popular and has been applied to many taxa and questions in conservation, ecology, and evolution including quantifying inbreeding, genomic diversity, effective population size (N e), and for discovery of adaptive genes and genome regions (reviewed in Andrews et al. 2016; see also Lowry et al. 2017; McKinney et al. 2017a; Catchen et al. 2017; Hohenlohe et al. 2010b; Nadeau et al. 2014; Benestan et al. 2016; Candy et al. 2015; Sovic et al. 2016; and also subsequent chapter by Holliday et al. (2018) in this volume).
4.1.2 Targeted Sequence Capture
Sequence capture allows targeted sequencing of any region of a genome for which DNA sequence information exists. Sequence capture is often called “exon capture” because it is often used to sequence coding regions of the genome, including candidate adaptive genes (Flanagan et al. 2018). It is more expensive than RAD but a cheaper and more efficient alternative to whole-genome sequencing and results in more uniform sequencing of individuals and loci (and therefore less missing data) than restriction enzyme-based methods. It can be scaled to sequence hundreds to tens of thousands of genes (Hodges et al. 2007; Jones and Good 2016). Another advantage of sequence capture is in the genotyping of degraded DNA such as ancient, historical, and fecal DNA (Castellano et al. 2014; Bi et al. 2012; Bos et al. 2015).
Targeted capture enriches for DNA of interest and washes away nontarget DNA, as mentioned. This is important for genotyping fecal DNA because a majority (>90%) of DNA can be from bacteria (e.g., Perry et al. 2010). Recent examples of sequence capture include a wide range of question from phylogenetics to the detection of adaption signatures in humans, wolves, sharks, wild sheep, ungulates, birds, amphibians, trees, aquatic invertebrates, and host-parasites simultaneously (Cosart et al. 2011; Schweizer et al. 2016, Roffler et al. 2016; Gasc et al. 2016; Portik et al. 2016; McCartney-Melstad et al. 2016; Syring et al. 2016; Dowle et al. 2016: Campana et al. 2016; Manthey et al. 2016; Suren et al. 2016; Gauthier et al. 2017; see also Chapter 2 by Holliday et al. 2018).
4.1.3 RAD Capture
RAD capture (“Rapture”) combines the primary advantages of RADseq with advantages of targeted sequence capture. For example, the relatively inexpensive and rapid DNA library preparation methods of RADseq (Ali et al. 2015) are combined with the high specificity in targeting hundreds or thousands of loci. Loci are of high value (in genes, evenly spaced genome wide) for addressing nearly any questions of interest, focusing sequencing effort on those loci (Andrews et al. 2016; Jones and Good 2016; Hoffberg et al. 2016; Peek et al. 2018; see also Chiou and Bergey 2018). Another advantage is that a single Rapture array (e.g., for trout) works for genotyping in multiple divergent species such as salmon and trout (M. Miller, pers. comm., 2018).
The Rapture method was first used to successfully study SNP variation in lake trout (M. Miller, unpublished, 2018) and rainbow trout (Ali et al. 2015). This study used a capture array targeting 500 loci that were distributed across 29 chromosomes (Ali et al. 2015). All 1,440 individuals genotyped for the 500 loci were sequenced in a single Illumina HiSeq lane.
4.1.4 DArT
Diversity array technology (DArT) is another sequencing-based approach (a modification of GBS) allowing affordable discovery and genotyping of thousands of SNPs in hundreds of individuals (Elbasyoni et al. 2018). DArT has been used mainly in agriculturally important species and plants (Valdisser et al. 2017). This technology is similar to RADseq. Commercial companies exist, as for RADseq, to facilitate the discovery and application of genome-wide markers for population genomics approaches.
4.2 Reference Genomes
A reference genome sequence (i.e., genome assembly) is the portion of the genome that has been sequenced and assembled, i.e., pieced together, from short sequence reads. A reference genome is important in population genomics because it improves mapping of NGS reads to facilitate both the initial discovery of loci and the eventual genotyping of loci from many individuals. For example, if the reads from a RADseq project can be mapped to a reference genome, it can improve the detection of SNPs and duplicated genes or chromosomal regions that will be difficult to genotype because reads from duplicated regions often will stack up (align) together as if from a putative single locus (Hand et al. 2015a; Shafer et al. 2017). Shafer et al. (2017) observed large differences between reference-based and de novo approaches; use of a reference genome yielded more SNPs and reduced estimates of FIS and Ts/Tv.
Genome assembly is difficult in large genomes of plants where repetitive elements (e.g., retrotransposons) constitute >50% of the genome (Nystedt et al. 2013). In loblolly pine (Pinus taeda), 62% of the 22 Gb genome is made up of retrotransposons, and other conifers have similarly large repeat element content (De La Torre et al. 2014). Similarly, for genomes resulting from recent polyploidization events, as in many fish and plants, the assembly is difficult because, for example, in a tetraploid four similar copies exist for much of the genome. Most eukaryotic genomes contain complex repetitive sequences that are difficult to sequence and assemble as mentioned above (Ellegren 2014).
Assembly is becoming vastly easier thanks to new long-read technology as suggested by the following quote: “Long reads enable near reference-quality genome assemblies, discovery of novel disease-causing structural variation, and the ability to sequence through previously ‘unsequenceable’ repetitive DNA contents of clinical utility” (Ameur et al. 2018).
A reference genome sequence is not a standardized concept or item (Ellegren 2014). Even for well-characterized genomes, large parts are often not yet included in the genomic contigs (small assembled chromosomal regions) or the scaffolds (sets of contigs linked into larger regions) that have been ordered and linked into chromosomes. For example, the first published rainbow trout genome had only ~50% of sequences assembled and ordered into chromosomes; in fact one entire chromosome (#25) was unassembled such that no sequences were known from that chromosome (Berthelot et al. 2014). Similarly, chromosome 16 in the collared flycatcher genome is unassembled (Kawakami et al. 2014). In the rainbow trout and flycatcher examples, much of the one unassembled chromosome was likely sequenced and exists among the many contigs that have not been incorporated (assembled) into chromosomes. The quality and completeness of reference genomes vary widely among species.
Importantly, even partially assembled genomes are useful for many research questions. Partial genomes facilitate discovery of non-duplicated (versus duplicated) SNP loci for marker discovery. Partial (draft) genomes also increase quality of genotyping (e.g., with RADSeq or DNA capture data). Finally, draft genomes help design probes for exon sequence capture (e.g., when exons are identified from RNAseq data), and are useful for estimating the rate or distance of decay of gametic disequilibrium (Hand et al. 2015a; Shafer et al. 2017). Even a draft assembly (N50 >50 kb) usually contains well-assembled coding gene regions because coding genes have few repetitive elements and low heterozygosity, making a draft assembly relatively feasible and highly useful. In Tasmanian devils (Sarcophilus harrisii), researchers used a partially assembled draft genome (containing thousands of scaffolds not anchored on chromosomes) to successfully identify genomic regions and candidate genes underlying cancer risk, along with concordant signatures of selection including increased GD (gametic disequilibrium) and changes in allele frequencies (Epstein et al. 2016).
Another advantage of having at least a draft reference genome is that it allows estimation of the rate of decay of gametic disequilibrium, which is crucial for knowing the number of markers needed to adequately cover the genome to address particularly interesting or challenging research questions (narrow sense genomics). Having even only a hundred long scaffolds (>100 kb) with multiple DNA markers provides information on whether long stretches of GD exist genome wide, which is crucial for assessing the number of markers needed to achieve high density (Hendricks et al. 2018).
4.3 Whole-Genome Sequencing (WGS) and Resequencing
A main reason for sequencing (i.e., resequencing) entire genomes from many individuals is to maximize power to discover and localize DNA loci underlying fitness, adaptation, and phenotypic variation important for population persistence and growth (e.g., Kardos et al. 2016b). Increased power results from detecting most SNPs in the species and from being able to compute summary statistics (H, F ST, GD) for those SNPs and other polymorphisms (e.g., indels) in a sliding window across genomic regions (e.g., Box 3).
Having only one individual’s genome sequence (e.g., from one male) will not allow understanding of genome structural diversity or variation. This could bias subsequent comparisons of diversity among individuals (e.g., males and females), populations, and species, for example, when using GBS or RAD seq methods and mapping reads to the one genome reference sequence.
Box 3 Whole-Genome Sequencing Identifies Selective Sweeps and Candidate Genes
Researchers used whole-genome sequencing of wild Rocky Mountain bighorn sheep (Ovis canadensis) to identify 3.2 million SNPs and genomic regions with signatures of historical directional selection, i.e., selective sweeps (Kardos et al. 2015b). Sweeps were detected as chromosomal regions with low heterozygosity. Heterozygosity-based sweep analysis revealed evidence for strong historical selection at a gene (RXFP2) that affects horn size in domestic sheep, cattle, and goats (Johnston et al. 2011, 2013). The massive horns carried by bighorn sheep rams appear to have evolved in part via strong selection at the RXFP2 gene (Fig. 12).
Sequencing-based (pool-seq) genome-wide scan for selective sweeps that reduced heterozygosity in Montana and Wyoming populations (A) of bighorn sheep (B). Sliding window estimates of heterozygosity (C) across the bighorn sheep genome from an analysis of three populations pooled from Montana and Wyoming. Chromosomes (linkage groups) are arranged from 1 to 26 (left to right with alternative color (blue then orange) shading). The horizontal jagged red line represents the rolling mean across 100 adjacent sliding windows. The horizontal dashed line is 5 standard deviations below the mean heterozygosity. (D) Sweep on chromosome 10 spanning the RXFP2 gene (vertical black lines at 29.5 Mb near the x-axis are exons). Expected heterozygosity is plotted for individual SNPs (gray dots) located across 2 Mb on chromosome 10. The location of exons (vertical lines) of EEF1A1, RXFP2, and an uncharacterized predicted gene (“UNC”) is shown below the plot. Gene and exon positions were obtained from the Ensemble gene models generated during annotation of OARv3.1. The continuous horizontal jagged line shows mean expected heterozygosity calculated for nonoverlapping windows of 20 SNPs. The lowest genetic variation in the region occurred in a window centered at position 29,473,544 between exons 3 and 4 of RXFP2 (dashed line arrow). Reproduced with permission from Kardos et al. (2015b)
The authors also identified evidence for selection at genes affecting early body growth and cellular response to hypoxia which is consistent with adaptation to life at high altitude. These results provide examples of strong genomic signatures of selection identified at genes with known function in wild populations of a non-model species.
A comparison of SNP diversity between the X chromosome and the autosomes also indicated that bighorn males had a dramatically reduced long-term effective population size compared to females. This likely reflects a long history of intense sexual selection mediated by male-male competition for mates, which reduces the effective population.
The approach of heterozygosity-based sweep analysis had been previously used successfully in domestic animals where breed formation and subsequent strong artificial selection have generated selective sweeps for genes that influence a spectrum of phenotypic traits (Rubin et al. 2010, 2012; Axelsson et al. 2013). In wildlife, genome sequencing of gray wolves from the high altitude plateaus of western Asia recently detected selective sweeps surrounding genes involved with adaptation to hypoxia (Zhang et al. 2014). Together, these studies provide encouragement that genome sequencing in carefully selected wild populations will continue to yield valuable insights into the genetics of adaptation (Kardos et al. 2015b).
The results illustrate the value of quality reference genome assemblies from agricultural or model species for studies of the genomic basis of adaptation in closely related wild taxa (domestic sheep in this case). This study also illustrates the use of genome sequencing of pooled DNA from many individuals (per population). This saves money and can be an efficient way to estimate allele frequencies at nearly all SNPs in the genome. However, drawbacks include imprecision in estimates of allele frequencies arising from uneven contribution individuals to sequencing (pool-seq without barcoded individuals). For more information, see discussions by Schlötterer et al. 2014; Kardos et al. 2015b; Narum et al. 2018).
Certain questions can only be reliably addressed by using whole-genome sequencing. For example, structural polymorphisms such as gene duplications (copy number variants) cannot be reliably detected with GBS (e.g., RADseq) or sequence capture but can be detected by whole-genome assemblies and ideally with a linkage map (Wellenreuther and Bernatchez 2018). Additionally, adequately covering the genome for applications, such as GWAS, will sometimes require whole-genome sequencing for populations in which gametic disequilibrium is low and decays rapidly along chromosomes, e.g., in populations with very large N e or high recombination rates (Kardos et al. 2016b; Miles et al. 2017; Table 2).
Nonetheless, most questions in population genetics, molecular ecology, and conservation genetics can be addressed sufficiently without whole-genome sequencing and by using a population genomics approach (Allendorf et al. 2010). These include estimating individual inbreeding, detecting hybridization, quantifying population structure, and inferring gene flow. Whole-genome or exome resequencing is most useful for questions, such as determining the genomic basis (architecture) of local adaption or fitness when only a limited amount of gametic disequilibrium exists along chromosomes and thus millions of SNPs are required, for example.
4.4 Population Transcriptomics, Gene Expression, and Adaptation
Transcriptomics is the study of all RNA transcripts (transcriptome) that are produced by the genome. Population transcriptomics is the use of transcriptome-wide data to study variation in gene expression within and among populations to understand mechanisms underlying evolutionary change, for example, in response to environmental change. Such mechanisms can include plasticity in gene expression if it underlies adaptive evolutionary responses to new environments (Ghalambor et al. 2015) or if the amount or nature of plasticity itself evolves in response to selection. Here, we discuss the two main tools of population transcriptomics, microarray analysis and RNA sequencing (RNAseq), with examples of applications to natural populations.
cDNA microarrays and oligonucleotide microarrays can measure expression of thousands of genes simultaneously by quantifying levels of mRNA present in different tissues or individuals. Thousands or tens of thousands of different short DNA fragments are spotted onto a glass slide or other template, and cDNA from the individuals being studied, labeled with fluorescent dyes or other markers, is hybridized with that array. The intensity of fluorescence provides a quantification of the relative expression levels of targeted genes. Results are often validated with more precise estimates of RNA abundance (expression) using quantitative PCR for a subset of genes.
Gene expression profiles can be viewed as phenotypes because they are the product of both genetic and environmental variation (Hansen 2010). To assess genetic differences underlying gene expression, individuals can be reared in a common environment. Information on gene expression differences among populations can be used to complement data on neutral or adaptive genetic markers and adaptive traits for circumscribing conservation units. For example, Vandersteen Tymchuk et al. (2010) quantified gene expression for populations of Atlantic salmon in and around the Bay of Fundy, Newfoundland, using a 16,000 gene cDNA microarray. They found consistent year-to-year population differences in the expression of 389 genes when fish were reared in common environments. Population differentiation for gene expression was stronger, and patterns were somewhat different than those observed for seven microsatellite loci.
RNAseq (also called whole transcriptome shotgun sequencing) is replacing hybridization-based microarray technologies for many applications thanks to lowering costs of next-generation sequencing (Ozsolak and Milos 2011; Wang et al. 2009; Oomen and Hutchings 2017). RNAseq can more comprehensively assess the entire repertoire of RNA molecules expressed from genomes over a wider range of expression levels than can microarrays. We note that RNAseq can also be used for SNP discovery, for SNP genotyping, or for probe design for exon capture. For example, Bi et al. (2012) use RNAseq to discover SNPs within coding genes. They then used the gene sequences to design DNA sequence capture baits to test for SNPs associated with adaptive differentiation in chipmunks.
RNAseq and RADseq were used by Chen et al. (2018) to test for genetic variation in thermal adaptation in redband trout populations (Oncorhynchus mykiss gairdneri) from warm versus cool environments. In a common garden, fish from a desert climate had significantly higher thermal tolerance and aerobic scope (>3°C) for higher cardiac performance (e.g., without arrhythmia) than fish from the cooler montane climate. In addition, the desert fish had the highest maximum heart rate during warming, indicating improved capacity to deliver oxygen to internal tissues. Following heat stress, distinct sets of cardiac genes were induced, which helped explain the differences in cardiorespiratory function. Candidate RADseq SNP markers and nearby genes underlying these physiological adaptations were identified, including genes involved in metabolic activity and stress response (such as heat shock genes hsp40, ldh-b, and camkk2). This kind of study is rare in that it identified both transcriptomic and genomic mechanisms of evolutionary adaptation that allow populations to persist in the difficult environmental conditions of desert streams.
5 Bioinformatics for Filtering, Genotyping, and Data Analyses
Bioinformatics skills and understanding are crucial to analyze the increasingly massive DNA sequence datasets. Bioinformatics involves intensive computations to analyze DNA, RNA, and protein sequence datasets. The field of bioinformatics underwent explosive growth starting in the mid-1990s, driven largely by the Human Genome Project and rapid advances in DNA sequencing technology. Thus, the need for bioinformatics training and approaches has increased greatly in the last decade as the data produced by massive parallel sequencing approaches has grown exponentially. However, while the costs of genome sequencing are plummeting, time and money spent on bioinformatic data filtering and analysis (and production of bioinformatics platforms) have increased more slowly over time (Sboner et al. 2011). Given the many advantages and increasing ease of generating massively parallel sequencing (MPS) data, it has become crucial for population geneticists to be trained in computer programming and scripting to take full advantage of the growing catalog of bioinformatics tools (Andrews and Luikart 2014).
There are four major bioinformatics steps, often referred to as a bioinformatics pipeline, that occur in most population genomics studies including (1) sequence read filtering, (2) assignment of reads to loci (e.g., alignment to a reference genome or de novo loci assembly), (3) genotype calling, and (4) final filtering for problematic loci that do not meet biological expectations (e.g., Hardy-Weinberg proportions, high numbers of SNPs per locus (or 100 bp) usually resulting from alignment error, high observed heterozygosity, or more than two observed alleles) (Benestan et al. 2016). A major challenge in bioinformatic analysis is the creation of standardized pipelines (e.g., see the Broad Institute webpage for best practices – software.broadinstitute.org) that would improve consistency and comparison of results among species (and studies within species) but also even within the same species. Worrisome is the fact that different pipelines often result in very different results (for a given dataset) such that the number SNPs discovered and basic summary statistics and conclusions can change between pipelines (Shafer et al. 2017).
Analysis of up to entire genomes (millions of SNPs) presents challenges in filtering out loci that could lead to erroneous results and conclusions. There are no concrete rules for what criteria should be used for filtering loci from genomic datasets. The current state of filtering in population genomics has led to some colorful terms for filtering such as labeling filtering as the “F-word” or that filtering of genomic data is the “wild west” of population genomics (Benestan et al. 2016). Indeed, the potential effects of locus filtering approaches on downstream analyses and research conclusions have only recently started to be investigated (e.g., Lowry et al. 2017; Rodríguez-Ezpeleta et al. 2016; Shafer et al. 2017). However, it has also been suggested and shown empirically that filtering is helped greatly by the existence of a reference genome (Ellegren 2014; Hand et al. 2015a, b; Shafer et al. 2017).
Despite recent attempts to build conceptual and practical frameworks for MPS data analysis, a standardized pipeline remains elusive, and perhaps infeasible, given the nature of data variability present in most genomic datasets (Benestan et al. 2016). There has also been a move toward web-based platform analysis and filtering tools such as Galaxy which has gained users and popularity in recent years (Giardine et al. 2005; Afgan et al. 2016). Galaxy offers a more user friendly graphical interface for easy visualization and reproducibility of results through the tracking (logging) of all bioinformatic analysis steps and user-created and shared workflows. Workflows are flowchart-style representations of bioinformatics pipelines with drag and drop functionality that allows for easy customization, reproduction, and even publication of bioinformatics pipelines (Catchen et al. 2013; Eaton 2014). Galaxy also offers tools across a range of datatypes including RAD and RNAseq, WGS, and exon capture (Blankenberg et al. 2010; Pogorelcnik et al. 2018; Tranchant-Dubreuil et al. 2018). See the chapter “Computational Tools for Population Genomics” by Salojärvi (2018) in this book for more information.
6 Emerging Population Genomics Approaches
Here, we discuss emerging approaches that will become more widely used as costs decrease and technologies improve. These include population metagenomics, transcriptomics, epigenomics, proteomics, and paleogenomics.
6.1 Metagenomics
Metagenomics is the sequencing and analysis of DNA from all species in an environmental or gut sample (Srivathsan et al. 2016; Stat et al. 2017; Laforest-Lapointe et al. 2017; Waite et al. 2018). Metagenomics has usually been defined more narrowly as the study of DNA from microbial communities in environmental samples, perhaps because the initial studies were in microbes (Venter et al. 2004; Garcia et al. 2018). Metagenomics can be used to describe the diversity and relative abundance of taxonomic groups present within a single sample, experiment, or local population (DeLong 2009). These techniques have been applied widely to microbes in environmental samples, including water, soil, fecal, or gut samples, and subjected to high-throughput sequencing. Further, analysis of the functional groups of genes and their relative abundance, without requiring knowledge of which organism each sequence fragment came from, can provide a functional metabolic profile of the microbial community (Dinsdale et al. 2008).
From a population genomics perspective, metagenomics can allow the application of population genomics approaches (e.g., Fig. 1 or Fig. 2) on each of multiple microbial species, simultaneously. Further, if the microbial species are sampled from across a heterogeneous environment (or gradient), it facilitates the application of a landscape community genomics approach to improve understanding of eco-evolution interactions (Sect. 2.4; Hand et al. 2015b). Another application of metagenomic data is to describe a microbial community as an essential part of an individual host’s phenotype, influencing the health and fitness of the host. The application of metagenomics in ecology, evolution, and conservation is in its early stages, but a few specific areas show promise for the future. A chapter in this book series volume describes how population genomics approaches can be applied to metagenomic data to delineate microbial populations in the environment and to study evolutionary processes within them (Denef 2018).
Metagenomic surveillance systems are increasingly being used to improve monitoring and determine mechanisms driving the spread of infectious diseases. Portable genomic sequencers provide rapid near real-time diagnostics that can resolve important epidemiological and genomic characteristics of an outbreak or epidemic’s dynamics. As pathogens replicate and spread, mutations accumulate in their genomes. The whole-genome sequencing of spatially referenced samples allows researchers to track and reconstruct geo-spatial pathways of spread. Genomic epidemiology surveillance and rapid response programs can now take a more anticipatory approach to outbreak prevention and control.
Genomics-informed DNA detection assays have been developed to track a wide range of important fungal plant pathogens, including introduced, invasive species causing widespread diseases and mortality in natural populations and crop species (Feau et al. 2018). Monitoring and understanding which strains are emerging and associated with different environments and species (including humans) would also help to model, predict, and manage outbreaks and spread of pathogens. Whole-genome data from individual pathogen species in each of many host individuals can be used in population genomics approaches (or landscape community genomics approaches) to better understand the genomic basis of adaptation to hosts and local environments and to predict the effects of environmental change on a pathogen population and microbial community (Hand et al. 2015b).
Another application of metagenomics is to monitor or predict physiological condition, health, or fitness of individual organisms. For instance, Vega Thurber et al. (2009) have found shifts in the endosymbiont community of corals in response to stressors, such as reduced pH, increased nutrients, and increased temperature. Such shifts in the endosymbiont community could serve as indicators or predictors of reef health, and they could also suggest mechanisms by which coral condition affects other taxa in the reef ecosystem (Roitman et al. 2018; Leite et al. 2018).
Finally, a large-scale study used metagenomic techniques on fecal samples to catalog 3.3 million microbial genomes in the human gut fauna (Qin et al. 2010). The study found significant differences in the microbial metagenome between healthy individuals and those with two types of inflammatory bowel disease (Qin et al. 2010). In the future metagenomic techniques will be applied to noninvasively-collected fecal samples from wildlife species to assess their health status, such as starvation or disease infection, and to understand mechanisms underlying host and microbe interactions, population genomics, and coevolution (e.g., Beja-Pereira et al. 2009; Chiou and Bergey 2018; Waite et al. 2018).
6.2 Metatranscriptomics
While metagenomics focuses on detecting the presence of microbial species, metatranscriptomics investigates their gene expression profiles to address questions such as which genes are expressed in different environments or conditions. Thus, metatranscriptomics investigates the function and activity of the entire set of transcripts (RNAseq) from environmental, fecal, gut, or other samples. It is often used to identify sequences of genes expressed within natural microbial communities to advance understanding of microbial ecology and drivers of gene expression variation.
Assessing all the microbial community transcripts from a particular time and location, including bacteria, archaea, or small eukaryotes in the ocean, soil, or an organism’s gut, can help understand the complex microbial processes simultaneously occurring in natural or disturbed environments. This allows “eavesdropping on microbial ecology,” a promising new approach for researchers in ecosystem ecology, animal health, and functional biodiversity monitoring (Moran 2009).
From a population genomics perspective, metatranscriptomics – like metagenomics – can facilitate landscape community genomics approaches to improve understanding of eco-evolutionary processes (Hand et al. 2015b). Transcriptomic and metatranscriptomic data can detect gene expression shifts in both host and microbes simultaneously (e.g., lung tissue and lung parasites, gut tissue and gut parasites, blood and malaria, etc.) and thus can help understand, model, and predict host-parasite interactions (e.g., Matthews et al. 2018; Lee et al. 2018; Campbell et al. 2018).
Metatranscriptomics and metagenomics together can provide entire transcriptome and genome repertoires of microorganisms through sequencing total DNA/RNA from samples; this provides taxonomic and also functional information with high resolution. These two approaches together with new bioinformatics tools can help us better understand mechanisms of adaption, coevolution, and processes like rumen fermentation, digestion, and community adaption to environmental change. A challenge for “meta” approaches is that only a small percentage of the many ecologically important genes has been annotated or identified. Sequence datasets often contain only the abundant genes from a limited number of natural microbial communities (Moran 2009).
6.3 Population Epigenomics
While epigenetic inheritance is well documented the adaptive significance, if any, of such a complementary inheritance system remains enigmatic (Lind and Spagopoulou 2018).
Among the most intriguing and perhaps controversial areas of population genomics research involves understanding the role of transgenerational epigenetic inheritance in adaptive evolution. Can a strong environment change produce transgenerational epigenetic adaptation? Epigenetics has been defined as the study of heritable changes in a trait or phenotype caused by mechanisms other than DNA mutation. We focus here on transgenerational epigenetic inheritance, which is defined as changes in gene expression and resulting phenotypic variation that are transmitted between generations through germline, but do not involve changes in the underlying DNA sequence (Horsthemke 2018).
If environmentally caused shifts in gene expression are adaptive and transmitted to subsequent generations, it could represent a Lamarckian-type mechanism facilitating adaptation to environmental challenges, such as climate warming (e.g., Christie et al. 2016; Lind and Spagopoulou 2018; Horsthemke 2018). This idea could perhaps provide hope to conservation biologists that rapid adaption to climate warming is more likely than previously thought based on adaptation through natural selection. This idea is perhaps intriguing but still farfetched given the lack of evidence. The explosive growth in research on this topic results in part from the question of whether “epigenetic mechanisms might provide a basis for the inheritance of acquired traits” (Horsthemke 2018).
Charlesworth et al. (2017) state that “allele frequency change caused by natural selection is the only credible process underlying the evolution of adaptive organismal traits.” Similarly, Horsthemke (2018) states that the evidence for transgenerational epigenetic inheritance, “is not (yet) conclusive,” in mammals, even though “it has been observed in plants, nematodes and fruit flies.” While there is strong evidence for environmentally induced transgenerational inheritance of epigenetic gene expression changes that influence fitness traits, there is not yet evidence that such epigenetic changes persist in the longer term (many generations) or that they influence population genetic or evolutionary processes.
Questions outlined by Charlesworth et al. (2017) can help guide future research to investigate the potential role of transgenerational epigenetic inheritance in evolutionary adaptation. These questions include the following: How many generations do inherited epigenetic marks persist, and do they spread within and among populations? Also, are transgenerational epigenetics changes an important source of adaptive change, relative to DNA sequence change (Charlesworth et al. 2017)? These are population epigenetics questions, which now can be addressed using densely distributed epigenetic marks genome wide, thereby representing “narrow sense” population epigenomics.
Here we discuss recent evidence for environmentally induced multigenerational epigenetic inheritance. We also discuss the role or importance of this inheritance in population genomics research and understanding.
Evidence is growing rapidly for multigenerational transmission of environmentally induced epigenetic changes that influence fitness traits. Environmental factors observed to cause transgenerational epigenetic inheritance of phenotypic variation include heat shock or other thermal stresses, drought, salt stress, low-calorie diet, high-fat diet, smoking, and exposure to toxins, such as hydrocarbons from plastics, atrazine, tributyltinthe, pesticide DDT (dichlorodiphenyltrichloroethane), and the agricultural fungicide vinclozolin. Many of these stressors have caused transgenerational epigenetic inheritance in humans, fish, birds, plants, and insects.
Genome-wide environmentally induced transgenerational epigenetic inheritance of disease was documented in a recent study in rats. Ben Maamar et al. (2018) exposed one generation of gestating female rats to DDT or alternatively vinclozolin. The offspring (F1 generation) were bred to generate the F2 generation that was then bred to generate the F3 generation (keeping separate the populations exposed – in the F0 generation – to vinclozolin, DDT, or control treatments). The F3 generation males’ sperm revealed persistent environmentally induced histone modification genome wide (Fig. 13), which influences gene expression to cause disease. The fact that two different environmental toxins, each promoted transgenerational epigenetic (histone) changes, suggest that histone sites have a role in epigenetic transgenerational inheritance.
Sperm histone site differences (site retention) caused by DDT (dichlorodiphenyltrichloroethane) and transmitted over multiple generations. Red arrowheads are individual chromosome locations of histone differences in sperm. DDT-induced histone differences cause transgenerational epigenetic inheritance of disease. Purified cauda epididymal sperm were collected from the transgenerational F3 generation male rats for histone analysis. Reproduced with permission from Ben Maamar et al. (2018)
A particularly interesting study of epigenetic changes suggested that a single generation in an extreme environment (captivity, in a hatchery) can translate into heritable differences in expression at hundreds of genes. Christie et al. (2016) measured differential gene expression in the offspring of wild and first-generation hatchery steelhead trout (Oncorhynchus mykiss) and found 723 differentially expressed genes in the two groups of offspring reared in the same common environment. Functional analyses of the 723 genes revealed that most genes involved responses in immunity, wound healing, and metabolism. The large proportion of immunity and healing genes being differentially methylated suggest that the high density, rapid growth (and diet change), and aggression among fish in captivity lead to disease and wounds. Finally, wild-born fish that had only one hatchery parent had much lower reproductive success in the wild (compared to fish with two wild parents), suggesting that adaptation to captivity leads to transmission of maladaptive gene expression to wild-born offspring. These findings suggest that rapid environmental adaptation is possible and might be transmitted to offspring through “heritable” (transmitted) epigenetic changes.
It is becoming clear that multiple ancestral environmental influences, such as toxins, stress, or unusual nutrition, can sometimes induce germline epigenome changes called epimutations that are transmitted to descendants. These epimutations often occur in the germline and thus are transmitted (Gapp and Bohacek 2018). The germline epigenetic changes are often imprinted, and avoid epigenetic reprogramming (resetting/removal), and thus transgenerational inheritance occurs. Sperm RNAs are a mechanism for transfer of acquired complex phenotypes from father to offspring (Gapp et al. 2014). Stressful experiences were shown to cause metabolic and behavioral changes in mice that can be transmitted through RNAs in sperm to the offspring (Gapp and Bohacek 2018). Long-term studies are needed in natural populations to understand if inherited epigenetic marks persist across enough generations to significantly affect evolutionary processes, such as individual fitness, local adaption, gene flow, and population persistence.
6.3.1 Epigenetic Variation and Mechanisms
Here we discuss epigenetic variation that is potentially important evolutionarily but for which limited transgenerational inheritance information exists. Epigenomic variation is widespread in wild populations of plants (Schmitz et al. 2013a, b; Niederhuth et al. 2016) and animals (review in Hu and Barrett 2017). Epigenetic mechanisms causing gene expression shifts include DNA methylation, histone modifications, as well as variation in small RNAs. DNA methylation is the most frequently studied and best-understood epigenetic process to date. With the development of massive parallel sequencing techniques to examine genome-wide epigenetic marks, such as bisulfite DNA sequencing, epigenomics has progressed from investigating individual epigenomes to studying epigenomic variation across populations and species (e.g., Gavery and Roberts 2017).
The sources of epigenetic/epigenomic variation include genetic factors, environmental factors, or stochastic epimutations (reviews in Taudt et al. 2016; Yi 2017; Richards et al. 2017; Martin and Fry 2018). Recent studies have identified both the cis and trans regulatory genetic mechanisms conditioning population epigenomic variation at individual epigenetic marks to integrated chromatin state maps in a wide variety of species (review in Taudt et al. 2016). A number of methylation quantitative trait loci (meQTL) and histone quantitative trait loci (hQTL) have been identified in humans, plants, and animals (Taudt et al. 2016). Most of the work has been done on understanding the association of genetic (SNP, meQTL) and epigenetic variants for DNA methylation (DMR, differentially methylated region; DMP, differentially methylated polymorphism; SMV, single methylation variant; SMP, single methylation polymorphism). Nearly all of the detected meQTL in human mapped in cis association (review in Taudt et al. 2016).
Schmitz et al. (2013a), in the first plant population epigenomics study, examined the genome-wide DMRs in natural accessions of Arabidopsis worldwide and integrated these data with the whole-genome DNA sequences of the same accessions. They reported that 35% of the DMRs could be associated with meQTL, and 26% of the associations could be mapped to methylation changes in cis. In maize (Zea mays) about 50% of DMRs were associated in cis, with SNPs found within or near the DMR (Eichten et al. 2013). Similarly, cis meQTL-DMR associations were widespread in soybean (Glycine max) (Schmitz et al. 2013b). Heritable variation in methylation can be genetically based (and not sensitive to the environment), or environmentally induced, or a combination of both. Additionally, random epimutations can cause epigenetic variation as well.
6.3.2 Associations Between Epigenomic Variation and Phenotypic, Ecological, and Disease Traits
There is growing evidence that epigenetic mechanisms and epigenomic variation contribute significantly to phenotypes, abiotic and biotic stress responses, disease conditions, adaptation to habitat, and range distributions in a variety of organisms (review in Richards et al. 2017). This has significance in the context of acclimation and adaptation to climate change. Epigenomic differences are often correlated with ecological and environmental factors (see Richards et al. 2017). For example, DNA methylation patterns were found to be associated with a climate gradient in Quercus lobata (Gugger et al. 2016).
Recent population epigenomics studies have concentrated on associations between epigenomic variation and phenotypic, ecological, disease, and other traits in humans, plants, and animals through epigenome-wide association studies (EWAS) and epigenome environment association analysis (epiEAA), and a number of significant associations have been identified. In particular, substantial EWAS work has been done in the past few years to identify the association of DNA methylation with common human disease conditions.
DNA methylation has been found to be significantly associated with kidney function (Chu et al. 2017), type 2 diabetes (Meeks et al. 2017), panic disorder (Shimada-Sugimoto et al. 2017), cardiovascular diseases (Nakatochi et al. 2017), cancer (Xu et al. 2013), chronic obstructive pulmonary disease and lung function (Lee et al. 2017), and other conditions. Population epigenomics has a role to play in pharmacogenomics and personal medicine (see Kabekkodu et al. 2017). In plants epigenetic variation has been associated with various phenotypic, phenological, and disease and adaptive traits, such as salt tolerance (Foust et al. 2016), disease susceptibility (Sollars and Buggs 2018), and flowering time (Aller et al. 2018).
Population epigenomics, as such, is an emerging approach in population genomics. The detailed discussion of various aspects of population epigenomics is presented in the chapter by Moler et al. (2018) later in this book. This includes the molecular basis of epigenetic mechanism, sources and evolution of population epigenomic variation, intra- and interspecific epigenomic variation, molecular and bioinformatics methods in population epigenomics, and association of epigenomic variation with phenotypic, ecological, and disease traits and pharmacogenomics. See also recent reviews (e.g., Gapp and Bohacek 2018) and the special edition set of papers on the evolutionary consequences of epigenetic inheritance (Lind and Spagopoulou 2018).
6.4 Population Proteomics
Population proteomics is the study of structural and functional variation (qualitative and quantitative) in proteins within and among populations to better understand their role in individual fitness, phenotypic variation, local adaption, and population performance (see also Biron et al. 2006; Nedelkov et al. 2006; Nedelkov 2008). Enzyme protein polymorphisms (isoenzymes, isozymes, allozymes) provided the first molecular markers for population genetic studies. Protein electrophoresis studies were widely conducted for several decades before DNA markers became available (Charlesworth et al. 2016).
Although population proteomics gained attention around 2005 (e.g., Biron et al. 2006; Nedelkov et al. 2006; Nedelkov 2008), especially for biomarker discovery for human disease conditions, it has not kept pace with population genomics owing to the rapid advances in high-throughput DNA and RNA sequencing technologies. However, the development of 2D gel electrophoresis, mass spectrophotometry methodologies (such as MALDI TOF), and shotgun proteomics methods has made high-throughput protein analysis possible. This has accelerated population proteomics studies across different species (e.g., Ma et al. 2015; Armengaud 2016; Di et al. 2016; Hidalgo-Galiana et al. 2016; Colinet et al. 2017; Gamboa et al. 2017; Suhre et al. 2017).
Since proteins influence important phenotypes and are the products of genes and epigenetic or posttranslational mechanisms, population proteomics has the potential to provide key insights into functional and metapopulation ecology, adaptation, and acclimation processes under various climate and environment conditions (e.g., Biron et al. 2006; Karr 2008; Di et al. 2016; Colinet et al. 2017; Gamboa et al. 2017; Trapp et al. 2018). Population proteomics approaches also help identify genetic loci underlying risk of disease and for clinical biomarkers for many human disease conditions (Nedelkov et al. 2006; Suhre et al. 2017).
Most population proteomics studies to date have been focused on humans, especially for discovering and validating biomarkers for clinical disease conditions. High levels of protein diversity have been reported in humans. For example, a total of 76 structural forms variants were observed for the 25 plasma proteins (an average of 3 variants per protein) in a cohort of 96 individuals (Nedelkov et al. 2005). Proteomics-based genome-wide association studies have identified many associations between protein levels and gene variants (protein QTLs, pQTLs) in different population cohorts (summary provided in the supplementary table in Suhre et al. 2017 and updated on http://www.metabolomix.com/a-table-of-all-published-gwas-with-proteomics/). For example, Suhre et al. (2017) reported 539 pQTLs in German, Asian, and Arab cohorts, and associations overlapped with 57 genetic risk loci for 42 unique diseases.
Proteomics approaches have also been useful in nonhuman systems. For example, clear ecotype-specific protein variation was found among eight Arabidopsis ecotypes that were related to their physiological status (Chevalier et al. 2004). Rees et al. (2011) reported significant within and among population variation in proteins in three species of the teleost fish Fundulus; The authors suggested that the patterns of protein expression have evolved by natural selection.
Gamboa et al. (2017) investigated protein expression in five stream stonefly species (Plecoptera) from four geographic regions along a latitudinal gradient in Japan with varying climatic conditions. They found high spatial variation in protein expression among four geographic regions that were positively correlated with water temperature. However, low interspecific variation was observed in proteins within geographical regions, suggesting regulation of protein expression varied with environment and relates to local adaptation.
In Drosophila, Colinet et al. (2017) studied the regulatory mechanisms involved in the acquisition of thermal tolerance. They note that reversible phosphorylation is a common posttranslational modification that can rapidly alter proteins functions. They conducted a large-scale comparative study of phosphorylation networks in control versus cold-acclimated adult Drosophila and found that acclimation evoked a strong phosphoproteomic signal characterized by large sets of unique and differential phosphoproteins. In diving beetles (Agabus ramblae and A. brunneus), Hidalgo-Galiana et al. (2016) found protein expression parallels thermal tolerance and ecological conditions in the diversification of these two Agabus species.
These studies suggest that research on proteomic variation among natural populations along environmental gradients can provide insights into mechanisms underlying eco-evolutionary processes such as local adaptation, diversification, range shifts, and speciation. Future studies including genome-wide proteome data combined with population and landscape genomics approaches on multiple species (e.g., landscape community proteogenomics) will be especially helpful for understanding and predicting adaptive evolution, population performance, coevolution, and adaptive divergence.
6.5 Paleogenomics
Paleogenomics is the study of genomes of ancient organisms from fossil remains or specimen excavated from caves, permafrost, ice cores, or archeological or paleontological sites or stored in museum and herbarium collections (Heintzman et al. 2015; Lan and Lindqvist 2018). Paleogenetics and paleogenomics are recent fields of research relying on the extraction and analysis of preserved ancient DNA (aDNA). Early paleogenetics research was based on sequencing of mitochondrial DNA (mtDNA) fragments because of high copy numbers of the mitochondrial genomes in a cell. This research has provided quite useful information on phylogenetic relationships and timing of divergence among organisms and biographical patterns (Lan and Lindqvist 2018).
Paleogenomic studies are providing insights into complex evolutionary histories of ancient and extinct organisms, including humans (Homo sapiens) (Rasmussen et al. 2010; Meyer et al. 2012; Prüfer et al. 2014), phylogenetic and evolutionary relationships of extinct organisms with living species and populations (e.g., Prüfer et al. 2014; Heintzman et al. 2015; Lan and Lindqvist 2018), inferences of demographic patterns and ancient admixtures in human and other organisms (Meyer et al. 2012; Prüfer et al. 2014; Shapiro and Hofreiter 2014; Lan and Lindqvist 2018), reconstruction of ancient adaptive phenotypes and inferences of extinction causes, such as in wooly mammoth (Mammuthus primigenius) (Palkopoulou et al. 2015; Rogers and Slatkin 2017), and causal agents and evolutionary history of ancient pandemics, such as Black Death (bubonic plague), small pox, tuberculosis and leprosy (reviewed in Lan and Lindqvist 2018), ancient pathogens through human history (Marciniak and Poinar 2018), and structural variants in ancient genomes (Resendez et al. 2018).
Paleogenomic investigations have provided key insights into the origin and history or domestication of crop plants (reviewed in Lan and Lindqvist 2018) and animals, such as dogs (Canis lupus familiaris) (Frantz et al. 2016; Thalmann and Perri 2018), cats (Felis catus) (Geigl and Grange 2018), and horses (Equus caballus) (Orlando et al. 2013; Orlando 2018), origins and genetic legacy of Neolithic farmers and human settlement in Europe (Skoglund et al. 2012), reconstruction of ancient plant communities (Parducci et al. 2018), and epigenomics of ancient species (Hanghøj et al. 2018). Most of the above paleogenomics aspects are discussed later in this book in the chapter “Paleogenomics: Genome-scale Analysis of Ancient DNA and Population and Evolutionary Genomic Inferences” by Lan and Lindqvist (2018).
One of the most studied topics in paleogenomics is the evolution of human species and its phylogenetic and evolutionary relationships with its closest evolutionary relatives. The first ancient human genome was sequenced by Rasmussen et al. (2010) from permafrost-preserved hair of a ~4-kyr-old Paleo-Eskimo. Then paleogenomes from archaic hominins, Neanderthal and Denisovan, were sequenced and published (Meyer et al. 2012; Prüfer et al. 2014). These paleogenomics studies suggested that that Neanderthal and Denisovan populations shared a common origin, that their common ancestor diverged from the ancestors of modern humans, and that admixture had taken place between archaic hominins and the ancestors of modern humans most likely after the dispersal of modern non-African humans out of Africa (Meyer et al. 2012; Prüfer et al. 2014). The analysis also indicated that this gene flow was from Neanderthal into the common ancestor of modern Eurasians.
Another example of paleogenomics applications is the inferences of the causes of extinction of the iconic ancient animal wooly mammoth, which was an abundant megafaunal species of the Northern Hemisphere. As mentioned above (Sect. 2.6), paleogenomics studies provided evidence that genetic stochasticity due to small population size could have contributed to the extinction of this species (Palkopoulou et al. 2015; Rogers and Slatkin 2017).
7 Does the Field of Population Genomics Promise More Than It Can Deliver?
Population genomics holds a great deal of promise for increasing our understanding of the genetic basis of phenotypic variation and adaptation in natural populations. However, population genomics is not a panacea for addressing the outstanding fundamental questions in many areas of biology. Genomes are tremendously complex, and traits related to fitness are often highly polygenic. Researchers need to better recognize the limitations of some methods and the opportunities for misleading or misinterpreted results (e.g., false positives and false negatives for selection tests). For example, the hallmark genomic signatures of positive selection (e.g., highly reduced genetic variation, shifted site frequency spectrum, or alleles associated with environmental variation) can arise from forces other than positive selection.
False signatures of positive selection can occur where purifying (background) selection has reduced genetic variation, particularly in genomic regions with low recombination (Charlesworth et al. 1993; Wolf and Ellegren 2017). Regions with low genetic variation can be caused by a locally low mutation rate, or where large haplotypes have drifted to high frequency or fixation in populations with small N e (Nielsen et al. 2005; Kardos et al. 2015b). Regions with very high F ST relative to the genome-wide background can occur between insipient species as a result of selection within lineages (e.g., background selection or recent selective sweeps), rather than via divergent selection during speciation (Burri et al. 2015; Charlesworth et al. 1993; Cruickshank and Hahn 2014; Payseur and Rieseberg 2016; Wolf and Ellegren 2017). Thus, genomic signatures of positive selection, including selective sweep signals and F ST outlying regions must be interpreted cautiously.
Population genomics studies can have low power to detect loci related to adaptation or variation in phenotypes among individuals, especially for highly polygenic traits. The relatively low density of SNPs generated, in certain species, when using some technologies (e.g., some RADseq or sequence capture) means that selective sweeps, F ST outliers, associations between markers and environmental variables, and QTLs may be missed because of low or no gametic disequilibrium between the genotyped SNPs and causal loci (Kardos et al. 2016a; (Catchen et al. 2017; McKinney et al. 2017a). Associations and outliers can also be missed by genotyping only a limited number of SNPs from an adaptive gene or a selected genome region (Fig. 6).
Additionally, the relatively low sample sizes that are frequent in studies of non-model organisms in the wild means that power to detect loci with relatively large effects may often be low, even when whole-genome sequencing is used in natural populations (Kardos et al. 2015a; Lotterhos and Whitlock 2014; Hunter et al. 2018; Flanagan et al. 2018). Finally, to help increase the understanding of the genetic basis of ecological and evolutionary traits and processes, we recommend applying multiple population genomics and related approaches (at different functional levels from DNA to RNA and proteins), as in Vasemagi and Primmer (2005).
8 Future Perspectives and Needs
Among the most exciting advances from “neutral” marker studies will be our improved understanding of inbreeding depression and genetic rescue in natural and managed populations. This will result from the fact that only 5000–10000 SNP loci are required to vastly improve precision of estimation of individual inbreeding compared to traditional marker-based and pedigree approaches (Kardos et al. 2016a). There will soon be many publications that use genomic data to estimate inbreeding depression (and genetic rescue) in many populations, which could change our view of the importance of inbreeding in conservation and evolution. Interestingly, most publications in the vast inbreeding literature had low power and precision to estimate inbreeding and inbreeding depression effects.
Even more exciting will be the use of novel, more informative statistical estimators such as ROH (runs of homozygosity), which measures inbreeding and effective population size change (Palkopoulou et al. 2015; Kardos et al. 2018; Grossen et al. 2018). The bioinformatic prediction of deleterious alleles from sequence data will also increase our ability to understand the genomic architecture of inbreeding depression and to predict and compare populations for genetic load.
An interesting advance will be the improved understanding of the importance of transgenerational epigenetic inheritance in adaptive traits (Charlesworth et al. 2017). Advances are likely to give the explosion of research and publications, following the controversy and calls to test the relevance of epigenetic “inheritance” in evolutionary processes and given lower costs for next-generation (bisulfite) sequencing (Christie et al. 2016; Le Luyer et al. 2017; Nilsson et al. 2018; Horsthemke 2018). Can environmentally induced transgenerational epigenetic inheritance contribute substantially to adaption to changing environments?
Another general advancement in power and precision will result from calling of microhaplotypes from short-read data. Most publications that use next-generation short-read data (e.g., RADseq) have not called haplotypes but rather scored only one SNP (or two independent SNPs) per locus, even though multiple SNPs exist per locus, e.g., RAD loci (Hendricks et al. 2018). Haplotype calling will yield more alleles (haplotypes), additional genealogical or phylogenetic information, and thus more power for many applications in population genetics (Sunnucks 2000). Longer single-end and paired-end reads and new software for haplotype calling will also improve power (Baetscher et al. 2018).
Understanding of the importance of structural polymorphisms in fitness and adaptation will increase soon (Wellenreuther and Bernatchez 2018). Genotyping and detection of inversions and copy number variants are becoming more feasible thanks to longer-read sequencing, reference genomes, linkage maps, and improved software for discovering and genotyping structural polymorphisms (e.g., Farek et al. 2018). This will help population genomics move beyond SNPs. This is an important advancement because structural variations are often involved with fitness-related phenotypic variation (e.g., Küpper et al. 2015) and are thought to play a key role in sex chromosome evolution, local adaptation, and speciation (Kirkpatrick 2010; Wellenreuther and Bernatchez 2018).
Many studies will estimate gametic disequilibrium along chromosomes (or contigs) using draft genome assemblies, thereby allowing more informative “narrow sense” population genomics studies with mapped high-density markers. Even a few hundred contigs of 50–500 kb and 1,000s of marker loci will provide quantification of genome-wide GD (gametic disequilibrium) required for some narrow sense genomics approaches. Depending on the genome size and complexity, an investment of $10k to $20k can achieve a useful draft reference genome with an N50 of >50 kb for many species (Catchen et al. 2017; McKinney et al. 2017a; Hendricks et al. 2018).
There is a need to train researchers and students in data analysis including the initial filtering, genotyping, and data interpretation steps which requires an understanding of population genetics theory (Andrews and Luikart 2014; Allendorf 2017; Shafer et al. 2015; Hendricks et al. 2018). The trend toward learning the latest molecular techniques (RAD approaches, DNA capture, pool-seq, etc.) at the expense of a solid grounding in population genetics theory is worrisome (Allendorf 2017). Training in theoretical and conceptual aspects of population genetics enables researcher to ask good questions and to adequately test and interpret the massive and growing datasets against appropriate null models (Benestan et al. 2016; Allendorf 2017).
There is an urgent need for understanding the effects of data analysis choices on downstream biological inferences (Farek et al. 2018), because these choices can dramatically influence downstream statistical results and inferences (Shafer et al. 2017; Hendricks et al. 2018). We need to validate pipelines and downstream genomic statistical estimators, ensuring they are unbiased, by analyzing raw simulated and empirical data from populations with known genotypes and evolutionary parameters (N e, Nm, S) in order to verify that we can recover or estimate the true (known) genotypes and parameters. Related to this, the field needs to develop a set of best practices for identifying possible genotyping errors, quantifying error rates, and quantifying effects of data analysis choices on downstream results and conclusions. The most rigorous approach for ensuring data quality can vary substantially from dataset to dataset and will change through time as the structure and quality or data change; thus we need the next generation of population genomicists to be well trained in bioinformatics and programming (Andrews and Luikart 2014).
Finally, new computational approaches and modeling made easy by ABC (approximate Bayesian computation) will vastly improve data analysis and inference from population genomic data (Cabrera and Palsbøll 2017; Elleouet and Aitken 2018). However, extensive model performance evaluations are required to ensure computational approaches are applied reliably and competently to natural populations (e.g., Lotterhos and Whitlock 2014; Forester et al. 2018; see Appendix in Allendorf et al. 2013).
9 Conclusions
Population genomics is transforming many sub-disciplines in biology and vastly improving our understanding of nature (Schlötterer 2004; Hohenlohe et al. 2018). The greatest advances in our fundamental understanding of populations and the translation of that knowledge to decisions around managing and conserving populations will result from applications of conceptually novel “narrow sense” genomics studies. This revolution will continue to accelerate for many years as more studies combine population genomics, transcriptomics, transgenerational epigenomics, and proteomics approaches simultaneously to multiple species co-distributed across environments (Chen et al. 2018; De Kort et al. 2018). This increase in strategic applications of narrow sense and multiple omics approaches combined with phenotypic and environmental data (e.g., from sensor networks and remote sensing) will ensure we will soon be answering long-standing questions along with novel questions yet to be imagined by humanity. It is an exciting time to be a population genomicist!
References
Afgan E, Baker D, van den Beek M, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016;44:W3–W10.
Ahrens CW, Rymer PD, Stow A, et al. The search for loci under selection: trends, biases and progress. Mol Ecol. 2018;27:1342–56.
Alachiotis N, Pavlidis P. RAiSD detects positive selection based on multiple signatures of a selective sweep and SNP vectors. Commun Biol. 2018;1:79.
Ali OA, O’Rourke SM, Amish SJ, et al. RAD capture (rapture): flexible and efficient sequence-based genotyping. BioRxiv. 2015;52:4–7.
Allendorf FW. Genetics and the conservation of natural populations: allozymes to genomes. Mol Ecol. 2017;26:420–30.
Allendorf FW, Hohenlohe PA, Luikart G. Genomics and the future of conservation genetics. Nat Rev Genet. 2010;11:697–709.
Allendorf FW, et al. Conservation and the genetics of populations. Hoboken: Wiley; 2013.
Aller EST, Jagd LM, Kliebenstein DJ, Burow M. Comparison of the relative potential for epigenetic and genetic variation to contribute to trait stability. G3. 2018;8:1733–46.
Amaral AJ, Megens H-J, Crooijmans RPMA, Heuven HCM, Groenen MAM. Linkage disequilibrium decay and haplotype block structure in the pig. Genetics. 2008;179:569–79.
Ameur A, Kloosterman WP, Hestand MS. Single-molecule sequencing: towards clinical applications. Trends Biotechnol. 2018. In press.
Amores A, Catchen J, Ferrara A, Fontenot Q, Postlethwait JH. Genome evolution and meiotic maps by massively parallel DNA sequencing: spotted gar, an outgroup for the teleost genome duplication. Genetics. 2011;188:799–808.
Andrews KR, Luikart G. Recent novel approaches for population genomics data analysis. Mol Ecol. 2014;23:1661–7.
Andrews KR, Good JM, Miller MR, Luikart G, Hohenlohe PA. Harnessing the power of RADseq for ecological and evolutionary genomics. Nat Rev Genet. 2016;17:81–92.
Armengaud J. Next-generation proteomics faces new challenges in environmental biotechnology. Curr Opin Biotechnol. 2016;38:174–82.
Armstrong C, Richardson DS, Hipperson H, et al. Genomic associations with bill length and disease reveal drift and selection across island bird populations. Evol Lett. 2018;2(1):22–36.
Axelsson E, Ratnakumar A, Arendt ML, et al. The genomic signature of dog domestication reveals adaptation to a starch-rich diet. Nature. 2013;495:360–4.
Baetscher DS, Clemento AJ, Ng TC, Anderson EC, Garza JC. Microhaplotypes provide increased power from short-read DNA sequences for relationship inference. Mol Ecol Resour. 2018;18:296–305.
Balkenhol N, Dudaniec RY, Krutovsky KV, Johnson JS, Cairns DM, Segelbacher G, et al. Landscape genomics: understanding relationships between environmental heterogeneity and genomic characteristics of populations. In: Om PR, editor. Population genomics: concepts, approaches and applications. Cham: Springer International Publishing AG; 2017. https://doi.org/10.1111/eva.12672.
Barson NJ, Aykanat T, Hindar K, et al. Sex-dependent dominance at a single locus maintains variation in age at maturity in salmon. Nature. 2015;528:405–8.
Beichman AC, Phung TN, Lohmueller KE. Comparison of single genome and allele frequency data reveals discordant demographic histories. G3. 2017;7:3605–20.
Beja-Pereira A, Luikart G, England PR, et al. Gene-culture coevolution between cattle milk protein genes and human lactase genes. Nat Genet. 2003;35:311–3.
Beja-Pereira A, et al. Advancing ecological understandings through technological transformations in noninvasive genetics. Mol Ecol Resour. 2009;9:1279–301.
Ben Maamar M, Sadler-Riggleman I, Beck D, Skinner MK. Epigenetic transgenerational inheritance of altered sperm histone retention sites. Sci Rep. 2018;8:5308.
Benestan LM, Ferchaud AL, Hohenlohe PA, et al. Conservation genomics of natural and managed populations: building a conceptual and practical framework. Mol Ecol. 2016;25:2967–77.
Bérénos C, Ellis PA, Pilkington JG, et al. Heterogeneity of genetic architecture of body size traits in a free-living population. Mol Ecol. 2015;24:1810–30.
Bérénos C, Ellis PA, Pilkington JG, Pemberton JM. Genomic analysis reveals depression due to both individual and maternal inbreeding in a free-living mammal population. Mol Ecol. 2016;25:3152–68.
Berg JJ, Coop G. A population genetic signal of polygenic adaptation. PLoS Genet. 2014;10:e1004412.
Berthelot C, Brunet F, Chalopin D, et al. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat Commun. 2014;5:3657.
Betts A, Gray C, Zelek M, MacLean RC, King KC. High parasite diversity accelerates host adaptation and diversification. Science. 2018;360:907–11.
Bi K, Vanderpool D, Singhal S, et al. Transcriptome-based exon capture enables highly cost-effective comparative genomic data collection at moderate evolutionary scales. BMC Genomics. 2012;13:403.
Biron D, et al. Population proteomics: an emerging discipline to study metapopulation ecology. Proteomics. 2006;6:1712–5.
Black WC, Baer CF, Antolin MF, DuTeau NM. Population genomics : genome-wide sampling of insect populations. Annu Rev Entomol. 2001;46:441–69.
Blankenberg D, Von KG, Coraor N, et al. Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol. 2010;89:1–21.
Blanquart F, Kaltz O, Nuismer SL, Gandon S. A practical guide to measuring local adaptation. Ecol Lett. 2013;16:1195–205.
Boitard S, Rodríguez W, Jay F, Mona S, Austerlitz F. Inferring population size history from large samples of genome-wide molecular data – an approximate Bayesian computation approach. PLoS Genet. 2016;12:e1005877.
Bonin A, Nicole F, Pompanon F, Miaud C, Taberlet P. Population adaptive index: a new method to help measure intraspecific genetic diversity and prioritize populations for conservation. Conserv Biol. 2007;21:697–708.
Bos K, et al. Parallel detection of ancient pathogens via array-based DNA capture. Philos Trans R Soc Lond B Biol Sci. 2015;370:20130375.
Bourret V, Dionne M, Bernatchez L. Detecting genotypic changes associated with selective mortality at sea in Atlantic salmon: polygenic multilocus analysis surpasses genome scan. Mol Ecol. 2014;23:4444–57.
Boyle EA, Li YI, Pritchard JK. An expanded view of complex traits: from polygenic to omnigenic. Cell. 2017;169:1177–86.
Brauer CJ, Unmack PJ, Smith S, Bernatchez L, Beheregaray LB. On the roles of landscape heterogeneity and environmental variation in determining population genomic structure in a dendritic system. Mol Ecol. 2018;27:3484–97.
Brelsford A, Toews DPL, Irwin DE. Admixture mapping in a hybrid zone reveals loci associated with avian feather coloration. Proc Roy Soc B Biol Sci. 2017;284:20171106.
Brieuc MSO, Ono K, Drinan DP, Naish KA. Integration of random forest with population-based outlier analyses provides insight on the genomic basis and evolution of run timing in Chinook salmon (Oncorhynchus tshawytscha). Mol Ecol. 2015;24:2729–46.
Browning SR, Browning BL. Accurate non-parametric estimation of recent effective population size from segments of identity by descent. Am J Hum Genet. 2015;97:404–18.
Bürger R, Akerman A. The effects of linkage and gene flow on local adaptation: a two-locus continent-island model. Theor Popul Biol. 2011;80:272–88.
Burri R, Nater A, Kawakami T, et al. Linked selection and recombination rate variation drive the evolution of the genomic landscape of differentiation across the speciation continuum of Ficedula flycatchers. Genome Res. 2015;25:1656–65.
Burri R, Antoniazza S, Gaigher A, et al. The genetic basis of color-related local adaptation in a ring-like colonization around the Mediterranean. Evolution. 2016;70:140–53.
Cabrera AA, Palsbøll PJ. Inferring past demographic changes from contemporary genetic data: a simulation-based evaluation of the ABC methods implemented in diyabc. Mol Ecol Resour. 2017;17:e94–e110.
Cammen KM, Schultz TF, Don Bowen W, et al. Genomic signatures of population bottleneck and recovery in Northwest Atlantic pinnipeds. Ecol Evol. 2018;8:6599–614.
Campana MG, Hawkins MTR, Henson LH, et al. Simultaneous identification of host, ectoparasite and pathogen DNA via in-solution capture. Mol Ecol Resour. 2016;16:1224–39.
Campbell LJ, Hammond SA, Price SJ, et al. A novel approach to wildlife transcriptomics provides evidence of disease-mediated differential expression and changes to the microbiome of amphibian populations. Mol Ecol. 2018;27:1413–27.
Campos JL, Halligan DL, Haddrill PR, Charlesworth B. The relation between recombination rate and patterns of molecular evolution and variation in drosophila melanogaster. Mol Biol Evol. 2014;31:1010–28.
Candy JR, Campbell NR, Grinnell MH, et al. Population differentiation determined from putative neutral and divergent adaptive genetic markers in Eulachon (Thaleichthys pacificus, Osmeridae), an anadromous Pacific smelt. Mol Ecol Resour. 2015;15:1421–34.
Carneiro M, Albert FW, Afonso S, et al. The genomic architecture of population divergence between subspecies of the European rabbit. PLoS Genet. 2014;10:e1003519.
Castellano S, Parra G, Sanchez-Quinto FA, et al. Patterns of coding variation in the complete exomes of three Neanderthals. Proc Natl Acad Sci. 2014;111:6666–71.
Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA. Stacks: an analysis tool set for population genomics. Mol Ecol. 2013;22:3124–40.
Catchen JM, Hohenlohe PA, Bernatchez L, et al. Unbroken: RADseq remains a powerful tool for understanding the genetics of adaptation in natural populations. Mol Ecol Resour. 2017;17:362–5.
Ceballos FC, Joshi PK, Clark DW, Ramsay M, Wilson JF. Runs of homozygosity: windows into population history and trait architecture. Nat Rev Genet. 2018;19:220–34.
Charlesworth B. Fundamental concepts in genetics: effective population size and patterns of molecular evolution and variation. Nat Rev Genet. 2009;10:195–205.
Charlesworth B. Molecular population genomics: a short history. Genet Res. 2010;92:397–411.
Charlesworth D, Willis JH. The genetics of inbreeding depression. Nat Rev Genet. 2009;10:783–96.
Charlesworth B, Morgan MT, Charlesworth D. The effect of deleterious mutations on neutral molecular variation. Genetics. 1993;134:1289–303.
Charlesworth B, Charlesworth D, Coyne JA, Langley CH. Hubby and Lewontin on protein variation in natural populations: when molecular genetics came to the rescue of population genetics. Genetics. 2016;203:1497–503.
Charlesworth D, Barton NH, Charlesworth B. The sources of adaptive variation. Proc Roy Soc B Biol Sci. 2017;284:20162864.
Chen Z, Farrell AP, Matala A, Hoffman N, Narum SR. Physiological and genomic signatures of evolutionary thermal adaptation in redband trout from extreme climates. Evol Appl. 2018. https://doi.org/10.1111/eva.12672.
Chevalier F, Martin O, Rofidal V, et al. Proteomic investigation of natural variation between Arabidopsis ecotypes. Proteomics. 2004;4:1372–81.
Chiou KL, Bergey CM. Methylation-based enrichment facilitates low-cost, noninvasive genomic scale sequencing of populations from feces. Sci Rep. 2018;8:1975.
Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31:2745–7.
Christie MR, Marine ML, Fox SE, French RA, Blouin MS. A single generation of domestication heritably alters the expression of hundreds of genes. Nat Commun. 2016;7:10676.
Chu AY, Tin A, Schlosser P, et al. Epigenome-wide association studies identify DNA methylation associated with kidney function. Nat Commun. 2017;8:1286.
Colinet H, Pineau C, Com E. Large scale phosphoprotein profiling to explore Drosophila cold acclimation regulatory mechanisms. Sci Rep. 2017;7:1713.
Conte GL, Hodgins KA, Yeaman S, et al. Bioinformatically predicted deleterious mutations reveal complementation in the interior spruce hybrid complex. BMC Genomics. 2017;18:970.
Cooke NP, Nakagome S. Fine-tuning of approximate Bayesian computation for human population genomics. Curr Opin Genet Dev. 2018;53:60–9.
Coop G, Witonsky D, Di Rienzo A, Pritchard JK. Using environmental correlations to identify loci underlying local adaptation. Genetics. 2010;185:1411–23.
Corbett-Detig RB, Hartl DL, Sackton TB. Natural selection constrains neutral diversity across a wide range of species. PLoS Biol. 2015;13:e1002112.
Cornuet JM, Luikart G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics. 1996;144:2001–14.
Cosart T, Beja-Pereira A, Chen S, et al. Exome-wide DNA capture and next generation sequencing in domestic and wild species. BMC Genomics. 2011;12:347–55.
Cruickshank TE, Hahn MW. Reanalysis suggests that genomic islands of speciation are due to reduced diversity, not reduced gene flow. Mol Ecol. 2014;23:3133–57.
Cutter AD, Payseur BA. Genomic signatures of selection at linked sites: unifying the disparity among species. Nat Rev Genet. 2013;14:262–74.
Dalongeville A, Benestan L, Mouillot D, Lobreaux S, Manel S. Combining six genome scan methods to detect candidate genes to salinity in the Mediterranean striped red mullet (Mullus surmuletus). BMC Genomics. 2018;19:217.
De Kort H, Baguette M, Prunier JG, et al. Genetic costructure in a meta-community under threat of habitat fragmentation. Mol Ecol. 2018;27:2193–203.
De La Torre AR, Birol I, Bousquet J, et al. Insights into conifer giga-genomes. Plant Physiol. 2014;166:1724–32.
De Mita S, Thuillet AC, Gay L, et al. Detecting selection along environmental gradients: analysis of eight methods and their effectiveness for outbreeding and selfing populations. Mol Ecol. 2013;22:1383–99.
Degiorgio M, Huber CD, Hubisz MJ, Hellmann I, Nielsen R. SweepFinder2: increased sensitivity, robustness and flexibility. Bioinformatics. 2016;32:1895–7.
DeLong EF. The microbial ocean from genomes to biomes. Nature. 2009;459:200–6.
Denef VJ. Peering into the genetic makeup of natural microbial populations using metagenomics. In: Polz MF, Om PR, editors. Population genomics: microorganisms. Cham: Springer International Publishing AG; 2018. https://doi.org/10.1007/13836_2018_14.
Di G, Miao X, Ke C, et al. Protein changes in abalone foot muscle from three geographical populations of Haliotis diversicolor based on proteomic approach. Ecol Evol. 2016;6:3645–57.
Dinsdale EA, Edwards RA, Hall D, et al. Functional metagenomic profiling of nine biomes. Nature. 2008;452:629–32.
Do C, Waples RS, Peel D, et al. NeEstimator v2: re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol Ecol Resour. 2014;14:209–14.
Dobrynin P, Liu S, Tamazian G, et al. Genomic legacy of the African cheetah, Acinonyx jubatus. Genome Biol. 2015;16:277.
Dowle EJ, Pochon X, C Banks J, Shearer K, Wood SA. Targeted gene enrichment and high-throughput sequencing for environmental biomonitoring: a case study using freshwater macroinvertebrates. Mol Ecol Resour. 2016;16:1240–54.
Dupuis JR, Oliver JC, Brunet BMT, et al. Genomic data indicate ubiquitous evolutionary distinctiveness among populations of California metalmark butterflies. Conserv Genet. 2018. In press.
Duranton M, Allal F, Fraïsse C, et al. The origin and remolding of genomic islands of differentiation in the European sea bass. Nat Commun. 2018;9:2518.
Eaton DAR. PyRAD: assembly of de novo RADseq loci for phylogenetic analyses. Bioinformatics. 2014;30:1844–9.
Edwards SV, Potter S, Schmitt CJ, Bragg JG, Moritz C. Reticulation, divergence, and the phylogeography–phylogenetics continuum. Proc Natl Acad Sci. 2016;113:8025–32.
Eichten SR, Briskine R, Song J, et al. Epigenetic and genetic influences on DNA methylation variation in maize populations. Plant Cell. 2013;25:2783–97.
Elbasyoni IS, Lorenz AJ, Guttieri M, et al. A comparison between genotyping-by-sequencing and array-based scoring of SNPs for genomic prediction accuracy in winter wheat. Plant Sci. 2018;270:123–30.
Ellegren H. Genome sequencing and population genomics in non-model organisms. Trends Ecol Evol. 2014;29:51–63.
Ellegren H, Smeds L, Burri R, et al. The genomic landscape of species divergence in Ficedula flycatchers. Nature. 2012;491:756–60.
Elleouet JS, Aitken SN. Exploring approximate Bayesian computation for inferring recent demographic history with genomic markers in nonmodel species. Mol Ecol Resour. 2018;18:525–40.
Epstein B, et al. Rapid evolutionary response to a transmissible cancer in Tasmanian devils. Microbiome. 2016;6(1):168.
Farek J, Hughes D, Mansfield A, et al. xAtlas: scalable small variant calling across heterogeneous next-generation sequencing experiments. BioRxiv. 2018:295071.
Faria NR, Kraemer MUG, Hill S, et al. Genomic and epidemiological monitoring of yellow fever virus transmission potential. BioRxiv. 2018:299842.
Fay JC, Wu CI. Hitchhiking under positive Darwinian selection. Genetics. 2000;155:1405–13.
Feau N, Beauseigle S, Bergeron M-J, et al. Genome-enhanced detection and identification (GEDI) of plant pathogens. PeerJ. 2018;6:e4392.
Feder JL, Egan SP, Nosil P. The genomics of speciation-with-gene-flow. Trends Genet. 2012;28:342–50.
Ferrer-Admetlla A, Liang M, Korneliussen T, Nielsen R. On detecting incomplete soft or hard selective sweeps using haplotype structure. Mol Biol Evol. 2014;31:1275–91.
Fisher R. The theory of inbreeding. 2nd ed. Edinburgh: Oliver & Boyd; 1965.
Flanagan SP, Forester BR, Latch EK, Aitken SN, Hoban S. Guidelines for planning genomic assessment and monitoring of locally adaptive variation to inform species conservation. Evol Appl. 2018;11:1035–52.
Foll M, Fischer MC, Heckel G, Excoffier L. Estimating population structure from AFLP amplification intensity. Mol Ecol. 2010;19:4638–47.
Forester BR, Jones MR, Joost S, Landguth EL, Lasky JR. Detecting spatial genetic signatures of local adaptation in heterogeneous landscapes. Mol Ecol. 2016;25:104–20.
Forester BR, Lasky JR, Wagner HH, Urban DL. Comparing methods for detecting multilocus adaptation with multivariate genotype-environment associations. Mol Ecol. 2018;27:2215–33.
Forstmeier W, Schielzeth H, Mueller JC, Ellegren H, Kempenaers B. Heterozygosity-fitness correlations in zebra finches: microsatellite markers can be better than their reputation. Mol Ecol. 2012;21:3237–49.
Foust CM, Preite V, Schrey AW, et al. Genetic and epigenetic differences associated with environmental gradients in replicate populations of two salt marsh perennials. Mol Ecol. 2016;25:1639–52.
Fraïsse C, Roux C, Gagnaire P-A, et al. The divergence history of European blue mussel species reconstructed from approximate Bayesian computation: the effects of sequencing techniques and sampling strategies. PeerJ. 2018;6:e5198.
Franklin IR. The distribution of the proportion of the genome which is homozygous by descent in inbred individuals. Theor Popul Biol. 1977;11:60–80.
Frantz LAF, Mullin VE, Pionnier-Capitan M, et al. Genomic and archaeological evidence suggests a dual origin of domestic dogs. Science. 2016;352:1228–31.
Frichot E, Schoville SD, Bouchard G, François O. Testing for associations between loci and environmental gradients using latent factor mixed models. Mol Biol Evol. 2013;30:1687–99.
Funk WC, McKay JK, Hohenlohe PA, Allendorf FW. Harnessing genomics for delineating conservation units. Trends Ecol Evol. 2012;27:489–96.
Funk WC, Forester BR, Converse SJ, Darst C, Morey S. Improving conservation policy with genomics: a guide to integrating adaptive potential into U.S. Endangered Species Act decisions for conservation practitioners and geneticists. Conserv Genet. 2018. In press.
Gamboa M, Tsuchiya MC, Matsumoto S, Iwata H, Watanabe K. Differences in protein expression among five species of stream stonefly (Plecoptera) along a latitudinal gradient in Japan. Arch Insect Biochem Physiol. 2017;96:e21422.
Gapp K, Bohacek J. Epigenetic germline inheritance in mammals: looking to the past to understand the future. Genes Brain Behav. 2018;17:e12407.
Gapp K, Jawaid A, Sarkies P, et al. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat Neurosci. 2014;17:667–9.
Garcia SL, Stevens SLR, Crary B, Martinez-Garcia M, Stepanauskas R, et al. Contrasting patterns of genome-level diversity across distinct co-occurring bacterial populations. ISME J. 2018;12:742–55. https://doi.org/10.1038/s41396-017-0001-0.
Garner BA, Hand BK, Amish SJ, et al. Genomics in conservation: case studies and bridging the gap between data and application. Trends Ecol Evol. 2016;31:81–2.
Gasc C, Peyretaillade E, Peyret P. Sequence capture by hybridization to explore modern and ancient genomic diversity in model and nonmodel organisms. Nucleic Acids Res. 2016;44:4504–18.
Gauthier J, Mouden C, Suchan T, et al. DiscoSnp-RAD: de novo detection of small variants for population genomics. BioRxiv. 2017:216747.
Gavery MR, Roberts SB. Epigenetic considerations in aquaculture. PeerJ. 2017;5:e4147.
Geigl E-M, Grange T. Of cats and men: ancient dNA reveals how the cat conquered the ancient world. In: Lindqvist C, Om PR, editors. Paleogenomics. Cham: Springer International Publishing AG; 2018. https://doi.org/10.1007/13836_2018_26.
Ghalambor CK, Hoke KL, Ruell EW, et al. Non-adaptive plasticity potentiates rapid adaptive evolution of gene expression in nature. Nature. 2015;525:372–5.
Giardine B, Riemer C, Hardison RC, et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–5.
Gibson G. Population genetics and GWAS: a primer. PLoS Biol. 2018;16:e2005485.
Gilbert KJ, Whitlock MC. Evaluating methods for estimating local effective population size with and without migration. Evolution. 2015;69:2154–66.
Gompert Z. A continuous correlated beta process model for genetic ancestry in admixed populations. PLoS One. 2016;11:e0151047.
Goudet J, Kay T, Weir BS. How to estimate kinship. Mol Ecol. 2018. In press.
Gray MM, Granka JM, Bustamante CD, et al. Linkage disequilibrium and demographic history of wild and domestic canids. Genetics. 2009;181:1493–505.
Grossen C, Biebach I, Angelone-Alasaad S, Keller LF, Croll D. Population genomics analyses of European ibex species show lower diversity and higher inbreeding in reintroduced populations. Evol Appl. 2018;11:123–39.
Gruber B, Unmack PJ, Berry OF, Georges A. dartr: an r package to facilitate analysis of SNP data generated from reduced representation genome sequencing. Mol Ecol Resour. 2018;18:691–9.
Guan Y. Detecting structure of haplotypes and local ancestry. Genetics. 2014;196:625–42.
Gugger PF, Fitz-Gibbon S, Pellegrini M, Sork VL. Species-wide patterns of DNA methylation variation in Quercus lobata and their association with climate gradients. Mol Ecol. 2016;25:1665–80.
Gunther T, Coop G. Robust identification of local adaptation from allele frequencies. Genetics. 2013;195:205–20.
Gur A, Tzuri G, Meir A, et al. Genome-wide linkage-disequilibrium mapping to the candidate gene level in melon (Cucumis melo). Sci Rep. 2017;7:9770.
Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 2009;5:e1000695.
Hackinger S, Kraaijenbrink T, Xue Y, et al. Wide distribution and altitude correlation of an archaic high-altitude-adaptive EPAS1 haplotype in the Himalayas. Hum Genet. 2016;135:393–402.
Hancock AM, et al. Adaptation to climate across the Arabidopsis thaliana genome. Science. 2011;334:83–6.
Hand BK, Hether TD, Kovach RP, et al. Genomics and introgression: discovery and mapping of thousands of species-diagnostic SNPs using RAD sequencing. Curr Zool. 2015a;61:146–54.
Hand BK, Lowe WH, Kovach RP, Muhlfeld CC, Luikart G. Landscape community genomics: understanding eco-evolutionary processes in complex environments. Trends Ecol Evol. 2015b;30:161–8.
Hanghøj K, Orlando L, Hanghøj K, Orlando ÁL. Ancient epigenomics. In: Lindqvist C, Om PR, editors. Paleogenomics. Cham: Springer International Publishing AG; 2018. https://doi.org/10.1007/13836_2018_18.
Hansen MM. Expression of interest: transcriptomics and the designation of conservation units. Mol Ecol. 2010;19:1757–9.
Hare MP, Nunney L, Schwartz MK, et al. Understanding and estimating effective population size for practical application in marine species management. Conserv Biol. 2011;25:438–49.
Harr B. Genomic islands of differentiation between house mouse subspecies. Genome Res. 2006;16:730–7.
Harris K, Nielsen R. Inferring demographic history from a spectrum of shared haplotype lengths. PLoS Genet. 2013;9:e1003521.
Harris C, Rousset F, Morlais I, Fontenille D, Cohuet A. Low linkage disequilibrium in wild Anopheles gambiae s.l. populations. BMC Genet. 2010;11:81.
Harrisson KA, Amish SJ, Pavlova A, et al. Signatures of polygenic adaptation associated with climate across the range of a threatened fish species with high genetic connectivity. Mol Ecol. 2017;26:6253–69.
Haussler D, O’Brien SJ, Ryder OA, et al. Genome 10K: a proposal to obtain whole-genome sequence for 10000 vertebrate species. J Hered. 2009;100:659–74.
Hedrick PW, Garcia-Dorado A. Understanding inbreeding depression, purging, and genetic rescue. Trends Ecol Evol. 2016;31:940–52.
Heintzman PD, Soares AER, Chang D, Shapiro B. Paleogenomics. Rev Cell Biol Mol Med. 2015;1:243–67.
Hendricks S, Anderson EC, Antao T, et al. Recent advances in conservation and population genomics data analysis. Evol Appl. 2018;11:1197–211.
Hermisson J, Pennings PS. Soft sweeps: molecular population genetics of adaptation from standing genetic variation. Genetics. 2005;169:2335–52.
Hermisson J, Pennings PS. Soft sweeps and beyond: understanding the patterns and probabilities of selection footprints under rapid adaptation. Methods Ecol Evol. 2017;8:700–16.
Hidalgo-Galiana A, Monge M, Biron DG, et al. Protein expression parallels thermal tolerance and ecologic changes in the diversification of a diving beetle species complex. Heredity. 2016;116:114–23.
Hoban S. Integrative conservation genetics: prioritizing populations using climate predictions, adaptive potential and habitat connectivity. Mol Ecol Resour. 2018;18:14–7.
Hoban SM, Gaggiotti OE, Bertorelle G. The number of markers and samples needed for detecting bottlenecks under realistic scenarios, with and without recovery: a simulation-based study. Mol Ecol. 2013;22:3444–50.
Hoban S, Kelley JL, Lotterhos KE, et al. Finding the genomic basis of local adaptation: pitfalls, practical solutions, and future directions. Am Nat. 2016;188:379–97.
Hodel RG, Chandler LM, Fahrenkrog AM, et al. Linking genome signatures of selection and adaptation in non-model plants: exploring potential and limitations in the angiosperm Amborella. Curr Opin Plant Biol. 2018;42:81–9.
Hodges E, Xuan Z, Balija V, et al. Genome-wide in situ exon capture for selective resequencing. Nat Genet. 2007;39:1522–7.
Hoffberg SL, Kieran TJ, Catchen JM, et al. RADcap: sequence capture of dual-digest RADseq libraries with identifiable duplicates and reduced missing data. Mol Ecol Resour. 2016;16:1264–78.
Hoffman JI, Simpson F, David P, et al. High-throughput sequencing reveals inbreeding depression in a natural population. Proc Natl Acad Sci. 2014;111:3775–80.
Hogg JT. Mating in bighorn sheep: multiple creative male strategies. Science. 1984;225:526–9.
Hohenlohe PA, Bassham S, Etter PD, et al. Population genomics of parallel adaptation in threespine stickleback using sequenced RAD tags. PLoS Genet. 2010a;6:e1000862.
Hohenlohe PA, Phillips PC, Cresko WA. Using population genomics to detect selection in natural populations: key concepts and methodological considerations. Int J Plant Sci. 2010b;171:1059–71.
Hohenlohe PA, Hand BK, Andrews KR, Luikart G. Population genomics provides key insights in ecology and evolution. In: Om PR, editor. Population genomics: concepts, approaches and applications. Cham: Springer International Publishing AG; 2018. https://doi.org/10.1007/13836_2018_20.
Holliday JA, Ritland K, Aitken SN. Widespread, ecologically relevant genetic markers developed from association mapping of climate-related traits in Sitka spruce (Picea sitchensis). New Phytol. 2010;188:501–14.
Holliday JA, Wang T, Aitken S. Predicting adaptive phenotypes from multilocus genotypes in Sitka spruce (Picea sitchensis) using random forest. G3. 2012;2:1085–93.
Holliday JA, Hallerman EM, Haak DC. Genotyping and sequencing technologies in population genetics and genomics. In: Om PR, editor. Population genomics: concepts, approaches and applications. Cham: Springer International Publishing AG; 2018. https://doi.org/10.1007/13836_2017_5.
Horsthemke B. A critical view on transgenerational epigenetic inheritance in humans. Nat Commun. 2018;9:2973.
Howard JT, Haile-Mariam M, Pryce JE, Maltecca C. Investigation of regions impacting inbreeding depression and their association with the additive genetic effect for United States and Australia Jersey dairy cattle. BMC Genomics. 2015;16:813.
Hu J, Barrett RDH. Epigenetics in natural animal populations. J Evol Biol. 2017;30:1612–32.
Huber B, Whibley A, Poul YL, et al. Conservatism and novelty in the genetic architecture of adaptation in Heliconius butterflies. Heredity. 2015;114:515–24.
Huerta-Sánchez E, Jin X, Asan, et al. Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature. 2014;512:194–7.
Huisman J, Kruuk LEB, Ellis PA, Clutton-Brock T, Pemberton JM. Inbreeding depression across the lifespan in a wild mammal population. Proc Natl Acad Sci U S A. 2016;113:3585–90.
Humble E, Dasmahapatra KK, Martinez-Barrio A, et al. RAD sequencing and a hybrid antarctic fur seal genome assembly reveal rapidly decaying linkage disequilibrium, global population structure and evidence for inbreeding. G3. 2018;8:2709–22.
Hunter ME, Hoban SM, Bruford MW, Segelbacher G, Bernatchez L. Next-generation conservation genetics and biodiversity monitoring. Evol Appl. 2018;11:1029–34.
Husby A, Kawakami T, Rönnegård L, et al. Genome-wide association mapping in a wild avian population identifies a link between genetic and phenotypic variation in a life-history trait. Proc Biol Sci. 2015;282:20150156.
Jensen JD, Foll M, Bernatchez L. The past, present and future of genomic scans for selection. Mol Ecol. 2016;25:1–4.
Johnson EC, Evans LM, Keller MC. Relationships between estimated autozygosity and complex traits in the UK Biobank. PLoS Genet. 2018a;14:e1007556.
Johnson JS, Krutovsky KV, Rajora OP, Gaddis KD, Cairns DM. Advancing biogeography through population genomics. In: Om PR, editor. Population genomics: concepts, approaches and applications. Cham: Springer International Publishing AG; 2018b. https://doi.org/10.1007/13836_2018_39.
Johnston SE, McEwan JC, Pickering NK, et al. Genome-wide association mapping identifies the genetic basis of discrete and quantitative variation in sexual weaponry in a wild sheep population. Mol Ecol. 2011;20:2555–66.
Johnston SE, Gratten J, Berenos C, et al. Life history trade-offs at a single locus maintain sexually selected genetic variation. Nature. 2013;502:93–5.
Jones MR, Good JM. Targeted capture in evolutionary and ecological genomics. Mol Ecol. 2016;25:185–202.
Jones MR, Scott Mills L, Alves PC, et al. Adaptive introgression underlies polymorphic seasonal camouflage in snowshoe hares. Science. 2018;360:1355–8.
Joost S, Bonin A, Bruford MW, et al. A spatial analysis method (SAM) to detect candidate loci for selection: towards a landscape genomics approach to adaptation. Mol Ecol. 2007;16:3955–69.
Kabekkodu SP, Chakrabarty S, Ghosh S, Brand A, Satyamoorthy K. Epigenomics, pharmacoepigenomics, and personalized medicine in cervical cancer. Public Health Genomics. 2017;20:100–15.
Kardos M, Shafer ABA. The peril of gene-targeted conservation. Trends Ecol Evol. 2018. https://doi.org/10.1016/j.tree.2018.08.011.
Kardos M, Luikart G, Allendorf FW. Measuring individual inbreeding in the age of genomics: marker-based measures are better than pedigrees. Heredity. 2015a;115:63–72.
Kardos M, Luikart G, Bunch R, et al. Whole-genome resequencing uncovers molecular signatures of natural and sexual selection in wild bighorn sheep. Mol Ecol. 2015b;24:5616–32.
Kardos M, Husby A, Mcfarlane SE, Qvarnstrom A, Ellegren H. Whole-genome resequencing of extreme phenotypes in collared flycatchers highlights the difficulty of detecting quantitative trait loci in natural populations. Mol Ecol Resour. 2016a;16:727–41.
Kardos M, Taylor HR, Ellegren H, Luikart G, Allendorf FW. Genomics advances the study of inbreeding depression in the wild. Evol Appl. 2016b;9:1205–18.
Kardos M, Qvarnström A, Ellegren H. Inferring individual inbreeding and demographic history from segments of identity by descent in Ficedula flycatcher genome sequences. Genetics. 2017;205:1319–34.
Kardos M, Åkesson M, Fountain T, et al. Genomic consequences of intensive inbreeding in an isolated wolf population article. Nat Ecol Evol. 2018;2:124–31.
Karr TL. Application of proteomics to ecology and population biology. Heredity. 2008;100:200–6.
Kawakami T, Smeds L, Backström N, et al. A high-density linkage map enables a second-generation collared flycatcher genome assembly and reveals the patterns of avian recombination rate variation and chromosomal evolution. Mol Ecol. 2014;23:4035–58.
Keller LF, Waller DM. Inbreeding effects in wild populations. Trends Ecol Evol. 2002;17:230–41.
Keller MC, Simonson MA, Ripke S, et al. Runs of homozygosity implicate autozygosity as a schizophrenia risk factor. PLoS Genet. 2012;8:e1002656.
Kelso J, Prüfer K. Ancient humans and the origin of modern humans. Curr Opin Genet Dev. 2014;29:133–8.
Kijas JW. Detecting regions of homozygosity to map the cause of recessively inherited disease. Methods Mol Biol. 2013;1019:331–45.
Kim Y, Stephan W. Detecting a local signature of genetic hitchhiking along a recombining chromosome. Genetics. 2002;160:765–77.
Kirin M, McQuillan R, Franklin CS, et al. Genomic runs of homozygosity record population history and consanguinity. PLoS One. 2010;5:e13996.
Kirkpatrick M. How and why chromosome inversions evolve. PLoS Biol. 2010;8:e1000501.
Knaus BJ, Grünwald NJ. vcfr: a package to manipulate and visualize variant call format data in R. Mol Ecol Resour. 2017;17:44–53.
Knief U, Kempenaers B, Forstmeier W. Meiotic recombination shapes precision of pedigree- and marker-based estimates of inbreeding. Heredity. 2017;118:239–48.
Kovach RP, Hand BK, Hohenlohe PA, et al. Vive la résistance: genome-wide selection against introduced alleles in invasive hybrid zones. Proc Roy Soc B Biol Sci. 2016;283:20161380.
Kozakiewicz CP, Burridge CP, Funk WC, et al. Pathogens in space: advancing understanding of pathogen dynamics and disease ecology through landscape genetics. Evol Appl. 2018. In press.
Kreiner JM, Stinchcombe JR, Wright SI. Population genomics of herbicide resistance: adaptation via evolutionary rescue. Annu Rev Plant Biol. 2018;69:611–35.
Küpper C, Stocks M, Risse JE, et al. A supergene determines highly divergent male reproductive morphs in the ruff. Nat Genet. 2015;48:79–83.
Laforest-Lapointe I, Paquette A, Messier C, Kembel SW. Leaf bacterial diversity mediates plant diversity and ecosystem function relationships. Nature. 2017;546(7656):145. https://doi.org/10.1038/nature22399.
Lamichhaney S, Fan G, Widemo F, et al. Structural genomic changes underlie alternative reproductive strategies in the ruff (Philomachus pugnax). Nat Genet. 2015;48:84–8.
Lan T, Lindqvist C. Paleogenomics: genome-scale analysis of ancient DNA and population and evolutionary genomic inferences. In: Om PR, editor. Population genomics: concepts, approaches and applications. Cham: Springer International Publishing AG; 2018. https://doi.org/10.1007/13836_2017_7.
Lander ES, Botstein D. Homozygosity mapping: a way to map human recessive traits with the DNA of inbred children. Science. 1987;236:1567–70.
Landry PA, Koskinen MT, Primmed CR. Deriving evolutionary relationships among populations using microsatellites and (δμ)2: all loci are equal, but some are more equal than others. Genetics. 2002;161:1339–47.
Laporte M, Pavey SA, Rougeux C, et al. RAD sequencing reveals within-generation polygenic selection in response to anthropogenic organic and metal contamination in North Atlantic Eels. Mol Ecol. 2016;25:219–37.
Larson WA, Seeb LW, Everett MV, et al. Genotyping by sequencing resolves shallow population structure to inform conservation of Chinook salmon (Oncorhynchus tshawytscha). Evol Appl. 2014;7:355–69.
Larson WA, Limborg MT, McKinney GJ, et al. Genomic islands of divergence linked to ecotypic variation in sockeye salmon. Mol Ecol. 2017;26:554–70.
Lawson DJ, Hellenthal G, Myers S, Falush D. Inference of population structure using dense haplotype data. PLoS Genet. 2012;8:e1002453.
Le Luyer J, Laporte M, Beacham TD, et al. Parallel epigenetic modifications induced by hatchery rearing in a Pacific salmon. Proc Natl Acad Sci. 2017;114:12964–9.
Lee MK, Hong Y, Kim S-Y, Kim WJ, London SJ. Epigenome-wide association study of chronic obstructive pulmonary disease and lung function in Koreans. Epigenomics. 2017;9:971–84.
Lee HJ, Georgiadou A, Otto TD, et al. Transcriptomic studies of malaria: a paradigm for investigation of systemic host-pathogen interactions. Microbiol Mol Biol Rev. 2018;82:e00071–17.
Leite DCA, Salles JF, Calderon EN, et al. Coral bacterial-core abundance and network complexity as proxies for anthropogenic pollution. Front Microbiol. 2018;9:833.
Leitwein M, Gagnaire P-A, Desmarais E, Berrebi P, Guinand B. Genomic consequences of a recent three-way admixture in supplemented wild brown trout populations revealed by local ancestry tracts. Mol Ecol. 2018;27:3466–83.
Lewontin RC, Krakauer J. Distribution of gene frequency as a test of the theory of the selective neutrality of polymorphisms. Genetics. 1973;74:175–95.
Li H, Durbin R. Inference of human population history from individual whole-genome sequences. Nature. 2011;475:493–6.
Lind MI, Spagopoulou F. Evolutionary consequences of epigenetic inheritance. Heredity. 2018;121:205–9.
Lorenzo FR, Huff C, Myllymäki M, et al. A genetic mechanism for Tibetan high-altitude adaptation. Nat Genet. 2014;46:951–6.
Lotterhos KE, Whitlock MC. Evaluation of demographic history and neutral parameterization on the performance of FST outlier tests. Mol Ecol. 2014;23(9):2178–92.
Lowry DB, Hoban S, Kelley JL, et al. Breaking RAD: an evaluation of the utility of restriction site-associated DNA sequencing for genome scans of adaptation. Mol Ecol Resour. 2017;17:142–52.
Luikart G, England PR, Tallmon D, Jordan S, Taberlet P. The power and promise of population genomics: from genotyping to genome typing. Nat Rev Genet. 2003;4:981–94.
Luikart G, Ryman N, Tallmon DA, Schwartz MK, Allendorf FW. Estimation of census and effective population sizes: the increasing usefulness of DNA-based approaches. Conserv Genet. 2010;11:355–73.
Ma L, Sun X, Kong X, et al. Physiological, biochemical and proteomics analysis reveals the adaptation strategies of the alpine plant Potentilla saundersiana at altitude gradient of the Northwestern Tibetan Plateau. J Proteomics. 2015;112:63–82.
Malécot G. The mathematics of heredity. San Francisco: W.H. Freeman; 1970.
Manthey JD, Campillo LC, Burns KJ, Moyle RG. Comparison of target-capture and restriction-site associated DNA sequencing for phylogenomics: a test in cardinalid tanagers (Aves, Genus: Piranga). Syst Biol. 2016;65:640–50.
Marciniak S, Poinar H. Ancient pathogens through human history: a paleogenomic perspective. In: Lindqvist C, Rajora OP, editors. Paleogenomics. Cham: Springer International Publishing AG; 2018. https://doi.org/10.1007/13836_2018.
Marques DA, Lucek K, Meier JI, et al. Genomics of rapid incipient speciation in sympatric threespine stickleback. PLoS Genet. 2016;12:e1005887.
Marques DA, Jones FC, Di Palma F, Kingsley DM, Reimchen TE. Experimental evidence for rapid genomic adaptation to a new niche in an adaptive radiation. Nat Ecol Evol. 2018;2:1128–38.
Marsden CD, Lee Y, Kreppel K, et al. Diversity, differentiation, and linkage disequilibrium: prospects for association mapping in the malaria vector Anopheles arabiensis. G3. 2014;4:121–31.
Martin EM, Fry RC. Environmental influences on the epigenome: exposure-associated DNA methylation in human populations. Annu Rev Public Health. 2018;39:309–33.
Martin SH, Dasmahapatra KK, Nadeau NJ, et al. Genome-wide evidence for speciation with gene flow in Heliconius butterflies. Genome Res. 2013;23:1817–28.
Matthews B, Best RJ, Feulner PGD, Narwani A, Limberger R. Evolution as an ecosystem process: insights from genomics. Genome. 2018;61:298–309.
Maynard Smith J, Haigh J. The hitch-hiking effect of a favorable gene. Genet Res. 1974;23:23–35.
McCartney-Melstad E, Mount GG, Shaffer HB. Exon capture optimization in amphibians with large genomes. Mol Ecol Resour. 2016;16:1084–94.
McCoy RC, Akey JM. Selection plays the hand it was dealt: evidence that human adaptation commonly targets standing genetic variation. Genome Biol. 2017;18:139.
McKain MR, Johnson MG, Uribe-Convers S, Eaton D, Yang Y. Practical considerations for plant phylogenomics. Appl Plant Sci. 2018;6:e1038.
Mckinney GJ, Seeb LW, Larson WA, et al. An integrated linkage map reveals candidate genes underlying adaptive variation in Chinook salmon (Oncorhynchus tshawytscha). Mol Ecol Resour. 2016;16:769–83.
McKinney GJ, Larson WA, Seeb LW, Seeb JE. RADseq provides unprecedented insights into molecular ecology and evolutionary genetics: comment on Breaking RAD by Lowry et al. (2016). Mol Ecol Resour. 2017a;17:356–61.
McKinney GJ, Waples RK, Seeb LW, Seeb JE. Paralogs are revealed by proportion of heterozygotes and deviations in read ratios in genotyping-by-sequencing data from natural populations. Mol Ecol Resour. 2017b;17:656–69.
Mckown AD, Klápště J, Guy RD, et al. Genome-wide association implicates numerous genes underlying ecological trait variation in natural populations of Populus trichocarpa. New Phytol. 2014;203:535–53.
McMahon BJ, Teeling EC, Höglund J. How and why should we implement genomics into conservation? Evol Appl. 2014;7:999–1007.
Meeks KAC, Henneman P, Venema A, et al. An epigenome-wide association study in whole blood of measures of adiposity among Ghanaians: the RODAM study. Clin Epigenetics. 2017;9:103.
Meyer M, Kircher M, Gansauge MT, et al. A high-coverage genome sequence from an archaic Denisovan individual. Science. 2012;338:222–6.
Miao B, Wang Z, Li Y. Genomic analysis reveals hypoxia adaptation in the tibetan mastiff by introgression of the gray wolf from the Tibetan plateau. Mol Biol Evol. 2017;34:734–43.
Miles A, Harding NJ, Bottà G, et al. Genetic diversity of the African malaria vector anopheles gambiae. Nature. 2017;552:96–100.
Miller JM, Malenfant RM, David P, et al. Estimating genome-wide heterozygosity: effects of demographic history and marker type. Heredity. 2014;112:240–7.
Moler ERV, Abakir A, Eleftheriou M, Johnson JS, Krutovsky KV, Lewis LC, Ruzov A, Whipple AV, Rajora OP. Population epigenomics. In: Om PR, editor. Population genomics: concepts, approaches and applications. Cham: Springer International Publishing AG; 2018.
Moran MA. Metatranscriptomics: eavesdropping on complex microbial communities. Microbe Mag. 2009;4:329–35.
Muhlfeld CC, Kalinowski ST, McMahon TE, et al. Hybridization rapidly reduces fitness of a native trout in the wild. Biol Lett. 2009;5:328–31.
Nadeau NJ, Kawakami T. Population genomics of speciation and admixture. In: Om PR, editor. Population genomics: concepts, approaches and applications. Cham: Springer International Publishing AG; 2018. https://doi.org/10.1007/13836_2018_24.
Nadeau NJ, Ruiz M, Salazar P, et al. Population genomics of parallel hybrid zones in the mimetic butterflies, H. melpomene and H. erato. Genome Res. 2014;24:1316–33.
Nakatochi M, Ichihara S, Yamamoto K, et al. Epigenome-wide association of myocardial infarction with DNA methylation sites at loci related to cardiovascular disease. Clin Epigenetics. 2017;9:54.
Narum SR, Buerkle CA, Davey JW, Miller MR, Hohenlohe PA. Genotyping-by-sequencing in ecological and conservation genomics. Mol Ecol. 2013;22:2841–7.
Narum SR, Di Genova A, Micheletti SJ, Maass A. Genomic variation underlying complex life-history traits revealed by genome sequencing in Chinook salmon. Proc Roy Soc B Biol Sci. 2018;285:20180935.
Nash DR, Als TD, Maile R, Jones GR, Boomsma JJ. A mosaic of chemical coevolution in a large blue butterfly. Science. 2008;319:88–90.
Nazareno AG, Bemmels JB, Dick CW, Lohmann LG. Minimum sample sizes for population genomics: an empirical study from an Amazonian plant species. Mol Ecol Resour. 2017;17:1136–47.
Nedelkov D. Population proteomics: investigation of protein diversity in human populations. Proteomics. 2008;8:779–86.
Nedelkov D, Kiernan UA, Niederkofler EE, Tubbs KA, Nelson RW. Investigating diversity in human plasma proteins. Proc Natl Acad Sci. 2005;102:10852–7.
Nedelkov D, U A K, Niederkofler EE, Tubbs KA, Nelson RW. Population proteomics: the concept, attributes, and potential for cancer biomarker research. Mol Cell Proteomics. 2006;5:1811–8.
Niederhuth CE, Bewick AJ, Ji L, et al. Widespread natural variation of DNA methylation within angiosperms. Genome Biol. 2016;17:174.
Nielsen R, Williamson S, Kim Y, et al. Genomic scans for selective sweeps using SNP data. Genome Res. 2005;15:1566–75.
Nilsson E, et al. Environmental toxicant induced epigenetic transgenerational inheritance of ovarian pathology and granulosa cell epigenome and transcriptome alterations: ancestral origins of polycystic ovarian syndrome and primary ovarian insufiency. Epigenetics. 2018;13:875–95.
Noble TJ, Tao Y, Mace ES, et al. Characterization of linkage disequilibrium and population structure in a mungbean diversity panel. Front Plant Sci. 2018;8:2102.
Norris LC, Main BJ, Lee Y, et al. Adaptive introgression in an African malaria mosquito coincident with the increased usage of insecticide-treated bed nets. Proc Natl Acad Sci. 2015;112:815–20.
Nosil P, Egan SP, Funk DJ. Heterogeneous genomic differentiation between walking-stick ecotypes: “isolation by adaptation” and multiple roles for divergent selection. Evolution. 2008;62:316–36.
Nunziata SO, Weisrock DW. Estimation of contemporary effective population size and population declines using RAD sequence data. Heredity. 2018;120:196–207.
Nystedt B, Street NR, Wetterbom A, et al. The Norway spruce genome sequence and conifer genome evolution. Nature. 2013;497:579–84.
O’Quin KE, Yoshizawa M, Doshi P, Jeffery WR. Quantitative genetic analysis of retinal degeneration in the blind cavefish Astyanax mexicanus. PLoS One. 2013;8:e57281.
Oomen RA, Hutchings JA. Transcriptomic responses to environmental change in fishes: insights from RNA sequencing. FACETS. 2017;2:610–41.
Orlando L. An ancient DNA perspective on horse evolution. In: Lindqvist C, Om PR, editors. Paleogenomics. Cham: Springer International Publishing AG; 2018. https://doi.org/10.1007/13836_2018_23.
Orlando L, Ginolhac A, Zhang G, et al. Recalibrating equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature. 2013;499:74–8.
Ozerov MY, Gross R, Bruneaux M, et al. Genomewide introgressive hybridization patterns in wild Atlantic salmon influenced by inadvertent gene flow from hatchery releases. Mol Ecol. 2016;25:1275–93.
Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet. 2011;12:87–98.
Palkopoulou E, Mallick S, Skoglund P, et al. Complete genomes reveal signatures of demographic and genetic declines in the woolly mammoth. Curr Biol. 2015;25:1395–400.
Paradis E, Gosselin T, Goudet J, Jombart T, Schliep K. Linking genomics and population genetics with R. Mol Ecol Resour. 2017;17:54–66.
Pardo-Diaz C, Salazar C, Baxter SW, et al. Adaptive introgression across species boundaries in Heliconius butterflies. PLoS Genet. 2012;8:e1002752.
Parducci L, Nota K, Wood J. Reconstructing past vegetation communities using ancient DNA from lake sediments. In: Lindqvist C, Om PR, editors. Paleogenomics. Cham: Springer International Publishing AG; 2018. https://doi.org/10.1007/13836_2018_38.
Paris JR, Stevens JR, Catchen JM. Lost in parameter space: a road map for stacks. Meth Ecol Evol. 2017;8:1360–73.
Payseur BA, Rieseberg LH. A genomic perspective on hybridization and speciation. Mol Ecol. 2016;25:2337–60.
Peek RA, O’Rourke SM, Miller MR. Flow regulation associated with decreased genetic health of a river-breeding frog species. BioRxiv. 2018;316604.
Pemberton TJ, Absher D, Feldman MW, et al. Genomic patterns of homozygosity in worldwide human populations. Am J Hum Genet. 2012;91:275–92.
Pemberton JM, Ellis PE, Pilkington JG, Bérénos C. Inbreeding depression by environment interactions in a free-living mammal population. Heredity. 2017;118:64–77.
Pennings PS, Hermisson J. Soft sweeps II – molecular population genetics of adaptation from recurrent mutation or migration. Mol Biol Evol. 2006;23:1076–84.
Pérez O’Brien AM, Utsunomiya YT, Mészáros G, et al. Assessing signatures of selection through variation in linkage disequilibrium between taurine and indicine cattle. Genet Sel Evol. 2014;46:19.
Perry GH, Marioni JC, Melsted P, Gilad Y. Genomic-scale capture and sequencing of endogenous DNA from feces. Mol Ecol. 2010;19:5332–44.
Petkova D, Novembre J, Stephens M. Visualizing spatial population structure with estimated effective migration surfaces. Nat Genet. 2016;48:94–100.
Pickrell JK, Pritchard JK. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012;8:e1002967.
Pino Del Carpio D, Lozano R, Wolfe MD, Jannink J-L. Genome-wide associationstudies and heritability estimation in the functional genomics era. In: Rajora OP, editor. Population genomics: concepts, approaches and applications. Cham: Springer International Publishing AG; 2018. https://doi.org/10.1007/13836_2018_12.
Poelstra JW, Vijay N, Bossu CM, et al. The genomic landscape underlying phenotypic integrity in the face of gene flow in crows. Science. 2014;344:1410–4.
Pogorelcnik R, Vaury C, Pouchin P, Jensen S, Brasset E. SRNAPipe: a Galaxy-based pipeline for bioinformatic in-depth exploration of small RNAseq data. Mob DNA. 2018;9:25.
Portik DM, Smith LL, Bi K. An evaluation of transcriptome-based exon capture for frog phylogenomics across multiple scales of divergence (Class: Amphibia, Order: Anura). Mol Ecol Resour. 2016;16:1069–83.
Prince DJ, O’Rourke SM, Thompson TQ, et al. The evolutionary basis of premature migration in Pacific salmon highlights the utility of genomics for informing conservation. Sci Adv. 2017;3:e1603198.
Prüfer K, Racimo F, Patterson N, et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–9.
Pruisscher P, Nylin S, Gotthard K, Wheat CW. Genetic variation underlying local adaptation of diapause induction along a cline in a butterfly. Mol Ecol. 2018. In press.
Pryce JE, Haile-Mariam M, Goddard ME, Hayes BJ. Identification of genomic regions associated with inbreeding depression in Holstein and Jersey dairy cattle. Genet Sel Evol. 2014;46:71.
Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75.
Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65.
Racimo F, Sankararaman S, Nielsen R, Huerta-Sánchez E. Evidence for archaic adaptive introgression in humans. Nat Rev Genet. 2015;16:359–71.
Rajora OP, Eckert AJ, Zinck JWR. Single-locus versus multilocus patterns of local adaptation to climate in eastern white pine (Pinus strobus, Pinaceae). PLoS One. 2016;11:e0158691.
Rasmussen M, Li Y, Lindgreen S, et al. Ancient human genome sequence of an extinct Palaeo-Eskimo. Nature. 2010;463:757–62.
Rastas P, Calboli FCF, Guo B, Shikano T, Merilä J. Construction of ultradense linkage maps with Lep-MAP2: stickleback F2 recombinant crosses as an example. Genome Biol Evol. 2016;8:78–93.
Razgour O, Taggart JB, Manel S, et al. An integrated framework to identify wildlife populations under threat from climate change. Mol Ecol Resour. 2018;18:18–31.
Rees BB, Andacht T, Skripnikova E, Crawford DL. Population proteomics: quantitative variation within and among populations in cardiac protein expression. Mol Biol Evol. 2011;28:1271–9.
Rellstab C, Gugerli F, Eckert AJ, Hancock AM, Holderegger R. A practical guide to environmental association analysis in landscape genomics. Mol Ecol. 2015;24:4348–70.
Renaut S, Grassa CJ, Yeaman S, et al. Genomic islands of divergence are not affected by geography of speciation in sunflowers. Nat Commun. 2013;4:1827.
Resendez SD, Bradley JR, Xu D, Gokcumen O. Structural variants in ancient genomes. In: Lindqvist C, Om PR, editors. Paleogenomics. Cham: Springer International Publishing AG; 2018. https://doi.org/10.1007/13836_2018_34.
Richards CL, Alonso C, Becker C, et al. Ecological plant epigenetics: evidence from model and non-model species, and the way forward. Ecol Lett. 2017;20:1576–90.
Rieseberg L. Adaptive introgression: the seeds of resistance. Curr Biol. 2011;21:R581–3.
Rochus CM, Tortereau F, Plisson-Petit F, et al. Revealing the selection history of adaptive loci using genome-wide scans for selection: an example from domestic sheep. BMC Genomics. 2018;19:71.
Rodríguez-Ezpeleta N, Bradbury IR, Mendibil I, et al. Population structure of Atlantic mackerel inferred from RAD-seq-derived SNP markers: effects of sequence clustering parameters and hierarchical SNP selection. Mol Ecol Resour. 2016;16:991–1001.
Roffler GH, Amish SJ, Smith S, et al. SNP discovery in candidate adaptive genes using exon capture in a free-ranging alpine ungulate. Mol Ecol Resour. 2016;16:1147–64.
Rogers RL, Slatkin M. Excess of genomic defects in a woolly mammoth on Wrangel island. PLoS Genet. 2017;13:e1006601.
Roitman S, Joseph Pollock F, Medina M. Coral microbiomes as bioindicators of reef health. In: Population genomics. Cham: Springer; 2018. p. 1–19.
Rondeau EB, Minkley DR, Leong JS, et al. The genome and linkage map of the northern pike (Esox lucius): conserved synteny revealed between the salmonid sister group and the Neoteleostei. PLoS One. 2014;e102089:9.
Rougeux C, Gagnaire P-A, Praebel K, Seehausen O, Bernatchez L. Convergent transcriptomic landscapes under polygenic selection accompany inter- continental parallel evolution within a Nearctic Coregonus (Salmonidae) sister-species complex. BioRxiv. 2018. https://doi.org/10.1101/311464.
Rubin C-J, et al. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature. 2010;464:587.
Rubin C-J, Megens H-J, Barrio AM, et al. Strong signatures of selection in the domestic pig genome. Proc Natl Acad Sci. 2012;109:19529–36.
Sabeti PC, Reich DE, Higgins JM, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–7.
Sabeti PC, Varilly P, Fry B, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–8.
Saint-Pé K, Blanchet S, Tissot L, et al. Genetic admixture between captive-bred and wild individuals affects patterns of dispersal in a brown trout (Salmo trutta) population. Conserv Genet. 2018;5:1269–79.
Salmona J, Heller R, Lascoux M, Shafer A. Inferring demographic history using genomic data. In: Rajora OP, editor. Population genomics: concepts, approaches and applications. Cham: Springer International Publishing AG; 2017. https://doi.org/10.1007/13836_2017_1.
Salojärvi J. Computational tools for population genomics. In: Om PR, editor. Population genomics: concepts, approaches and applications. Cham: Springer International Publishing AG; 2018. https://doi.org/10.1007/13836_2018_57.
Santure AW, Garant D. Wild GWAS-association mapping in natural populations. Mol Ecol Resour. 2018;18:729–38.
Sboner A, Mu XJ, Greenbaum D, Auerbach RK, Gerstein MB. The real cost of sequencing: higher than you think! Genome Biol. 2011;12:125.
Schlötterer C. The evolution of molecular markers – just a matter of fashion? Nat Rev Genet. 2004;5:63–9.
Schlötterer C, Tobler R, Kofler R, Nolte V. Sequencing pools of individuals-mining genome-wide polymorphism data without big funding. Nat Rev Genet. 2014;15:749–63.
Schmidt TL, Filipovi I, Hoffmann AA, Rašić G. Fine-scale landscape genomics of Aedes aegypti reveals loss of Wolbachia transinfection, dispersal barrier and potential for occasional long distance movement. BioRxiv. 2017. https://doi.org/10.1101/103598.
Schmitz RJ, He Y, Valdés-López O, et al. Epigenome-wide inheritance of cytosine methylation variants in a recombinant inbred population. Genome Res. 2013a;23:1663–74.
Schmitz RJ, Schultz MD, Urich MA, et al. Patterns of population epigenomic diversity. Nature. 2013b;495:193–8.
Schrider DR, Kern AD. Soft sweeps are the dominant mode of adaptation in the human genome. Mol Biol Evol. 2017;34:1863–77.
Schweizer RM, VonHoldt BM, Harrigan R, et al. Genetic subdivision and candidate genes under selection in North American grey wolves. Mol Ecol. 2016;25:380–402.
Sedlazeck FJ, Lee H, Darby CA, Schatz MC. Piercing the dark matter: bioinformatics of long-range sequencing and mapping. Nat Rev Genet. 2018;19:329–46.
Shafer ABA, Wolf JBW, Alves PC, et al. Genomics and the challenging translation into conservation practice. Trends Ecol Evol. 2015;30:78–87.
Shafer ABA, Peart CR, Tusso S, et al. Bioinformatic processing of RAD-seq data dramatically impacts downstream population genetic inference. Methods Ecol Evol. 2017;8:907–17. https://doi.org/10.1111/2041-210X.12700.
Shapiro B, Hofreiter M. A paleogenomic perspective on evolution and gene function: new insights from ancient DNA. Science. 2014;343
Shimada-Sugimoto M, Otowa T, Miyagawa T, et al. Epigenome-wide association study of DNA methylation in panic disorder. Clin Epigenetics. 2017;9:6.
Shin D, Kim S-H, Park J, Lee H-K, Song K-D. Extent of linkage disequilibrium and effective population size of the Landrace population in Korea. Asian Australas J Anim Sci. 2018;31:1078–87.
Simons YB, Bullaughey K, Hudson RR, Sella G. A population genetic interpretation of GWAS findings for human quantitative traits. PLoS Biol. 2018;16:e2002985.
Skoglund P, Malmström H, Raghavan M, et al. Origins and genetic legacy of Neolithic farmers and hunter-gatherers in Europe. Science. 2012;336:466–9.
Slatkin M. Linkage disequilibrium – understanding the evolutionary past and mapping the medical future. Nat Rev Genet. 2008;9:477–85.
Smith MW, O’Brien SJ. Mapping by admixture linkage disequilibrium: advances, limitations and guidelines. Nat Rev Genet. 2005;6:623–32.
Sollars ESA, Buggs RJA. Genome-wide epigenetic variation among ash trees differing in susceptibility to a fungal disease. BMC Genomics. 2018;19:502.
Song Y, Endepols S, Klemann N, et al. Adaptive introgression of anticoagulant rodent poison resistance by hybridization between old world mice. Curr Biol. 2011;21:1296–301.
Sork VL. Gene flow and natural selection shape spatial patterns of genes in tree populations: implications for evolutionary processes and applications. Evol Appl. 2016;9:291–310.
Sovic MG, Carstens BC, Gibbs HL. Genetic diversity in migratory bats: results from RADseq data for three tree bat species at an Ohio windfarm. PeerJ. 2016;4:e1647.
Speed D, Balding DJ. Relatedness in the post-genomic era: is it still useful? Nat Rev Genet. 2015;16:33–44.
Srivathsan A, Ang A, Vogler AP, Meier R. Fecal metagenomics for the simultaneous assessment of diet, parasites, and population genetics of an understudied primate. Front Zool. 2016;13:17.
Stam P. The distribution of the fraction of the genome identical by descent in finite random mating populations. Genet Res. 1980;35:131–55.
Stat M, Huggett MJ, Bernasconi R, et al. Ecosystem biomonitoring with eDNA: metabarcoding across the tree of life in a tropical marine environment. Sci Rep. 2017;7:12240.
Stetter MG, Thornton K, Ross-Ibarra J. Genetic architecture and selective sweeps after polygenic adaptation to distant trait optima. BioRxiv. 2018:313247.
Stölting KN, Paris M, Meier C, et al. Genome-wide patterns of differentiation and spatially varying selection between postglacial recolonization lineages of Populus alba (Salicaceae), a widespread forest tree. New Phytol. 2015;207:723–34.
Storz JF, Beaumont MA, Alberts SC. Genetic evidence for long-term population decline in a savannah-dwelling primate: inferences from a hierarchical Bayesian model. Mol Biol Evol. 2002;19:1981–90.
Sugden LA, Atkinson EG, Fischer AP, et al. Localization of adaptive variants in human genomes using averaged one-dependence estimation. Nat Commun. 2018;9:703.
Suhre K, Arnold M, Bhagwat AM, et al. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat Commun. 2017;8:14357.
Sunnucks P. Efficient genetic markers for population biology. Trends Ecol Evol. 2000;15:199–203.
Suren H, Hodgins KA, Yeaman S, et al. Exome capture from the spruce and pine giga-genomes. Mol Ecol Resour. 2016;16:1136–46.
Syring JV, Tennessen JA, Jennings TN, et al. Targeted capture sequencing in whitebark pine reveals range-wide demographic and adaptive patterns despite challenges of a large, repetitive genome. Front Plant Sci. 2016;7:484.
Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–95.
Tallmon DA, Luikart G, Waples RS. The alluring simplicity and complex reality of genetic rescue. Trends Ecol Evol. 2004;19:489–96.
Taudt A, Colomé-Tatché M, Johannes F. Genetic sources of population epigenomic variation. Nat Rev Genet. 2016;17:319–32.
Terhorst J, Kamm JA, Song YS. Robust and scalable inference of population history from hundreds of unphased whole genomes. Nat Genet. 2017;49:303–9.
Teshima KM, Coop G, Przeworski M. How reliable are empirical genomic scans for selective sweeps? Genome Res. 2006;16:702–12.
Thalmann O, Perri AR. Paleogenomic inferences of dog domestication. In: Lindqvist C, Om PR, editors. Paleogenomics. Cham: Springer International Publishing AG; 2018. https://doi.org/10.1007/13836_2018_27.
Thompson EA. Identity by descent: variation in meiosis, across genomes, and in populations. Genetics. 2013;194:301–26.
Thurber RV, Willner-Hall D, Rodriguez-Mueller B, et al. Metagenomic analysis of stressed coral holobionts. Environ Microbiol. 2009;11:2148–63.
Tishkoff SA, Reed FA, Ranciaro A, et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet. 2007;39:31–40.
Tranchant-Dubreuil C, Ravel S, Monat C, et al. TOGGLe, a flexible framework for easily building complex workflows and performing robust large-scale NGS analyses. BioRxiv. 2018. https://doi.org/10.1101/245480.
Trapp J, Gouveia D, Almunia C, et al. Digging deeper into the pyriproxyfen-response of the amphipod gammarus fossarum with a next-generation ultra-high-field orbitrap analyser: new perspectives for environmental toxicoproteomics. Front Environ Sci. 2018;6:54.
Turner TL, Hahn MW, Nuzhdin SV. Genomic islands of speciation in Anopheles gambiae. PLoS Biol. 2005;3:1572–8.
Valdés A, Ehrlén J. Caterpillar seed predators mediate shifts in selection on flowering phenology in their host plant. Ecology. 2017;98:228–38.
Valdisser PAMR, Pereira WJ, Almeida Filho JE, et al. In-depth genome characterization of a Brazilian common bean core collection using DArTseq high-density SNP genotyping. BMC Genomics. 2017;18:423.
Vallejo RL, Silva RMO, Evenhuis JP, et al. Accurate genomic predictions for BCWD resistance in rainbow trout are achieved using low-density SNP panels: evidence that long-range LD is a major contributing factor. J Anim Breed Genet. 2018;135:263–74.
van Dijk EL, Jaszczyszyn Y, Naquin D, Thermes C. The third revolution in sequencing technology. Trends Genet. 2018;34:666–81.
Vandersteen Tymchuk W, O’Reilly P, Bittman J, MacDonald D, Schulte P. Conservation genomics of Atlantic salmon: variation in gene expression between and within regions of the Bay of Fundy. Mol Ecol. 2010;19:1842–59.
Vasemagi A, Primmer CR. Expressed sequence tag-linked microsatellites as a source of gene-associated polymorphisms for detecting signatures of divergent selection in Atlantic salmon (Salmo salar L.). Mol Biol Evol. 2005;22:1067–76.
Vattathil S, Akey JM. Small amounts of archaic admixture provide big insights into human history. Cell. 2015;163:281–4.
Venter J, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74.
Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e154.
VonHoldt BM, Pollinger JP, Earl DA, et al. A genome-wide perspective on the evolutionary history of enigmatic wolf-like canids. Genome Res. 2011;21:1294–305.
Waite DW, Dsouza M, Sekiguchi Y, Hugenholtz P, Taylor MW. Network-guided genomic and metagenomic analysis of the faecal microbiota of the critically endangered kakapo. Sci Rep. 2018;8:8228.
Wallberg A, Han F, Wellhagen G, et al. A worldwide survey of genome sequence variation provides insight into the evolutionary history of the honeybee Apis mellifera. Nat Genet. 2014;46:1081–8.
Wang J. Estimation of effective population sizes from data on genetic markers. Phil Trans Roy Soc B Biol Sci. 2005;360:1395–409.
Wang Z, Gerstein M, Snyder M. RNA-seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63.
Wang J, Street NR, Scofield DG, Ingvarsson PK. Variation in linked selection and recombination drive genomic divergence during allopatric speciation of European and American aspens. Mol Biol Evol. 2016;33:1754–67.
Waples RS, Do C. Linkage disequilibrium estimates of contemporary Ne using highly variable genetic markers: a largely untapped resource for applied conservation and evolution. Evol Appl. 2010;3:244–62.
Waples RK, Larson WA, Waples RS. Estimating contemporary effective population size in non-model species using linkage disequilibrium across thousands of loci. Heredity. 2016;117:233–40.
Waterhouse MD, Erb LP, Beever EA, Russello MA. Adaptive population divergence and directional gene flow across steep elevational gradients in a climate-sensitive mammal. Mol Ecol. 2018;27:2512–28.
Wecek K, Hartmann S, Paijmans JLA, et al. Complex admixture preceded and followed the extinction of wisent in the wild. Mol Biol Evol. 2017;34:598–612.
Wellenreuther M, Bernatchez L. Eco-evolutionary genomics of chromosomal inversions. Trends Ecol Evol. 2018;33:427–40.
Wellenreuther M, Hansson B. Detecting polygenic evolution: problems, pitfalls, and promises. Trends Genet. 2016;32:155–64.
Wessinger CA, Kelly JK, Jiang P, Rausher MD, Hileman LC. SNP-skimming: a fast approach to map loci generating quantitative variation in natural populations. Mol Ecol Resour. 2018. https://doi.org/10.1111/1755-0998.12930.
Whiteley AR, Bhat A, Martins EP, et al. Population genomics of wild and laboratory zebrafish (Danio rerio). Mol Ecol. 2011;20:4259–76.
Whiteley AR, Fitzpatrick SW, Funk WC, Tallmon DA. Genetic rescue to the rescue. Trends Ecol Evol. 2015;30:42–9.
Wilson G, Rannala B. Bayesian inference of recent migration rates using multilocus genotyoes. Genetics. 2003;163:1177–91.
Wolf JBW, Ellegren H. Making sense of genomic islands of differentiation in light of speciation. Nat Rev Genet. 2017;18:87–100.
Wright S. Coefficients of inbreeding and relationship. Am Nat. 1922;56:330–8.
Xu Z, Bolick SCE, Deroo LA, et al. Epigenome-wide association study of breast cancer using prospectively collected sister study samples. J Natl Cancer Inst. 2013;105:694–700.
Xue Y, Prado-Martinez J, Sudmant PH, et al. Mountain gorilla genomes reveal the impact of long-term population decline and inbreeding. Science. 2015;348:242–5.
Yeaman S, Whitlock MC. The genetic architecture of adaptation under migration-selection balance. Evolution. 2011;65:1897–911.
Yeaman S, Hodgins KA, Lotterhos KE, et al. Convergent local adaptation to climate in distantly related conifers. Science. 2016;353:1431–3.
Yi SV. Insights into epigenome evolution from animal and plant methylomes. Genome Biol Evol. 2017;9:3189–201.
Zhang W, Fan Z, Han E, et al. Hypoxia adaptations in the grey wolf (Canis lupus chanco) from Qinghai-Tibet plateau. PLoS Genet. 2014;10:e1004466.
Zhang W, Zhang H, Yang H, et al. Computational resources associating diseases with genotypes, phenotypes and exposures. Brief Bioinform. 2018:bby071.
Acknowledgments
We thank G. McKinney for helpful comments and information on linkage mapping and Fred Allendorf for discussions and ideas regarding population genomics concepts and definitions. GL, MK, and BKH were supported in part by funding from US National Science Foundation grants DEB-1258203 and DoB-1639014. Montana Fish Wildlife and Parks provided supported GL and MK through contract #199101903. GL and BKH were also supported in part by funding from NASA grant number NNX14AB84G. OPR received support from a Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant RGPIN 2017-04589. PAH received support from National Science Foundation grants DEB-1316549 and DEB-1655809.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer International Publishing AG, part of Springer Nature
About this chapter
Cite this chapter
Luikart, G., Kardos, M., Hand, B.K., Rajora, O.P., Aitken, S.N., Hohenlohe, P.A. (2018). Population Genomics: Advancing Understanding of Nature. In: Rajora, O. (eds) Population Genomics. Population Genomics. Springer, Cham. https://doi.org/10.1007/13836_2018_60
Download citation
DOI: https://doi.org/10.1007/13836_2018_60
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-04587-6
Online ISBN: 978-3-030-04589-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)