Abstract
Only a few human and animal pathogenic viruses are known that have a single-stranded DNA genome. The members of the family Parvoviridae have a linear genome, whereas the genome of the members of the family Circoviridae and that of the recently created family Anelloviridae have a circular structure. The members of the family Geminiviridae are also characterized by a circular, single-stranded DNA genome, but they infect only plants. Circoviruses are pathogens in both plants and various animal species (monkeys, swine, poultry, etc.). A circular, single-stranded DNA virus, torque teno virus, was first isolated from humans in 1997. Several types of torque teno virus have been detected and classified into the family Anelloviridae; all persist, like the later discovered torque teno mini viruses and the torque teno midi viruses, in most people and various animals without causing any apparent disease.
Access provided by Autonomous University of Puebla. Download reference work entry PDF
Similar content being viewed by others
Keywords
- Inverted Terminal Repeat
- Torque Teno Virus
- Helper Virus
- Chicken Anaemia Virus
- Postweaning Multisystemic Wasting Syndrome
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
Only a few human and animal pathogenic viruses are known that have a single-stranded DNA genome. The members of the family Parvoviridae have a linear genome, whereas the genome of the members of the family Circoviridae and that of the recently created family Anelloviridae have a circular structure. The members of the family Geminiviridae are also characterized by a circular, single-stranded DNA genome, but they infect only plants. Circoviruses are pathogens in both plants and various animal species (monkeys, swine, poultry, etc.). A circular, single-stranded DNA virus, torque teno virus, was first isolated from humans in 1997. Several types of torque teno virus have been detected and classified into the family Anelloviridae; all persist, like the later discovered torque teno mini viruses and the torque teno midi viruses, in most people and various animals without causing any apparent disease.
1 Parvoviruses

Parvoviruses (from Latin parvus, meaning “small”) belong to the smallest viruses known to date. They are extremely resistant to external influences and detergents, have a narrow host range and a strong tropism for infection of dividing cells. Today, three human pathogenic species are known among parvoviruses, namely human parvovirus B19, human bocavirus and parvovirus 4. In addition, there are a large number of animal pathogenic parvoviruses, which are of great importance in both domestic and livestock husbandry. The reproduction capability of adeno-associated viruses is dependent on the presence of helper viruses, but their infections seem to be asymptomatic.
1.1 Classification and Characteristic Prototypes
The family Parvoviridae encompasses two subfamilies (Table 20.1): Parvovirinae and Densovirinae. The subfamily Parvovirinae comprises five genera:
-
1.
The genus Dependovirus includes the adeno-associated viruses and its members also infect humans. They are only able to replicate and to induce a productive infection cycle when the cells are concomitantly infected with a helper virus (adenovirus, vaccinia virus or herpesvirus). Thus, their productive multiplication is dependent on co-infection with these viruses. Alternatively, dependoviruses are capable of establishing a latent infection by integrating their genomes into the genome of the host cell.
-
2.
The genus Erythrovirus comprises autonomous parvoviruses that do not require a helper virus, and have a pronounced tropism for erythroid progenitor cells. Human pathogenic parvovirus B19 causes fifth disease (erythema infectiosum) and belongs to this genus.
-
3.
The genus Parvovirus contains animal pathogenic viruses, which can cause serious diseases in domestic and farm animals. They are usually very host-specific, and transmissions to humans are not observed. In contrast to the erythroviruses, they principally cause enteritis and myocarditis. In addition to canine parvovirus, which causes the most important infectious disease of dogs – and apart from the pathogen of feline panleucopenia – porcine parvovirus warrants special mention. It is the cause of economically important fertility disorders in swine. Other members of the genus Parvovirus are of minor significance, even though in individual cases they can induce a clinically apparent infection in the corresponding animal species. Minute virus of mice is a well-studied prototype. Most molecular biological processes concerning replication and transcription have been elucidated in this model virus, which does not play a role as an animal pathogen. This viral species comprises several strains which predominantly cause asymptomatic infections in mice.
-
4.
The genus Amdovirus also comprises solely animal pathogenic viruses: the prototype is Aleutian mink disease virus, which infects minks, causing Aleutian mink disease. It is considered a serious threat in regions where fur farming is of economic importance.
-
5.
Infections with viruses belonging to the genus Bocavirus cause respiratory and gastrointestinal disorder in animals and humans. In addition to some animal pathogenic species, such as bovine parvovirus and canine minute virus, human bocavirus has also been discovered recently, and its infections in humans may be associated with disease, especially in young children.
The genus Partetetravirus has been proposed for human parvovirus 4, which has been discovered in human blood and blood products. Symptoms or diseases have not been correlated with the acute infection. Similar viruses have been isolated from several mammals, e.g. chimpanzees, gorillas, pigs, wild boars (Hong Kong virus) and sheep.
Integration of viral DNA into the genome of the host cell has not been found in any members of the subfamily Parvovirinae except the dependoviruses; all are able to replicate independently of helper viruses.
The subfamily Densovirinae is divided into four genera, which include the parvoviruses of insects.
1.2 Structure
1.2.1 Virus Particle
Viral capsids have an icosahedral structure and a diameter of 18–26 nm (Fig. 20.1). In some cases, the capsid structure has been determined by X-ray crystallography. With the exception of parvovirus B19, they possess protein projections at their vertices, which are about 7 nm long. The particles of autonomous parvoviruses consist of 60 capsomers composed of proteins VP2 and VP1 in a ratio of 95 % and 5 %, respectively. VP2 is identical to the carboxy-terminal region of VP1. Some parvoviruses contain a third capsid protein (VP3) in differing amounts. It is generated by proteolytic cleavage of VP2 during maturation of minute virus of mice. In adeno-associated viruses, VP3 is an amino-terminal truncated version of VP1, which is synthesized by using the rare ACG codon as a translation start site (Fig. 20.3c). The virions are non-enveloped. The viral genome resides inside the capsid. It is associated through 11 nucleotides with the internal moiety of VP2 proteins.
1.2.2 Genome Organization and Structure
Parvoviruses have a single-stranded, linear DNA genome with a length of 4,860, 5,217 and 5,594 nucleotides in adeno-associated virus 2 (AAV-2), human bocavirus and parvovirus B19, respectively. Whereas the capsids of dependoviruses and erythroviruses (parvovirus B19) can contain DNA strands with both polarities in about equal ratio, members of the genus Parvovirus preferentially package genomes, which are complementary to the messenger RNAs (mRNAs) that are synthesized during infection. It is still unknown whether bocavirus and amdovirus particles contain single-stranded genomes with two polarities or only one type. Palindromic sequences are located at both termini, they flank the genome and are referred to as inverted terminal repeats (ITR). These sequences can fold into T- or Y-shaped structures, which facilitate the formation of double-stranded hairpin loop structures at the ends of the genome (Fig. 20.2a). In parvovirus B19 and adeno-associated viruses, both ITR sequences are complementary to each other, and have a length of 383 and 145 nucleotides in parvovirus B19 and AAV-2, respectively. They can hybridize with each other and keep the genome in a quasi-circular structure containing a panhandle-like double-stranded region (Fig. 20.2b). The ITR regions of members of the genera Parvovirus and Bocavirus have a length of 115–300 nucleotides, and their 3′- and 5′-terminal sequences are not complementary. For example, the 5′ ITR of minute virus of mice contains 207 nucleotides, whereas the 3′ ITR contains only 115 nucleotides.
The genome of parvovirus B19. (a) The single-stranded, linear DNA genomes are flanked by inverted terminal repeats (ITR) of 383 nucleotides in length (they are 145 nucleotides long in adeno-associated virus 2). The upper-case letters and lower-case letters represent DNA sequences that are complementary to each other. (b) Inasmuch as not only the sequences of the terminal 3′ and 5′ ITRs are complementary to each other, but also short regions within the repeats, T-shaped hairpin loops can be formed by the generation of double-stranded regions. Alternatively, the genomes can also adopt panhandle-like structures
The nucleotide sequence of parvoviruses is less variable than that of other viruses. Sequence differences found in different parvovirus B19 isolates are clustered in the regions encoding the carboxy-terminal end of the NS1 protein and the amino-terminal domain of VP1. The genomic arrangement is similar in all parvoviruses (Fig. 20.3): Two large open reading frames are located in the coding DNA strand, which is complementary to the mRNA. The rep gene locus, which is located in the 3′ half of the viral genome, encodes non-structural proteins involved in both viral DNA replication and regulation of gene expression. These proteins are the non-structural NS1 protein in parvovirus B19, two differently sized forms of a non-structural protein, NS1 and NS2, in animal pathogenic parvoviruses and a family of four Rep proteins in adeno-associated viruses. Apart from the gene encoding the non-structural protein NS1, bocaviruses contain additional reading frames that direct the synthesis of NS2 and NP1 proteins using alternative start codons. The genes encoding structural proteins are situated in the 5′ half of the viral genome. In parvovirus B19, there are two additional, short open reading frames in the central region and at the 5′ end of the genome which code for small non-structural proteins (7.5 kDa, 11 kDa/NS2).
Genome organization transcription and translation in parvoviruses. The genomes, including the positions of ITR elements and promoters that regulate transcription, are shown in the upper part of (a–d). Underneath, the different transcripts from which the viral proteins are translated are depicted (red structural proteins, grey non-structural proteins). The exons are indicated by thick lines, the excised introns by thin lines and the 3′ poly (A) tail by serrated lines. The regions of messenger RNAs coding for proteins are shown as bars. The 7.5- and 11-kDa proteins of parvovirus B19 have not yet been definitively demonstrated. (a) Erythrovirus – parvovirus B19. (b) Parvovirus – minute virus of mice (MVM), (c) Dependovirus – adeno-associated virus 2 (AAV-2), (d) Bocavirus – bovine parvovirus type 1 (BPV)
The promoters that control gene transcription are arranged differently in the various viruses: in erythroviruses, bocaviruses and amdoviruses, all mRNA species are initiated from a common promoter at the 3′ end of the genome. The mRNA molecules, from which the various structural and non-structural proteins are translated, are formed by alternative splicing (Fig. 20.3a, d). There are two polyadenylation sites in the genome of these viruses, one of which is located at the 5′ terminus and the other is situated in the middle of the genome. In addition, splicing is regulated by cell-specific influences: e.g. non-permissive cells synthesize a bicistronic mRNA, which contains sequences upstream of the genes encoding structural proteins; it is generated by using unusual splice sites. This leads to the synthesis of a product in which the amino-terminal end of the NS1 protein is linked to the 11-kDa NS2 polypeptide. In contrast, the autonomous viruses of the genus Parvovirus (minute virus of mice) possess two promoters: one promoter is also located at the 3′ end of the genome, and controls the transcription of non-structural genes (NS or rep); the second, centrally located promoter (genome position 38) is responsible for transcribing the genes that encode structural proteins. These viruses have only one polyadenylation site at the end of the genome, and it is used for termination of all transcripts (Fig. 20.3b).
Adeno-associated viruses also use a common polyadenylation site at the end of the genome for all mRNA species. Their genome contains three promoters. The promoter at the 3′ end of the genome (position 5) regulates the synthesis of the mRNA for the Rep78 and Rep52 proteins. The latter is translated from a spliced mRNA. The promoter at position 19 controls the transcription of genes that encode the Rep68 and Rep40 proteins. Rep40 is also translated from a spliced mRNA. In both cases, the splicing process leads to excision of the same intron sequence in Rep52 and Rep40. The synthesis of mRNA species coding for capsid proteins is under the control of the third promoter, which is located at genome position 40 (Fig. 20.3c). Hence, gene expression in the individual virus types is regulated differentially and specifically by the different localization of transcriptional control elements in combination with alternative splicing, despite the similar gene arrangement found in all parvoviruses.
1.3 Viral Proteins
1.3.1 Structural Proteins
Two structural proteins compose the capsids of parvoviruses: VP1 (molecular mass 80–86 kDa) and VP2 (58–62 kDa). VP2 is identical to the carboxy-terminal region of VP1; the VP1 proteins of parvovirus B19 and human bocavirus have 227 and 129 additional amino acids at the amino terminus, respectively (VP1 unique region). For the synthesis of VP2, the VP1-specific mRNA is spliced. The excision of the several-hundred-nucleotide-long intron leads to removal of the start codon of the VP1 protein. Therefore, the translation of VP2 starts at another AUG codon in the same reading frame (Fig. 20.3a, d). The regions necessary for particle formation are located in the VP2 protein, and in the corresponding VP2-specific part of VP1. Therefore, both proteins are present as capsomers of the virions. The VP1 unique region of parvovirus B19 is exposed on the capsid surface. Amino acid sequence variations are frequently found in the amino-terminal region. Most neutralizing antibodies are directed against epitopes in this protein domain. However, part of the neutralizing immune response is also directed against the VP2 moiety. The VP2 protein of parvovirus B19 binds to globoside (erythrocyte P antigen), which is used as a cellular receptor. The carboxy-terminal domain of the VP1 unique region has a phospholipase A2 like enzyme activity. Table 20.2 summarizes the various functions and properties of parvovirus proteins.
The VP3 protein of minute virus of mice is a degradation product of VP2 and is generated by proteolytic cleavage of about 20 amino acids at the amino terminus. In the case of adeno-associated viruses, in the same reading frame there is a rare upstream start codon of the VP1-specific mRNA which is used for the synthesis of VP3 (73 kDa). Thus, its amino terminus is located between those of the VP1 and VP2 proteins.
1.3.2 Non-Structural Proteins
1.3.2.1 Minute Virus of Mice
The non-structural NS1 and NS2 proteins of parvoviruses are encoded by the rep gene cluster. The NS2 protein constitutes a heterogeneous protein group that is translated from spliced mRNA molecules. Starting from the splice donor site, these mRNAs are translated in all three possible reading frames (Fig. 20.3b). NS proteins have essential functions in viral DNA replication. NS1 is a sequence-specific endonuclease that is active during genome replication. It cleaves the double-stranded DNA within ITR elements, which serve as the origin of replication, thus generating a nick that provides a free 3′-OH end that serves as a primer for DNA synthesis. NS1 remains covalently linked to the 5′ termini of newly synthesized genome intermediates during replication. Furthermore, it has been shown that this protein acts as an ATP-dependent helicase in vitro. This enzyme activity is necessary for unwinding of DNA duplexes at the origin of replication. Moreover, the NS1 protein acts as transactivator of viral promoters, especially of the central promoter that controls the expression of capsid genes, e.g. in members of the genus Parvovirus. In addition, it also influences cellular promoters, whereas it does not bind to the sequence elements, but it activates or represses them by interacting with cellular factors. Both non-structural proteins possess nuclear transport signals, and the larger NS1 protein is phosphorylated at several positions by cellular protein kinase C. Point mutations in the non-structural genes altering the phosphorylation pattern lead to replication-defective viruses, decreased helicase activity and a reduction of the cytotoxic effect of NS1. The cytotoxic effect of the NS1 protein is probably associated with the ability to arrest the cell cycle at the G1, S or G2 phase. It has been demonstrated that NS1 interacts with the catalytic α subunit of casein kinase 2. This process interferes with intracellular signal transduction pathways, and gives rise to the development of the typical cytopathic effect. In particular, interruption of the cycle in the G2 phase seems to be caused by the ability of the NS1 protein to induce the expression of cellular protein p21 (Waf1/Cip1), which functions as an inhibitor of cyclin A dependent kinase. Furthermore, the NS1–casein kinase 2 complex is responsible for phosphorylation of capsid proteins.
The function of NS2 proteins is less well explored: they interact with the nuclear export protein Crm1, and have a very short half-life; furthermore, they are degraded by a ubiquitin-independent pathway through the proteasome. Probably, they act as helicases, and are involved in strand displacement during genome replication.
1.3.2.2 Parvovirus B19
Parvovirus B19 expresses only one non-structural protein, NS1 (71 kDa). It is translated from an unspliced mRNA that is terminated at the central polyadenylation signal. NS1 is a phosphorylated, multifunctional protein that exerts its activities in the nucleus. Accordingly, the NS1 protein carries a nuclear localization signal andis essential for genome replication. It is presumably responsible for the formation of the initiation complex at the origin of replication, which is composed of the cellular DNA polymerase and other additional host factors. Furthermore, the ATP-dependent helicase and endonuclease activities of NS1 are also involved in viral replication. In addition, NS1 is a transcriptional activating protein; it induces the parvovirus B19 specific promoter p6 by interacting with the cellular transcription factors Sp1, Sp3 and YY1 and possibly other cellular transcription factors. It is also capable of influencing cellular promoters such as the IL-6 promoter. It induces cell cycle arrest in the G1 phase, a process that is associated with an increased expression of p53 as well as the cyclin-dependent kinase inhibitors p21/Waf1 and p16Ink4. The NS1 protein also interacts with the cellular transcription factor Sp1 to upregulate the respective promoter activities. NS1 protein expression is toxic in eukaryotic cells. The toxic effect is probably associated with its ability to induce the synthesis of the proapoptotic factors Bax and Bad (members of the Bcl-2 protein family) as well as that of a variety of cellular caspases (caspases 3, 6, 8 and 9), and its ability to induce the TNF- and Fas-dependent apoptosis pathway in infected cells.
The function of the 11-kDa NS2 protein of parvovirus B19, which codes at the 5′ end of the genome and is translated from a twofold spliced mRNA, has not been finally resolved. It appears to be required in the production of infectious progeny viruses, and to influence both the synthesis and the intracellular transport of capsid proteins. When the synthesis of the 7.5-kDa protein, which is encoded by a short reading frame located upstream of the genes coding for structural proteins, is prevented, there is no reduced viral production. Thus, the function of the 7.5-kDa protein in the infection cycle remains mysterious.
1.3.2.3 Adeno-Associated Viruses
Adeno-associated viruses synthesize four different non-structural proteins. Rep78 and Rep52 differ by an internal deletion, which is based on the use of a spliced mRNA for translation (Fig. 20.3c). The same is true for Rep68 and Rep40, which are translated from transcripts whose synthesis is controlled by the promoter at genome position 19. They are amino-terminally truncated protein variants of Rep78 or Rep52, respectively. Therefore, all Rep proteins share large homology regions. It is believed that further splicing events may contribute to the generation of additional Rep protein variants.
Rep78 and Rep 68 are localized in the nucleus. Their functions in replication correspond in many respects to those of NS1 proteins of animal parvoviruses. The amino-terminal domains of Rep78 and Rep68 are essential for genome replication of adeno-associated viruses: the 208 amino-terminal residues harbour the endonuclease domain, which cleaves double-stranded DNA at the terminal resolution site (nucleotide 125), within ITR elements during replication. In contrast to the NS1 protein of minute virus of mice, Rep proteins of adeno-associated viruses do not remain associated with the nascent 5′ termini. In vitro, an ATP-dependent helicase activity has been demonstrated to be associated with the large Rep proteins, and is capable of unwinding not only DNA–DNA double strands, but also DNA–RNA heteroduplexes. Rep78 and Rep68 also transactivate viral promoters by interacting with cellular transcription factors – such as the cellular protein PC4 – and factors encoded by helper viruses, especially the immediate early proteins of herpesviruses or adenoviruses. In addition, Rep68 and Rep78 also recruit the single-strand-binding ICP8 protein of herpesviruses to the centres of adeno-associated virus replication, and thus presumably inhibit herpesvirus replication. Apart from adenoviruses and herpesviruses, human papillomaviruses can also provide helper functions: The E1 protein of human papillomavirus 16 – also a helicase – binds to Rep78, resulting in modulation of enzyme function. During latency of adeno-associated viruses, i.e. during non-productive infection in the absence of helper viruses, in which the viral genome is integrated in the chromosomal DNA of the cell, a repressing effect has been attributed to Rep proteins, especially on the promoter of capsid protein genes. The expression of cellular and other viral promoters is also negatively influenced by Rep78 and Rep68, which repress the activity of the long terminal repeat promoter of human immunodeficiency virus 1 and that of the c-H-ras promoter.
To what extent the functions of the large Rep proteins are related to the property of adeno-associated viruses to suppress uncontrolled cell proliferation of various origins, and thus act as tumour suppressors, has not yet been conclusively resolved. Independently of the presence of p53 in infected cells, Rep78 induces apoptosis by activating caspase 3. Recently, it has also been discovered that Rep78 prevents phosphorylation of retinoblastoma protein by a still unknown pathway, thereby stabilizing the RB105/E2F complex. This leads to an inhibition of E2F protein function, thereby blocking the induction of specific cellular proliferation genes. Moreover, Rep78 binds to EF2-dependent promoters, thus additionally impeding E2F binding to the promoters. As a result, the cells are arrested in the S phase of the cell cycle. In this regard, Rep78 acts as an antioncogene, functioning conversely to the oncogenic proteins of hepatitis B, polyomaviruses, papillomaviruses, adenoviruses and herpesviruses (Sects. 19.1–19.5).
In vitro, during establishment of the latent state in the absence of helper viruses, the genome of adeno-associated viruses integrates specifically into the 19q13.3-qter locus of human chromosome 19. The proteins Rep78 and Rep68 are involved in this site-specific integration. They interact concomitantly with Rep-binding sites (RBS) located in the ITR elements at the ends of the genome, and with RBS-like sequences at the integration site (AAVS1) on chromosome 19 via a second protein domain. They bring the viral genome close to the chromosomal DNA of the cell, and cleave one DNA strand by their endonuclease activity.
The two smaller Rep52 and Rep40 proteins are found predominantly in the cytoplasm. They also possess helicase activity, which is required during morphogenesis. They bind to the preassembled capsids of adeno-associated viruses, and bring about the introduction of the single-stranded DNA genomes in 3′ to 5′ direction. Hence, Rep proteins are versatile and multifunctional. Since these activities are performed in interaction with cellular and helper-virus-specific proteins, they are dependent on the type and differentiation state of the infected cell and the presence of certain functional activities of helper viruses.
1.4 Replication
1.4.1 Parvovirus B19 and Other Autonomous Parvoviruses
Parvoviruses can replicate only in cells that are in the S phase of the cell cycle. Unlike papillomaviruses, polyomaviruses or adenovirus (Sects. 19.2–19.4), they can not stimulate resting cells to divide. Expression and replication of the viral genome are highly dependent on host cell factors. This explains the narrow cell and host tropism of the virus. The various parvoviruses bind to different receptors on the cell surface of the respective target cells: parvovirus B19 infects erythroid progenitor cells in the bone marrow, and interacts with the erythrocyte P antigen, a glycosphingolipid that is exposed on the cell surface. This interaction is mediated by surface structures of the capsid protein VP2. Integrin α5β1 and the autoantigen Ku80 are being discussed as possible co-receptors. Animal pathogenic feline parvoviruses (feline panleucopenia virus and canine parvovirus) use the transferrin receptor for attachment. By contrast, glycophorin A has been identified as a receptor for bovine parvoviruses, which bind to terminal sialic acid residues that are present as a modification on this surface component of erythrocytes. Minute virus of mice also uses N-acetylneuraminic acid residues as a cellular interaction partner.
After attachment, bovine parvovirus particles enter the cells probably by receptor-mediated endocytosis. In feline parvovirus, it is considered as proven that the bound virus particles accumulate in areas containing clathrin-coated pits and clathrin-coated vesicles, and that dynamin, a component of intracellular filaments, is involved in the particle internalization process. The phospholipase A2 like activity of parvovirus VP1 proteins, which is necessary for their infectivity, facilitates the release of virus particles from the endosomes. There is only little information concerning the further transport of virus particles or viral genomes into the nucleus, where viral gene expression and genome replication occur.
In the nucleus, the single-stranded DNA genome is converted into a double-stranded molecule using the OH group at the 3′ ITR as a primer, and is transcribed by the cellular RNA polymerase II. In minute virus of mice, it has been shown that the promoter of NS genes is supported by binding of a viral structural component. It is postulated that this component, similar to the α-transinducing factor, a tegument protein of herpes simplex virus, acts in combination with cellular factors to facilitate transcription of immediate early genes (Sect. 19.5). The resulting mRNAs are capped, partially spliced and polyadenylated. After translation, NS proteins, as well as capsid proteins, are transported into the nucleus using their transport signals, where the NS proteins are required for both DNA replication and induction of transcription of genes encoding structural proteins. Thus, they are an essential prerequisite for the further replication process, which occurs in delimited nuclear areas in autonomous parvoviruses (parvovirus-associated replication bodies).
In contrast to cellular DNA synthesis, no RNA primers are needed for viral genome replication. This function is performed by the 3′-OH group of the ITR region at the 3′ terminus of the single-stranded DNA genome. A lagging-strand synthesis does not occur. The model described here has been developed for replication of the genome of minute virus of mice. Whether this model can be applied in all aspects to parvovirus B19 is not known. Because of its pronounced tropism for erythroid progenitor cells, it has not been possible to cultivate this virus, and therefore its genome replication mechanisms have not been investigated. The ITR sequences, which form double-stranded hairpin loop structures at the ends of the genome, are essential for initiation of DNA replication (Fig. 20.2). The 3′-OH group of the 3′ ITR serves as a primer for the first polymerization reactions, which are probably catalysed by the cellular DNA polymerase δ complex. The result is an intermediate product which is present as a nearly completely double-stranded DNA molecule, with the exception of a short single-stranded region at the T-shaped ITR structure that remains covalently closed. In the next step, the sequence-specific endonuclease activity of the NS1 protein generates a nick at the terminal resolution site within the 5′ palindrome. The NS1 protein of minute virus of mice remains covalently linked to the newly generated 5′ end of the nick. The secondary structures at the end of the genome are resolved by unwinding by the ATP-dependent helicase, and the 3′-OH end of the nick provides the necessary primer to start the synthesis of the complementary strand by a single-strand-displacement DNA replication mechanism (Fig. 20.4). The replication fork progresses along the template strand, thus generating a replication intermediate that comprises two single-stranded genomes in double-stranded configuration. The nuclease activity of the NS1 protein cleaves the dimeric genomes at the respective terminal resolution site, and the resulting 3′-OH ends are used for initiation of new polymerization and strand-displacement reactions. The displaced DNA strands interact with the structural proteins to shape precursor capsid forms.
Parvovirus genome replication model. DNA synthesis is catalysed by cellular enzymes and is initiated at the 3′-OH end of the ITR element, which is folded into a stem-loop structure. With progressive elongation, the secondary structure of the 5′-terminal palindrome is resolved and DNA synthesis continues until the end of the genome. The recognition site for the endonuclease activity of the NS1 protein resides at the transition of the double-stranded 3′ ITR to the single genome sequences; it generates a single-strand nick. This creates a 3′-OH end, which serves as a primer to initiate DNA synthesis, which continues until the other end of the genome. The result is a completely double-stranded DNA genome. Subsequently, the ITRs fold into stem-loop structures, thus providing new initiation sites for DNA replication. Newly synthesized sequences are depicted in red. TRS terminal resolution site
Viral morphogenesis has also been scarcely examined. It is not known whether the DNA is introduced into a preformed capsid structure or whether the proteins attach to a condensed form of the genome. The newly assembled capsids are detectable as nuclear inclusion bodies only a few hours after infection. Later, they are also found in the cytoplasm and in evaginations of the cell membrane through which the particles are partially released by the cell. In these vesicles, viruses are protected from attack by the immune system and can be easily spread throughout the body. However, most virions are released by apoptosis of infected cells.
1.4.2 Adeno-Associated Viruses
1.4.2.1 Productive Viral Replication
Adeno-associated viruses infect various human epithelial cells. The receptor of adeno-associated virus 5 is, like for certain influenza A viruses, N-acetylneuraminic acid (sialic acid), which is linked with oligosaccharides as a modification of cell surface proteins by α-2,3 glycosidic bonds (Sect. 16.3.3). Capsids of AAV-2 have been found to bind to heparan sulphate. There is evidence that αvβ5 integrins and/or the human fibroblast growth factor receptor act as co-receptors during attachment. Furthermore, an interaction of AAV-2 capsids with nucleolin has also been described. This protein (110 kDa) functions as a transport protein between the cytoplasm and the nucleolus during cellular processes such as ribosomal RNA synthesis and ribosome assembly. Therefore, nucleolin may be involved in the transport of virus particles into the nucleus. A similar interaction has also been described for capsid proteins of coxsackievirus B5 (Sect. 14.1). The productive replication process is very similar to that of autonomous parvoviruses. The 3′-OH ends of the terminal palindromes are also used as primers in adeno-associated viruses. Further steps are DNA polymerization, strand displacement and synthesis of a double-stranded replication intermediate.
The major difference from the replication of autonomous parvoviruses is the fact that adeno-associated virus replication is dependent on the simultaneous infection with a helper virus (adenovirus or herpesvirus). The presence of certain proteins, especially the immediate early gene products of helper viruses, is a prerequisite for DNA replication of adeno-associated viruses, and thus for production of infectious progeny viruses. Particularly, adenoviral E1A proteins are involved in transactivation of adeno-associated virus promoters. Possibly, the 35-kDa E4 protein is also involved, in complex with the E1B protein, in the process that regulates viral mRNA export from the nucleus to the cytoplasm (Sect. 19.4). This assistance seems to impair adenovirus replication. Even the oncogenic properties of E1A and E1B proteins are neutralized.
1.4.2.2 Latent Infection
In the absence of helper viruses, adeno-associated viruses can establish persistent infections and remain in the organism for life. In vitro, it has been found that the genome of adeno-associated viruses is integrated in human chromosome 19 by a site-specific recombination mechanism during latency. Two genome copies remain at the integration site and are present in tandem. Intact ITR elements and the large non-structural proteins Rep78 and Rep 68 are required for the integration process. The integrated genome can be reactivated from latency by infection with an appropriate helper virus. A similar site-specific integration of the adeno-associated virus genome does not seem to occur in vivo.
Gene Therapy Vectors Are Derived from Adeno-Associated Viruses
Since adeno-associated viruses possess the unique ability to integrate stably into the human genome, infection with them does not cause severe genetic consequences and their reactivation from latency is dependent on the presence of helper viruses, they appear to be suitable vectors for gene therapy approaches. In such cases, the genes for structural proteins or the entire coding region is replaced by the desired heterologous gene to be expressed. The chimeric viral genome is packaged into capsids in so-called helper cells, which constitutively synthesize the deleted viral functions, and are infected with a helper virus. These recombinant capsids are able to transduce the chimeric genome into cells, and it can be stably integrated into the human genome via ITR elements.
1.5 Human Pathogenic Parvoviruses
1.5.1 Parvovirus B19
1.5.1.1 Epidemiology and Transmission
In 1975, Yvonne Cossart and co-workers serendipitously discovered parvovirus B19 in blood samples. A few years later, the group of Sergeant found that parvovirus B19 is the causative agent of fifth disease (erythema infectiosum), a classic childhood disease that is associated with rash. Nowadays, three genotypes of this virus are known which cause the same disease, but exhibit a different regional distribution: genotype 1 predominates in central and southern Europe, Asia and the Americas, genotype 2 is frequently found in northern Europe, particularly in elderly people, whereas genotype 3 occasionally prevails in West Africa and France. The prevalence of parvovirus B19 is high in the human population. In Western countries, antibodies against the structural proteins VP1 and VP2 have been found in 40–70 % of adults examined; the seroprevalence is approximately 65–75 % among adults In the first few days of infection, before the onset of symptoms, very large amounts of virus particles are detectable in the blood of infected individuals (up to 1013 particles per millilitre). In this phase, the virus is excreted in the saliva. It is transmitted by droplet infection or through contaminated blood and blood products. Because of its high stability, the virus is difficult to inactivate. It can be detected as infectious virus even in purified factor VIII and factor IX preparations for blood clotting and in highly pure samples of human serum albumin. If parvovirus B19 infects pregnant women, it can be transmitted diaplacentally to the fetus, causing hydrops fetalis, with lethal consequences.
1.5.1.2 Clinical Features
The incubation period of parvovirus infection lasts on average 1–2 weeks. In this phase, the patient is already viraemic, and can transmit the virus. The infection is frequently asymptomatic in children. The commonest clinical picture is erythema infectiosum, also known as fifth disease. It is found mainly in childhood, and is characterized by flu-like symptoms with mild fever. It is associated with a rash, which initially appears on the cheeks concurrently with the first virus-specific antibodies, it spreads further to the inner sides of the arms and legs, and usually lasts 1–2 days. Because of the viral destruction of erythrocyte progenitor cells, all infected individuals develop a transient anaemia. Occasionally, acute parvovirus B19 infection may lead to the development of papular purpuric gloves and socks syndrome, thrombocytopenia and neutropenia or hepatitis and myocarditis; encephalitis has also been reported in individual cases. If individuals with impaired formation and maturation of red blood cells are infected (e.g. sickle cell anaemia, or thalassaemia), a severe life-threatening, aplastic crisis may be caused by the virus-induced destruction of erythrocyte progenitor cells.
Acute infections are frequently associated with arthralgia and severe inflammation of the joints. The forms of acute arthritis usually last a few weeks after infection. However, they may also cause problems for years and are often similar to rheumatoid arthritis. In these cases, a persistent parvovirus infection is the basis of protracted, parvovirus-associated arthritis, in which the viruses are present in low concentrations in the synovial fluid of inflamed joints for prolonged periods. In addition, it is found that other autoimmune diseases can be established in some patients after infection. These include various forms of vasculitis, Hashimoto’s thyroiditis and autoimmune anaemia, neutropenia and thrombocytopenia. Chronic persistent infections occur especially in immunocompromised patients: they usually develop long-lasting severe diseases, particularly chronic anaemia or thrombocytopenia. The viral DNA can be detected permanently in relatively large quantities in the blood.
Other complications are caused by parvovirus B19 infection during pregnancy. An acute parvovirus B19 infection can cause spontaneous abortions during the first trimester of pregnancy. As compared with non-infected pregnant women, in pregnant women with acute parvovirus B19 infection, the spontaneous abortion rate is increased by 5–6 %. Especially in the second trimester, the virus is transmitted diaplacentally to the embryo in about one third of infections, where it infects proerythroblasts in the liver. Acute infection of the expectant mother up to and including the 20th week of pregnancy can lead to the development of hydrops fetalis (an accumulation of large amounts of fluid in the embryo, which is associated with oedema, anaemia, hydraemia and liver failure). In the case of early diagnosis, blood transfusion through the umbilical vein can prevent abortion. If the fetus develops an additional myocarditis, it usually dies. Parvovirus B19 infections are possibly also associated with intrauterine fetal death in late pregnancy. There is no evidence that parvovirus infection causes malformations of embryos.
1.5.1.3 Pathogenesis
During transmission, parvovirus B19 reaches the mucous membranes of the mouth and throat area. What cells are primarily infected is unclear. The cellular receptor for parvovirus B19, the blood group P antigen, is expressed on many cells, including endothelial cells and megakaryocytes. However, a productive infection cycle cannot occur in them. It is also unknown in which way the virus reaches its target cells, the erythroid progenitor cells of the differentiation stages erythrocyte burst-forming unit, erythrocyte colony-forming unit and erythroblasts in the bone marrow. During the early phase of infection, in which the virus multiplies in erythrocyte progenitor cells, it is present in high concentrations in the peripheral blood (1011–1013 particles per millilitre). This viraemic phase is attenuated by two factors: first, through the production of neutralizing antibodies that prevent further spread of the virus, and second, by the virus-induced elimination of erythroid progenitor cells, which are permissive for viral infection. Patients are temporarily anaemic in this phase. The damage to this cell population can cause severe aplastic anaemia in individuals with genetic erythropoietic disorders because these patients cannot adequately regenerate erythrocytes.
The exanthem develops concomitantly with the virus-specific antibodies. Therefore, it is assumed that immune complexes attach to the endothelium of blood capillaries and cause inflammatory reactions that cause the manifestation of rash symptoms. Alternatively, there are also hypotheses suggesting that the rash is generated by non-permissive infection of endothelial cells.
About 20 % of immunocompetent patients cannot effectively control the virus: they produce viruses for months and occasionally even for years, and these can be detected in blood. Protracted arthritis is associated with such persistent parvovirus B19 infection. Viral DNA and virus particles complexed with antibodies can be detected in the synovial fluid of inflamed joints, which can cause immunological reactions and contribute to inflammation. The occurrence of NS1-specific antibodies that are increasingly found in these patients indicates a chronic, persistent infection with a delayed and incomplete control of virus production. It is believed that the continued presence of the virus results in infection of non-permissive cells, inducing an abortive infection cycle in them. These cells do not generate infectious progeny viruses, but may produce NS1 proteins. The cytotoxic effect of NS1 protein can be manifested as can its capability of transactivating cellular promoters which regulate the expression of inflammatory proteins such as IL-6 and TNF-α. Transgenic mice carrying the parvoviral NS1 gene in their genome are more susceptible to the development of arthritis than control mice. A central role in the induction of chronic inflammatory diseases can also be ascribed to the phospholipase A2 like activity in the VP1 unique region of viral capsids, which may contribute to the continuous production of prostaglandins, leukotrienes and inflammatory mediators in the joints, and thus to the maintenance of arthritis. The enzymatically active VP1 unique region can activate synoviocytes in vitro, leading to an increased production of prostaglandin E in them. On the other hand, the involvement of autoimmune mechanisms is also conceivable as a cause of prolonged inflammation: patients frequently produce antiphospholipid antibodies, whose development may be induced and triggered by the VP1 unique region and its similarity to cellular protein–lipid complexes.
Apart from the blood of patients with chronic persistent infectious, viral genomes are also present in various tissues and organs (liver, myocardium, skin, muscle, tonsils, bone marrow, synovial tissue) of healthy people; probably, low amounts of viral genomes remain latent in endothelial cells of all individuals with a past parvovirus B19 infection. However, the underlying mechanism of this DNA latency is unclear. Whether the latent parvovirus B19 genomes can be reactivated for renewed production of viruses by other diseases or immunosuppression is also unclear.
1.5.1.4 Immune Response and Diagnosis
The diagnosis is performed by detecting specific antibodies against viral structural proteins by means of ELISA or Western blot analyses (Fig. 20.5). The presence of IgG indicates a past infection. IgM antibodies are usually detectable only in fresh parvovirus B19 infections, and are preferentially directed against particulate forms of structural proteins. A renewed increase in IgM levels is occasionally found in patients with persistent infections. Viral DNA in the serum of patients with fresh or persistent infections can be detected by using polymerase chain reaction (PCR). Antibodies against the NS1 protein of parvovirus B19 can be found particularly in patients with persistent infections (protracted granulocytopenia, thrombocytopenia) and in individuals with parvovirus B19 associated persistent arthritis.
1.5.1.5 Therapy and Prophylaxis
There is no vaccine against parvovirus B19. In acutely infected pregnant women, the therapeutic focus is on the treatment of fetuses with intrauterine transfusions, which are applied when the fetal haemoglobin level decreases below 8–10 g/dl. This can generally prevent the development of hydrops fetalis in the embryo. The use of immunoglobulin products has yielded successful results in the treatment of immunocompromised patients (transplant patients) with chronic anaemia and erythroblastopenia caused by persistent parvovirus B19 infections.
1.5.2 Human Bocavirus
1.5.2.1 Epidemiology and Transmission
Human bocavirus was discovered by Tobias Allander in airway aspirates of children with acute respiratory disease in 2005. Sequence analysis revealed that this virus is a member of the parvoviruses, and that it exhibits similarities to bovine parvovirus type 1 and canine minute virus, which are members of the genus Bocavirus, which was been named after the bovine and canine viruses that it includes. The new virus has been denominated human bocavirus, and has been assigned to the genus Bocavirus; four genotypes have been characterized to date (genotypes 1–4); their nucleic acid sequences coding for the non-structural proteins are conserved to 70 %. Epidemiological studies have shown that human bocavirus has a worldwide distribution, and causes diseases of the respiratory and gastrointestinal tract primarily in infants aged less than 3 years. A seroprevalence of 95 % is found in children older than 6 years and in adults. The virus is probably transmitted by droplet infection.
1.5.2.2 Clinical Features
Little is known about the incubation period until the onset of symptoms. Probably, most human bocavirus infections are asymptomatic. Genotype 1 can be detected particularly in children with acute illnesses of the lower respiratory tract (bronchitis, pneumonia). By contrast, genotypes 2–4 are preferentially found in young children with gastrointestinal disorders (diarrhoea). It has not been conclusively clarified whether human bocavirus is causally associated with these disorders because other viruses (respiratory syncytial virus, human metapneumovirus, norovirus) and bacteria (streptococci) have also been found in many of the affected children. Nevertheless, many findings suggest that human bocavirus infections are associated with diseases of the lower respiratory tract in infants.
1.5.2.3 Pathogenesis
There are no data concerning the pathogenesis of human bocavirus infection; the target cells are also unknown. The virus is found in the sputum and pharyngeal lavage as well as in the blood of children with respiratory diseases; the latter is an indication that this is not a local, but a systemic infection. The pathogen is also detectable in the stool of children with diarrhoea.
1.5.2.4 Immune Response and Diagnosis
In individuals with past infection, IgG antibodies against the capsid VP2 protein can be detected by ELISA; furthermore, CD4+ cell-mediated cellular immune responses against the VP2 protein are also observable. VP2-specific IgM and viral DNA in blood or respiratory tract aspirates are indicative of acute infection.
1.5.2.5 Prophylaxis and Therapy
There is neither a vaccine nor an effective antiviral therapy.
Human Parvovirus 4
The virus was discovered in 2005. It was found in a patient who had symptoms of acute infection, and was hospitalized in a clinic in San Francisco. It was also detected by PCR in human blood plasma, especially in plasma pools of blood donors from the west coast of the USA. Analysis of the genome revealed that this virus is a parvovirus, and it received the provisional designation parvovirus 4 because it is the fourth member of this virus family which is capable of infecting humans. It has not yet been conclusively elucidated whether parvovirus 4 can be classified into the genus Erythrovirus. Two genotypes of this new virus have been described to date. It is unclear whether parvovirus 4 infection is associated with symptoms. Parvovirus 4 has been found sporadically in plasma pools and blood products (clotting factors) as well as in drug addicts and patients who are infected with hepatitis C virus. These data indicate a transmission by human blood.
1.5.3 Adeno-Associated Viruses
1.5.3.1 Epidemiology and Transmission
Adeno-associated virus 2, adeno-associated virus 3 and adeno-associated virus 5 can infect humans. They were found as a contaminant in adenovirus preparations, with the exception of adeno-associated virus 5, which was isolated from a flat penile condyloma. Adeno-associated viruses are likely transmitted along with adenoviruses by droplet infection during childhood. More than 90 % of adults are infected with these viruses, which are present in a latent stage after primary infection.
1.5.3.2 Clinical Features
Despite the high prevalence rate, no disease has been associated with adeno-associated virus infections so far.
1.5.3.3 Pathogenesis
In vivo, it is unknown what cells are infected by adeno-associated viruses, and in which cells they integrate their DNA into the genome of the host cell. In vitro, they can replicate in various epithelial cells with the assistance of a helper virus. Adeno-associated virus 2 has been detected by PCR in endometrial biopsies.
It is unclear how adeno-associated viruses inhibit tumour cell proliferation. This antioncogenic function is closely related to the activity of the proteins Rep78 and Rep68, and is probably associated with their repression effect on cellular promoters. There is evidence that adeno-associated viruses can also be reactivated by human papillomavirus, and that both virus types infect the same cells. Whether the antioncogenic effect of adeno-associated viruses plays a role in preventing papillomavirus-associated tumours is unclear. Overexpression of Rep proteins is toxic to cells, and it cannot be excluded that some of their oncolytic properties may be based on this phenomenon.
1.5.3.4 Immune Response and Diagnosis
IgG antibodies against structural proteins can be detected by ELISA in infected individuals. IgM antibodies are found after reactivation of the virus during pregnancy. Acute infections are rarely diagnosed because they are asymptomatic.
1.5.3.5 Prophylaxis and Therapy
There is neither a vaccine nor an effective antiviral drug. They seem to be unnecessary because infections are not associated with disorders.
1.6 Animal Pathogenic Parvoviruses
1.6.1 Feline and Canine Parvoviruses
1.6.1.1 Epidemiology and Transmission
The prototypes of feline parvoviruses – feline panleucopenia virus, mink enteritis virus and canine parvovirus – are very closely related to each other: their genome sequences exhibit a homology of more than 98 %. They cause almost identical diseases in their respective hosts, mainly a haemorrhagic gastroenteritis. The evolution of these viruses can be traced in detail because canine parvovirus emerged suddenly in 1978–1979, and all data suggest that it has developed directly or indirectly from the long-known feline panleucopenia virus, possibly via infections of foxes (Fig. 20.6, Chap. 12).
Evolution of animal pathogenic parvoviruses. The mutations in the amino acid sequence in the structural VP2 protein which have led to the host tropism change from feline panleucopenia virus to canine parvovirus type 2 are indicated. Foxes have been postulated as hypothetical intermediate hosts. Infections with canine parvovirus type 2 were associated with lethal diseases in dogs only. Additional mutations changed the amino acid sequence of this virus, leading to canine parvovirus types 2a and 2b, which infect both cats and dogs, replacing the highly virulent type 2
Feline parvoviruses are transmitted by direct animal contact or indirectly through contact with virus-contaminated materials (bedding, bowls, litter boxes, etc.). Humans are considered to be a frequent source of transmission, e.g. breeders or whelp buyers, who can introduce them via contaminated clothing or the soles of shoes into other animal populations. This indirect dissemination is facilitated by the high physical stability of parvoviruses. They are capable of remaining infectious in dried faeces for months. This property was responsible for the rapid global spread of canine parvovirus in 1979: it was introduced to continents such as Australia without delay, although strict quarantine requirements were imposed for importation of dogs. The virus is also excreted in very large quantities by infected animals (up to 109 infectious particles per gram of faeces). Approximately 102–103 infectious viruses are sufficient to elicit an infection. Therefore, a high infection pressure can be rapidly developed in infected farms or animal shelters. Feline panleucopenia virus and mink enteritis virus infect cats and raccoon-like animals. Both viruses are considered virulent variants of the same virus because they cannot be distinguished at the level of the DNA sequence. Dogs and other canids (wolves, jackals, foxes, raccoon dogs, etc.) do not seem to be susceptible to feline parvoviruses under natural conditions. Canine parvovirus type 2 was first isolated in 1978, and triggered a devastating worldwide pandemic with millions of deaths in the dog population. However, it seems to be limited to dogs and other canids. The emergence of the so-called new antigenic types CPV-2a, CPV-2b and CPV-2c changed this clear picture, as these viruses can infect dogs and cats and cause a disease in both types of animals.
1.6.1.2 Clinical Features
The symptoms of feline parvovirus infections are largely identical in cats and dogs; however, there are some differences. The main symptoms are acute haemorrhagic diarrhoea, lymphopenia and a generalized leucopenia, especially in cats. In addition to these classic manifestations, young kittens which are infected with feline parvovirus during the first few days of life develop an infection of Purkinje cells of the cerebellum, which leads to cerebellar hypoplasia. The result of this injury is the clinical picture of feline ataxia: it manifests itself in an age of 3–4 weeks, when the kittens begin to run. However, dogs infected with canine parvovirus develop no cerebellar infection. If dogs are infected during the first few days after birth, they develop a myocarditis, which can lead to a complete malfunction of the heart, with a fatal outcome.
1.6.1.3 Pathogenesis
After entering the body, the virus infects the epithelial cells of the nasopharyx and regional lymphoid organs, in which the virus initially replicates, spreading then to the lymphatic organs and bone marrow via lymphocyte-associated viraemia. The virus infects the cells of the Lieberkühn glands through the Peyer patches, i.e. the regeneration site of the intestinal epithelium. These cells are damaged by direct cell lysis, which leads to atrophy of the villi and impairment of intestinal function. The associated clinical symptom is acute haemorrhagic diarrhoea. Infected animals develop a generalized leucopenia owing to infection of the bone marrow, which is particularly pronounced in feline parvovirus infections of cats. The generalized infection of lymphoid organs leads to a marked relative lymphopenia.
1.6.1.4 Immune Response and Diagnosis
The diagnosis can be made with certainty because of the histological changes in the Lieberkühn glands. In living animals, the high-grade leucopenia and haemorrhagic diarrhoea provide clear clinical evidence. The virus can be detected by immunofluorescence of biopsy material from the infected organs, through isolation of the virus or by detecting viral DNA by PCR. Detection of the virus in living animals is easily performed by means of electron-microscopic or immunochromatographic analysis of samples of faeces. The different antigenic variants can be distinguished by haemagglutination-inhibition tests using type-specific monoclonal antibodies.
Neutralizing antibodies are the main component of the protective immunity; they function for all virus types. A prerequisite for infection of young animals is that pups originate from parvovirus-seronegative mother animals. Seropositive mothers pass on neutralizing antibodies to the pups via colostrum, and these protect the pups from infection in the first few weeks of life. A cellular immunity is proven, but the extant of its contribution to protection is unknown.
1.6.1.5 Control and Prophylaxis
Parvovirus infection in dogs and cats is controllable by vaccination. Different, highly effective attenuated live vaccines are available. Inactivated vaccines have proved to be less effective and are not used any more. The parvovirus component is contained in every standard combination vaccine.
Just a Few Amino Acid Residues of the Capsid Protein Determine the Host Range of Feline Parvoviruses
The parvovirus capsid consists of 60 proteins molecules, in which VP1 is represented by approximately six molecules and VP2 by about 54 molecules. The spatial capsid structure of both feline panleucopenia virus and canine parvovirus has been determined by X-ray crystallography. This allows the exact assessment of the location of individual amino acids in the capsid, and the determination of which amino acids are located on the surface of the virus, and what contacts individual residues have with amino acids of the same protein or with those of neighbouring proteins.
Experiments with recombinant viruses which were derived from infectious DNA clones of feline and canine parvoviruses have revealed that three amino acids are important in determining the feline host range, namely lysine at position 80, asparagine at position 564 and alanine at position 568. They reside in a region of the capsid protein which mediates the interaction of four neighbouring molecules. In contrast, the canine host range is defined by the asparagine at position 93, alanine at position 103 and asparagine at position 323. The new virus types CPV-2a and CPV-2b differ from the originally isolated canine parvovirus by the amino acid residues at positions 87, 300 and 305 (Fig. 20.6). The differences between CPV-2a, CPV-2b and CPV-2c concern the amino acid residue at position 426, which is an aspartic acid, an asparagine or a glutamic acid. The mutated amino acids are also located in the region responsible for the intermolecular interaction, and apparently also determine the feline host range. The mechanisms that underlie this phenomenon are unclear. On the one hand, a change in the capsid stability may be associated with them, which is especially important during the release of the DNA genomes after infection of the cell. On the other hand, they might cause a slightly altered binding to the feline or canine transferrin receptors, thus determining the host spectrum.
1.6.2 Porcine Parvovirus
1.6.2.1 Epidemiology and Transmission
Porcine parvovirus causes clinical pictures in swine which are quite different from those induced by feline parvoviruses. The commonest manifestations of porcine parvovirus infections are fertility disorders and fetal infections, which are summarized by the term “stillbirth, mummification, embryonic death and infertility” (SMEDI). The infection of seronegative, receptive sows is economically important. The virus is transmitted by direct contact among animals or through contaminated equipment and stall accessories. It is excreted by infected asymptomatic adult pigs in the faeces. The high physical stability of the parvovirus capsid is of epidemiological importance also in this case. However, the long persistence of maternal antibodies in young animals is significant as well. These immunoglobulins can be detected up to the age of 6 months, and effectively protect animals from infection during this period. However, they can interfere with vaccination, thus hindering its success.
1.6.2.2 Clinical Features
Adult pigs do not become sick by porcine parvovirus infection. The symptoms affect exclusively the fetuses of acutely infected seronegative pregnant sows. Depending on the gestation stage at the time of infection, different consequences can be observed: in the first trimester of pregnancy (up to about the 40th day), the infection leads to fetal death and resorption of the fetus. In the sow, this is clinically manifested by a return to heat (oestrous cycle). In the second trimester (until the 70th day), infected fetuses also die; however, they are usually not aborted, but mummified, and remain in the sow, and are finally farrowed at the expected date. Infection in the last trimester of gestation eventually leads to the generation of an immune response in the fetus, resulting in elimination of the virus. These piglets are born healthy and virus-free, but they are serologically positive before intake of colostrum.
1.6.2.3 Pathogenesis
After oral incorporation, the virus proliferates in the regional lymphoid organs, and reaches virtually almost all organs of the animal during a first viraemia, even the uterus of pregnant females and the accessory glands of the boar. As a result of infection of the uterus, the fetuses are infected. However, all fetuses of a litter are rarely affected.
1.6.2.4 Immune Response and Diagnosis
Induction of an immune tolerance in the fetuses has not been described for porcine parvovirus infections. Virological diagnosis is not easy because isolation of the virus from mummified piglets is rarely successful. Therefore, porcine parvovirus infections can be better diagnosed by detection of the viral genome using PCR technology.
1.6.2.5 Control and Prophylaxis
Effective inactivated vaccines are available; hence, vaccination management is crucial to prevent porcine parvovirus infection. Therefore, all gilts have to be immunized before insemination. However, the already mentioned long persistence of maternal antibodies in young animals can impede successful vaccination in these cases.
1.6.3 Aleutian Mink Disease Virus
1.6.3.1 Epidemiology and Transmission
A completely different clinical picture is caused by Aleutian mink disease virus. It principally causes immune-complex-induced glomerulonephritis and arteritis in minks. The development of symptoms depends mainly on the virulence of the pathogen, but also on the genotype of the minks. Mink carrying the Aleutian genotype, which are characterized by a specially desired blue-grey coat colour (sapphire minks), have Chediak–Higashi syndrome, a malfunction of mononuclear phagocytes, which affects the induction of both the specific and the non-specific immune response. The virus is primarily transmitted by direct contact among animals. However, because of the high physical stability of all parvoviruses, Aleutian mink disease virus can also be spread by contaminated equipment, stables, animal keepers and animal breeders.
1.6.3.2 Clinical Features
Overall, Aleutian mink disease is a very complex process. The fulminant disease develops only after infection of Aleutian minks with virulent strains of Aleutian mink disease virus. In such cases, they cause the development of a hypergammaglobulinaemia and the formation and deposition of immune complexes in the kidneys and arteries. The resulting glomerulonephritis is invariably fatal. If non-Aleutian-genotype minks are infected, then the disease is milder, and, under certain conditions, the minks can even eliminate the pathogen.
In young pups of all genetic variants, the virus infects type II pneumocytes, which are responsible for the synthesis of surfactant factors, and thus for the surface tension in the alveoli. This provokes an acute interstitial pneumonia, which also has an invariably fatal outcome.
1.6.3.3 Pathogenesis
The pathogenesis of Aleutian mink disease virus infection is unclear; it is currently assumed that the virus induces a dysregulation of cytokine synthesis. This leads to an increased synthesis of IL-6 in virus-infected macrophages, which secrete the cytokine. These elevated IL-6 concentrations lead to a hypergammaglobulinaemia and to a proliferative glomerulonephritis, the main symptoms of Aleutian mink disease.
1.6.3.4 Immune Response and Diagnosis
Even though virus-specific antibodies are produced during Aleutian mink disease virus infection, they have no neutralizing effect. The diagnosis can be performed by detecting these antibodies.
1.6.3.5 Control and Prophylaxis
Vaccination is not possible; therefore, the disease is controlled by elimination of infected animals or infected flocks.
1.6.4 Animal Bocaviruses: Bovine Parvovirus and Canine Minute Virus
1.6.4.1 Epidemiology and Transmission
Bovine parvovirus has a global distribution. It was first isolated from calves with diarrhoea in 1961. Infected animals excrete the virus in high titres in the faeces, and the virus persists for long periods in the environment owing to the strong physical stability of the capsid. Canine minute virus is also known as canine parvovirus type 1 because it was isolated before canine parvovirus, which it is also referred to as canine parvovirus type 2. It is widespread, and its transmission routes are similar to those of bovine parvovirus.
1.6.4.2 Clinical Features
Gastrointestinal symptoms are the primary manifestation of bovine parvovirus and canine minute virus infections. Animals infected experimentally with bovine parvovirus develop, besides diarrhoea, also respiratory symptoms and especially abortions. Infections during the first two trimesters of pregnancy can lead to abortion of fetuses, mummifications, malformations of the central nervous system and stunted growth; contrarily, infections at later embryonic development stages are subclinical. The clinical picture of canine minute virus infections is different: in adult dogs, infections are usually either subclinical or accompanied by mild diarrhoea. Even in this case, the main manifestation is infection of the fetus. Depending on the stage of gestation, it results in abortion, resorption, mummification and live birth of weak pups. Myocarditis and malformations (anasarca) have also been described.
1.6.4.3 Pathogenesis
There are only a few data regarding the pathogenesis of animal bocavirus infections. Following oronasal infection, bovine parvovirus, like all autonomous parvoviruses, initially replicates in the nasopharyngeal region, leading to a viraemia. Thenceforth, the virus can be detected in epithelial cells of the small intestine (Lieberkühn glands or intestinal follicles) and immune cells.
1.6.4.4 Immune Response and Diagnosis
Haemagglutinating antibodies with virus-neutralizing activity are detectable within a few days after bovine parvovirus infections. Since infected fetuses produce antibodies as early as the 140th day of gestation, and the pathogens are widespread, many commercial bovine fetal calf serum batches contain antibodies against bovine parvoviruses. Since fetal calf sera are routinely used for cell culture applications, it is difficult to isolate and cultivate the virus from biopsy samples. Diagnostic detection of the viral genome can be easily performed by PCR.
So far, canine minute virus can be grown only on a specific canine cell line (Walter Reed canine cell line). The reason for this restriction is not known. The serological detection is performed by haemagglutination-inhibition tests using erythrocytes from rhesus monkeys or by detection of viral proteins by immunofluorescence in infected cell cultures.
1.6.4.5 Prophylaxis and Therapy
There are neither vaccines nor effective antiviral drugs.
2 Circoviruses and Anelloviruses

2.1 Classification and Characteristic Prototypes
The circoviruses and anelloviruses comprise viruses containing a single-stranded DNA genome. However, the DNA genome is not linear like in parvoviruses, but it forms a covalently closed circular molecule. They are possibly a transition to geminiviruses: these plant viruses have circular, partially twofold segmented single-stranded DNA genomes, and show a significant homology with some circoviruses.
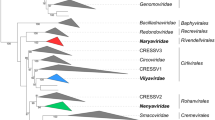
The family Circoviridae is divided into the genera Circovirus and Gyrovirus (Table 20.3). The genera Circovirus and Gyrovirus contain only animal pathogenic viruses. Whereas chicken anaemia virus, the characteristic member of the genus Gyrovirus, can be isolated only from chickens, circoviruses infect pigs – porcine circovirus 1 (PCV-1) and porcine circovirus 2 (PCV-2) – and various species of birds, e.g. parrots, canaries, finches, pigeons, ducks and geese. Sequence homologies have been found only in the genome sequences of beak and feather disease virus and porcine circovirus. None of them exhibit sequence similarity to chicken anaemia virus, which, not only for this reason, has been classified into the separate genus Gyrovirus. Despite their similar structure, circoviruses differ substantially in their respective replication strategy.
In 1997, torque teno virus was isolated from a Japanese patient with the initials TT who developed post-transfusion hepatitis, and was originally called TT virus. The name “torque teno” (Latin for “twisted ring”, “twisted cord”) is an allusion to the single-stranded, circular DNA genome of this virus, which is present in a pronounced secondary structure. Since the molecular characteristics are similar to those of circoviruses, torque teno viruses were classified initially into the family Circoviridae. Recently, a separate virus family Anelloviridae (Latin anellus, “ring”) was created for the multifarious types of torque teno virus. Twenty-nine types of torque teno virus have been isolated from humans and are classified into the genus Alphatorquevirus. They are related to the various types of human torque teno midi virus (genus Gammatorquevirus) and human torque teno mini virus (genus Betatorquevirus); related virus types can also be found in many animal species and have been classified into various additionally created genera (Table 20.4). All human virus types are present worldwide in most healthy people. So far, no illnesses have been associated with their infections. Perhaps, torque teno viruses, torque teno midi viruses and torque teno mini viruses are infectious agents which have adapted well to cohabit with their hosts without damaging them. Since the molecular characteristics of anelloviruses and circoviruses share many similarities, they are discussed together.
2.2 Structure
2.2.1 Virus Particle
The particles of circoviruses and anelloviruses are not surrounded by an envelope; they are probably composed of just one capsid protein (VP1), and have an icosahedral structure (Fig. 20.7). Their diameter ranges from 17 nm (porcine circovirus, beak and feather disease virus), to 22 nm (chicken anaemia virus) to 30 nm (torque teno virus). Therefore they are the smallest known viruses. Details of the particle structure are not known.
2.2.2 Genome Organization and Structure
The genomes of circoviruses and anelloviruses consist of a closed circular, single-stranded DNA with a length of 1,759 (porcine circovirus), 2,319 (chicken anaemia virus), 2,700–2,900 (torque teno mini virus) and 3,100–3,200 (torque teno midi virus) nucleotides. The genome of torque teno virus is significantly longer (3,500–3,900 nucleotides. The torque teno virus isolate TA278 comprises 3,825 nucleotides. The genome contains three (beak and feather disease virus, chicken anaemia virus) to four (porcine circovirus) open reading frames. Whereas all three reading frames of chicken anaemia virus are oriented in the same direction (Fig. 20.8a), they are arranged in opposite directions in the porcine circovirus and in beak and feather disease virus (Fig. 20.8b). Here, a hairpin loop of nine nucleotides is located at the origin of replication within the untranslated region, which does not occur in the genomes of gyroviruses and anelloviruses. These viruses follow an ambisense transcription strategy, which is usually only seen in arenaviruses and some bunyaviruses (Sects. 16.1 and 16.2). Like the animal pathogenic chicken anaemia virus, the genomes of torque teno viruses, torque teno midi viruses and torque teno mini viruses have negative polarity. Despite their divergence in length, their genome organization is similar. Their sequences comprise two large (ORF1, ORF2) and three small (ORF3, ORF4, ORF5) open reading frames, which partially overlap with each other and with ORF1. In torque teno viruses, a non-coding, untranslated region is located between ORF4 and ORF2, and encompasses approximately 1,200 nucleotides and has a high GC content. It probably contains the regulatory elements necessary for genome transcription and replication. Whereas the nucleotide sequence is conserved in animal pathogenic circoviruses, a high variability can be observed in torque teno viruses: Twenty-nine types of human torque teno virus have been identified so far, and their DNA sequences of ORF1 differ from each other by more than 20 %.
Genome organization of circoviruses. (a) Chicken anaemia virus. (b) Torque teno virus. The transcriptional regulatory sequences are located within the non-coding regions (at the top). In both viruses, only the genomic negative-sense DNA is transcribed, even after the postulated synthesis of the complementary DNA strand. The genome of chicken anaemia virus encodes up to three proteins, whose functions have hardly been explored. The same applies for torque teno virus. Under the assumption of alternative splicing, the sequences of ORF2 and/or ORF3 may be fused with those of ORF4 and ORF5. However, the corresponding non-structural protein versions have not yet been experimentally detected during infection, or such findings have not been reproduced
2.3 Viral Proteins
2.3.1 Structural Proteins
The capsids are made of the VP1 protein. The largest open reading frame, ORF2, of chicken anaemia virus encodes a precursor protein from which the capsid protein VP1 is cleaved. In contrast, the capsid protein of porcine circovirus is encoded by ORF2. In ORF1 of torque teno viruses it is believed that the capsid protein is encoded together with a polypeptide that is implicated in the replication of the viral DNA. Although other proteins are also packaged into the virion, their function and origin are unknown. Table 20.5 provides an overview of the proteins of circoviruses and anelloviruses.
2.3.2 Non-Structural Proteins
The largest reading frame (ORF1) of porcine circovirus is responsible for the synthesis of the Rep and Rep’ proteins. These are important for replication of the viral genome by a rolling-circle mechanism. By use of alternative splice sites, a shortened version of the Rep’ protein is synthesized, and this is also involved in replication of the single-stranded DNA genome. The function of the protein encoded by ORF3 of porcine circovirus is unknown.
ORF2 of both chicken anaemia virus and torque teno virus probably also encodes a non-structural protein involved in viral genome replication; Rep-protein-like functions have been attributed to this gene product. Considering genotype-specific differences, it has a length ranging from 120 to over 200 amino acids. In vitro, it has been shown that the proteins encoded in ORF2 of chicken anaemia virus and of torque teno viruses possess the activity of a dual-specificity protein phosphatase. In addition to phosphotyrosines and phosphoserines, this protein tyrosine phosphatase subfamily also dephosphorylates non-protein substrates. In torque teno viruses there may be additional variants of this non-structural protein which are generated by using alternative splice donor and acceptor sites. This alternative splicing may lead to fusion of ORF4 and ORF5 with ORF2.
Chicken anaemia virus directs the synthesis of a 13-kDa non-structural protein, apoptin (VP3). It is probably encoded by ORF3 (Fig. 20.8a), and is able to trigger a p53-independent apoptosis mechanism in a variety of transformed cells or tumour cells. The simultaneous synthesis of the cellular Bcl-2 protein seems to accelerate this process. The physiological function of apoptin is unknown. It is possible that the ORF3 gene product of torque teno virus has a similar function. Depending on the virus isolate, this protein comprises 57–105 amino acids. Expression of ORF3 induces apoptosis in hepatocellular carcinoma cell lines.
2.4 Replication
Torque teno viruses, torque teno midi viruses and torque teno mini viruses cannot be propagated in cell culture systems. There are virtually no data regarding their genome replication. It is believed to occur in the nucleus, presumably by a rolling-circle mechanism involving DNA-dependent DNA polymerase encoded by the host cell. A GC-rich sequence is located in the non-coding genome region, and is believed to be involved in regulating transcription and replication. Three transcripts have been detected in cells transfected with the genomic DNA of torque teno virus; there lengths are 3,000, 1,200 and 1,000 nucleotides.
Only little is known about the replication mechanism of animal pathogenic circoviruses. Viral genome replication occurs in the nucleus of infected cells, whereby circoviruses, like parvoviruses (Sect. 20.1), are dependent on the presence of cellular proteins, which are synthesized during the S phase of the cell cycle, e.g. DNA polymerases. Presumably, the single-stranded DNA genome is initially converted into double-stranded DNA. Chicken anaemia virus synthesizes a probably polycistronic transcript of 2,000 nucleotides in length. In contrast, porcine circovirus generates two mRNAs from the negative DNA strand (i.e. the genomic DNA strand) and a spliced mRNA from ORF1 by using the positive DNA strand as a template. There is evidence that the viral genome is replicated by a rolling-circle mechanism. The origin of replication of porcine circovirus is located in the untranslated region of the viral genome, whereby a hairpin loop forms the initiation structure. The genome of chicken anaemia virus lacks such a structure. Assembly of the various components into new virus particles occurs in the cytoplasm. They are probably released by cell lysis as a result of apoptotic processes.
Apoptin: A Novel Antitumour Therapeutic Approach?
The apoptin of chicken anaemia virus is able to induce apoptosis exclusively in tumour cells. It forms multimeric complexes that bind to DNA. In tumour cells, apoptin is present as a phosphoprotein mainly in the nucleus, whereas it is present as an unphosphorylated protein in the cytoplasm of normal untransformed cells, where it is rapidly degraded. The activity of apoptin, its modification and its nuclear transport can be induced temporarily by enabling the cells to express the T antigen of simian virus 40 (Sect. 19.2). In tumour cells, apoptin seems to identify “survival signals” such as DEDAF, Nur77, NMI, Hippi and APC1, leading to the development of its activity. In transgenic mice and in animal models, apoptin has proven to have safe and effective antitumour activity. Whether it can be used as a general apoptotic agent in tumour cells remains to be elucidated.
2.5 Human Pathogenic Anelloviruses
2.5.1 Torque Teno Virus
2.5.1.1 Epidemiology and Transmission
Torque teno virus was first isolated from a patient with post-transfusion hepatitis in 1997. Therefore, it was initially considered as a possible member of non-A, non-B hepatitis viruses. However, it has turned out that torque teno viruses, like torque teno mini viruses and torque teno midi viruses, are widespread in the human population throughout the world, and their infections do not appear to be associated with diseases. Both viruses persist in more than 90 % of examined subjects, and their genomes are found in blood, saliva and various tissues. The virus isolates are genetically highly heterogeneous, whereby the highly variable regions are concentrated in the central regions of ORF1. The protein domains encoded by this region exhibit differences of up to 70 % at the amino acid sequence level. To date, more than 20 genotypes have been discovered, and these are classified into five groups: these include SANBAN and SEN viruses. Phylogenetic analyses suggest that frequent genetic recombination additionally contributes to variability. Up to five different genotypes of torque teno virus and torque teno mini virus have been found concurrently in one individual. Torque teno virus sequences which are not distinguishable from human isolates have also been detected in the blood of different primates (chimpanzees, gorillas) and other animals (pigs, cattle, sheep and chickens),. Therefore, it is assumed that these viruses are transmitted as zoonotic pathogens from domestic animals to humans.
In humans and animals, these viruses establish persistent infections, and the viruses are excreted through saliva and faeces. Viral DNA has been found in blood and semen, as well as in lachrymal fluid and bile. Besides the transmission by droplet infection and smear infections, the physically very stable virus can also be transmitted by blood and blood products. This notion is additionally supported by the disproportionately high prevalence of torque teno virus in haemophilia and transplant patients as well as in drug addicts who use intravenous drugs.
2.5.1.2 Clinical Features
According to current knowledge, torque teno virus, torque teno midi and torque teno mini virus infections are asymptomatic. There is some evidence that increased genome copy numbers can be detected in patients with severe idiopathic myopathies, cancer and lupus erythematosus. However, it is likely that the increased viral DNA load is a consequence of immunosuppression and is not the cause. Signs of viral replication have been observed in children with acute respiratory infections.
2.5.1.3 Pathogenesis
Little is known about the pathogenesis of infection. The target cells of torque teno virus are unknown, and there is no cell culture system for cultivation of the virus in vitro. The viruses seem to infect mononuclear cells of the peripheral blood as well as certain cells in the liver and bone marrow. Intermediates of genome replication can be detected in the liver by PCR. In addition, this organ exhibits higher amounts of viral genomes than other tissues. However, an increase in transaminase levels or histological liver alterations have not been documented.
2.5.1.4 Immune Response and Diagnosis
Nothing is known about immunological responses to viral proteins. However, it has been reported that acutely and chronically infected individuals may spontaneously excrete torque teno virus. There is neither an ELISA nor a Western blot test for detection of the virus. The diagnostic detection of infection is performed using PCR, whereby the high genetic variability of the virus has to be taken into account.
2.5.1.5 Therapy and Prophylaxis
Owing to the asymptomatic course of infection, there is no need for the development of vaccines or antiviral drugs. Blood donors are not tested.
2.6 Animal Pathogenic Circoviruses
2.6.1 Chicken Anaemia Virus
2.6.1.1 Epidemiology and Transmission
Chicken anaemia virus has a worldwide distribution. It causes an economically important infectious disease in chickens. After experimental infection, it has been found that chickens excrete the virus for up to 30 days in faeces and saliva. The infection of adult animals is usually asymptomatic, although co-infections with other viruses (retroviruses, herpesviruses) may cause severe clinical courses. The classic clinical picture is found in the infection of 1-day-old chicks, which can be infected by virus-contaminated bedding or direct contact. Chicks from seropositive hens are infected, but the maternal antibodies usually prevent a disease. A vertical transmission is also possible when laying hens are in the viraemic phase. This explains why even specific-pathogen-free chickens initially excrete chicken anaemia virus.
2.6.1.2 Clinical Features
The infection leads to immunosuppression in 1-day-old chickens, and is characterized by anorexia, lethargy, anaemia and systemic bleeding. The mortality rate can be up to 50 %.
2.6.1.3 Pathogenesis
The virus infects the erythroid and lymphoid progenitor cells in the bone marrow and thymus of chickens. B lymphocytes are not infected by chicken anaemia virus. The infection results in destruction of erythroid progenitor cells, whereby the animals develop severe anaemia, thrombocytopenia and granulocytopenia. The destruction of T progenitor cells in the cortex of the thymus may be associated with the apoptotic effect of the VP3 protein (apoptin) and determines the depletion of mature cytotoxic T lymphocytes and T-helper cells, which leads to severe immunosuppression. Pathologically, the atrophy of all lymphoid organs, including bone marrow, is striking.
2.6.1.4 Immune Response and Diagnosis
Diagnosis can be performed by isolation of the virus in susceptible chicken cell lines. Diagnosis is verified by immunoflorescence or PCR. A past infection confers protective immunity.
2.6.1.5 Control and Prophylaxis
Oral vaccination based on live attenuated chicken anaemia virus is available and used in many countries.
Beak and Feather Disease Virus
Another scarcely understood avian circovirus infection affects parrot-like birds such as cockatoos, parrots and parakeets in Australia and more recently also in South America. The disease is characterized by a massive immunosuppression, feather loss, regrowth of deformed feathers in terms of size and structure, and less frequent disorders of the horn growth on the beak. The virus is transmitted by various secretions and excretions from infected birds. The disease is controlled by eliminating infected animals and strict hygiene measures.
2.6.2 Porcine Circoviruses
2.6.2.1 Epidemiology and Transmission
In 1974, a small non-enveloped virus with a circular single-stranded DNA genome was described as a contaminant of a porcine kidney cell line. It was named porcine circovirus (PCV-1). Further virological and serological studies have revealed that this virus is widespread in the swine population; however, it could not be associated with any disease. Around 1995, another circovirus was occasionally found in swine, and has been found more frequently since that time. It causes a non-specific disease that is called porcine postweaning multisystemic wasting syndrome (PMWS). The genome of this ubiquitous virus (PCV-2) is only 70 % homologous with that of PCV-1 from the 1970s. The host range seems to be restricted to swine. In recent years, extensive investigations have revealed that PCV-2 alone is not able to cause diseases, but other factors (including housing conditions, co-infection with porcine parvovirus; Sect. 20.1.6) must be involved in the development of PCV-2-associated diseases.
Circoviruses are physically very stable, and are excreted in the faeces of infected animals. Infection occurs through direct oral contact or via contaminated bedding or equipment. Vertical transmission from the mother to the piglets is possible. Whether piglets born with a persistent infection exhibit an immune tolerance is still unclear.
2.6.2.2 Clinical Features
Infections with PCV-2 cause two main diseases: PMWS and porcine dermatitis and nephropathy syndrome. The former is characterized by runtiness of individual piglets of a litter. The animals frequently show interstitial pneumonia and lymphadenopathy, but rarely diarrhoea or jaundice. Porcine dermatitis and nephropathy syndrome is a dramatic disease that resembles classical swine fever. The principal symptoms are petechial haemorrhages on the skin, which can develop into necrotizing lesions. The adrenal cortex also shows petechial haemorrhages and interstitial nephritis.
2.6.2.3 Pathogenesis
The pathogenesis of this disease is largely unknown. The virus is regularly detected in macrophages, and PCV-2 infection may lead to a generalized immunosuppression, in which just a few pathogens may cause an apparent disease. In particular, co-infections with porcine parvovirus (Sect. 20.1.6) or porcine reproductive and respiratory syndrome virus (an arterivirus; Sect. 14.5.5) are a matter of discussion.
2.6.2.4 Immune Response and Diagnosis
The infection usually leads to seroconversion and elimination of the virus. Infections with PCV-2 can be verified by detecting the viral genome in altered tissues. This can be performed by in situ hybridization or by detecting the viral DNA by PCR. PCV-1 infections can be reliably distinguished from PCV-2 infections by selection of appropriate primers.
2.6.2.5 Control and Prophylaxis
Recently, recombinant vaccines which are composed of VP1 proteins that are produced in different expression systems have become available. Although there were obvious doubts about their real effectiveness because of the multifactorial origin of the disease, the experience with the vaccines shows, however, that they are extremely effective. Immunization of piglets in experimental infections has revealed a significantly reduced viraemic phase and weakened PMWS-associated disease symptoms. However, particularly noteworthy is that both the frequency and the severity of other infections are also weakened and the animals exhibit better growth and increase in weight. In addition to vaccination, hygiene measures are of great importance for preventing viral transmission in livestock.
Further Reading
Adair BM (2000) Immunopathogenesis of chicken anemia virus infection. Dev Comp Immunol 24:247–255
Allan GM, Ellis JA (2000) Porcine circoviruses: a review. J Vet Diagn Invest 12:3–14
Allander T, Tammi MT, Eriksson M, Bjerkner A, Tiveljung-Lindell A, Andersson B (2005) Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci USA 102:12891–12896
Anderson LJ, Hurwitz ES (1988) Human parvovirus B19 and pregnancy. Clin Perinatol 15:273–286
Arthur JL, Higgins GD, Davidson GP, Givney RC, Ratcliff RM (2009) A novel bocavirus associated with acute gastroenteritis in Australian children. PLoS Pathog 5:e1000391
Backendorf C, Visser AE, de Boer AG, Zimmerman R, Visser M, Voskamp P, Zhang YH, Noteborn M (2008) Apoptin: therapeutic potential of an early sensor of carcinogenic transformation. Annu Rev Pharmacol Toxicol 48:143–169
Bashir T, Hörlein R, Rommelaere J, Willwand K (2000) Cyclin A activates the DNA polymerase δ-dependent elongation machinery in vitro: a parvovirus DNA replication model. Proc Natl Acad Sci USA 97:5522–5527
Batchu RB, Shammas MA, Wang JY, Munshi NC (2001) Dual level inhibition of E2F-1 activity by adeno-associated virus Rep78. J Biol Chem 276:24315–24322
Bendinelli M, Pistello M, Maggi F, Fornai C, Freer G, Vatteroni ML (2001) Molecular properties, biology, and clinical implications of TT virus, a recently identified widespread infectious agent of humans. Clin Microbiol Rev 14:98–113
Biagini P (2004) Human circoviruses. Vet Microbiol 98:95–101
Brown KE, Jonathan MD, Young NS (1993) Erythrocyte P-antigen: cellular receptor for parvovirus B19. Science 262:114–117
Brown KE, Young NS, Liu JM (1994) Molecular, cellular and clinical aspects of parvovirus B19 infection. Crit Rev Oncol Hematol 16:1–31
Chapman MS, Rossman MG (1993) Structure, sequence and function correlations among parvoviruses. Virology 194:491–508
Corbau R, Duverger V, Rommelaere J, Nüesch JP (2000) Regulation of MVM NS1 by protein kinase C: impact of mutagenesis at consensus phosphorylation sites on replicative functions and cytopathic effects. Virology 278:151–167
Cotmore SF, Tattersall P (2007) Parvoviral host range and cell entry mechanisms. Adv Virus Res 70:183–232
Cotmore SF, Gottlieb RL, Tattersall P (2007) Replication initiator protein NS1 of the parvovirus minute virus of mice binds to modular divergent sites distributed throughout duplex viral DNA. J Virol 81:13015–13027
Danen-Van Oorschot AA, van der Eb AJ, Noteborn MH (1999) BCL-2 stimulates apoptin-induced apoptosis. Adv Exp Med Biol 457:245–249
Davidson I, Shulman LM (2008) Unraveling the puzzle of human anellovirus infections by comparison with avian infections with the chicken anemia virus. Virus Res 137:1–15
Dorsch S, Liebisch G, Kaufmann B, von Landenberg P, Hoffmann JH, Drobnik W, Modrow S (2002) The VP1-unique region of parvovirus B19 and its constituent phospholipase A2-like activity. J Virol 76:2014–2018
Fryer JF, Delwart E, Bernardin F, Tuke PW, Lukashov VV, Baylis SA (2007) Analysis of two human parvovirus PARV4 genotypes identified in human plasma for fractionation. J Gen Virol 88:2162–2167
Gergely P Jr, Perl A, Poór G (2006) Possible pathogenic nature of the recently discovered TT virus: does it play a role in autoimmune rheumatic diseases? Autoimmun Rev 6:5–9
Hino S, Miyata H (2007) Torque teno virus (TTV): current status. Rev Med Virol 17:45–57
Hsu TC, Wu WJ, Chen MC, Tsay GJ (2004) Human parvovirus B19 non-structural protein (NS1) induces apoptosis through mitochondria cell death pathway in COS-7 cells. Scand J Infect Dis 36:570–577
Kakkola L, Hedman K, Qiu J, Pintel D, Söderlund-Venermo M (2009) Replication of and protein synthesis by TT viruses. Curr Top Microbiol Immunol 331:53–64
Kantola K, Hedman L, Allander T, Jartti T, Lehtinen P, Ruuskanen O, Hedman K, Söderlund-Venermo M (2008) Serodiagnosis of human bocavirus infection. Clin Infect Dis 46:540–546
Karalar L, Lindner J, Schimanski S, Kertai M, Segerer H, Modrow S (2010) Prevalence and clinical aspects of human bocavirus infection in children. Clin Microbiol Infect 16:633–639
Kaufmann B, Chipman PR, Kostyuchenko VA, Modrow S, Rossmann MG (2008) Visualization of the externalized VP2 N termini of infectious human parvovirus B19. J Virol 82:7306–7312
King JA, Dubielzig R, Grimm D, Kleinschmidt JA (2001) DNA helicase-mediated packaging of adeno-associated virus type 2 genomes into preformed capsids. EMBO J 20:3282–3291
Kleinschmidt JA, Mohler M, Weindler FW, Heilbronn R (1995) Sequence elements of the adeno-associated virus rep gene required for suppression of herpes-simplex-virus-induced DNA amplification. Virology 206:254–262
Kooistra K, Zhang YH, Henriquez NV, Weiss B, Mumberg D, Noteborn MH (2004) TT virus-derived apoptosis-inducing protein induces apoptosis preferentially in hepatocellular carcinoma-derived cells. J Gen Virol 85:1445–1450
Kotin RM, Linden RM, Berns KI (1992) Characterization of the preferred site on human chromosome 19q for integration of adeno-associated virus DNA by non-homologous recombination. EMBO J 11:5071–5078
Leary TP, Erker JC, Chalmers ML, Desai SM, Mushahwar IK (1999) Improved detection systems for TT virus reveal high prevalence in humans, non-human primates and farm animals. J Gen Virol 80:2115–2120
Lehmann HW, Knöll A, Küster RM, Modrow S (2003a) Frequent infection with a viral pathogen, parvovirus B19, in rheumatic diseases of childhood. Arthritis Rheum 48:1631–1638
Lehmann HW, von Landenberg P, Modrow S (2003b) Parvovirus B19 infection and autoimmune disease. Autoimmun Rev 2:218–223
Lin C-L, Kyono W, Tongson J, Chua PK, Easa D, Yanagihara R, Nerurkar VR (2000) Fecal excretion of a novel human circovirus, TT virus in healthy children. Clin Diagn Lab Immunol 7:960–963
Lin F, Guan W, Cheng F, Yang N, Pintel D, Qiu J (2008) ELISAs using human bocavirus VP2 virus-like particles for detection of antibodies against HBoV. J Virol Methods 149:110–117
Lindner J, Modrow S (2008) Human bocavirus – a novel parvovirus to infect humans. Intervirology 51:116–122
Lu J, Zhi N, Wong S, Brown KE (2006) Activation of synoviocytes by the secreted phospholipase A2 motif in the VP1-unique region of parvovirus B19 minor capsid protein. J Infect Dis 193:582–590
Mankertz A, Hillenbrand B (2001) Replication of porcine circovirus type 1 requires two proteins encoded by the viral rep gene. Virology 279:429–438
Mankertz A, Persson F, Mankertz J, Blaess G, Buhk HJ (1997) Mapping and characterization of the origin of DNA replication of porcine circovirus. J Virol 71:2562–2566
Manteufel J, Truyen U (2008) Animal bocaviruses: a brief review. Intervirology 51:328–334
Miller MM, Schat KA (2004) Chicken infectious anemia virus: an example of the ultimate host-parasite relationship. Avian Dis 48:734–745
Miyata H, Tsunoda H, Kazi A, Yamada A, Ali Khan M, Murakami J, Kamahora T, Shiraki K, Hino S (1999) Identification of a novel GC-rich 113 nucleotide region to complete the circular, single-stranded DNA genome of TT virus, the first human circovirus. J Virol 73:3582–3586
Morey A, Ferguson D, Fleming KA (1993) Ultrastructural features of fetal erythroid precursors infected with parvovirus B19 in vitro: evidence of cell death by apoptosis. J Pathol 169:213–220
Naides SJ, Karetnyi YV, Cooling LLW, Mark RS, Langnas AN (1996) Human parvovirus B19 infection and hepatitis. Lancet 347:1563–1564
Nakashima A, Morita E, Saito S, Sugamura K (2004) Human Parvovirus B19 nonstructural protein transactivates the p21/WAF1 through Sp1. Virology 329:493–504
Naoumov NV (2000) TT virus – highly prevalent, but still in search of a disease. J Hepatol 33:157–159
Nishizawa T, Okamoto H, Konishi K, Yoshizawa H, Miyakawa Y, Mayumi M (1997) A novel DNA virus (TTV) associated with elevated transaminase levels in posttransfusion hepatitis of unknown etiology. Biochem Biophys Res Commun 241:92–97
Norja P, Hokynar K, Aaltonen LM, Chen R, Ranki A, Partio EK, Kiviluoto O, Davidkin I, Leivo T, Eis-Hübinger AM, Schneider B, Fischer HP, Tolba R, Vapalahti O, Vaheri A, Söderlund-Venermo M, Hedman K (2006) Bioportfolio: lifelong persistence of variant and prototypic erythrovirus DNA genomes in human tissue. Proc Natl Acad Sci USA 103:7450–7453
Nüesch JP, Rommelaere J (2007) A viral adaptor protein modulating casein kinase II activity induces cytopathic effects in permissive cells. Proc Natl Acad Sci USA 104:12482–12487
Okamoto H, Fukuda M, Tawara M, Nishizawa T, Itoh Y, Hayasaka I, Tsuda F, Tanaka T, Miyakawa Y, Mayumi M (2000) Species-specific TT viruses and cross-species infection in non-human primates. J Virol 74:1132–1139
Parker JS, Murphy WJ, Wang D, O’Brien SJ, Parrish CR (2001) Canine and feline parvoviruses can use human or feline transferrin receptors to bind, enter, and infect cells. J Virol 75:3896–3902
Parrish CR, Aquadro CF, Strassheim ML, Evermann JF, Sgro JY, Mohammed HO (1991) Rapid antigenic-type replacement and DNA sequence evolution of canine parvovirus. J Virol 65:6544–6552
Qing K, Mah C, Hansen J, Zhou S, Dwarki V, Srivastava A (1999) Human fibroblast growth factor receptor 1 is a co-receptor for infection by adeno-associated virus 2. Nat Med 5:71–77
Qiu J, Brown KE (1999) A 110-kDa nuclear shuttle protein, nucleolin, specifically binds to adeno-associated virus type 2 (AAV-2) capsid. Virology 257:373–382
Röhrer C, Gärtner B, Sauerbrei A, Böhm S, Hottenträger B, Raab U, Thierfelder W, Wutzler P, Modrow S (2008) Seroprevalence of parvovirus B19 in the German population. Epidemiol Infect 16:1–12
Rovira A, Balasch M, Segales J, Garcia L, Plana-Duran J, Rosell C, Ellerbrok H, Mankertz A, Domingo M (2002) Experimental inoculation of conventional pigs with porcine reproductive and respiratory syndrome virus and porcine circovirus 2. J Virol 76:3232–3239
Saudan P, Vlach J, Beard P (2000) Inhibition of S-phase progression by adeno-associated virus Rep78 protein is mediated by hypophosphorylated pRb. EMBO J 19:4351–4361
Schmidt M, Afione S, Kotin RM (2000) Adeno-associated virus type 2 Rep78 induces apoptosis through caspase activation independently of p53. J Virol 74:9441–9450
Shackelton LA, Parrish CR, Truyen U, Holmes EC (2005) High rate of viral evolution associated with the emergence of carnivore parvovirus. Proc Natl Acad Sci USA 102:379–384
Smith RH, Kotin RM (2000) An adeno-associated virus (AAV) initiator protein, Rep78, catalyzes the cleavage and ligation of single-strand AAV ori DNA. J Virol 74:3122–3129
Steinel A, Parrish CR, Bloom ME, Truyen U (2001) Parvovirus infections in wild carnivores. J Wildl Dis 37:594–607
Summerford C, Bartlett JS, Samulski RJ (1999) AlphaVbeta5 integrin: a co-receptor for adeno-associated virus type 2 infection. Nat Med 5:78–82
Takahashi K, Hijikata M, Samokhvalov EI, Mishiro S (2000) Full or near full length nucleotide sequences of TT virus variants (types SANBAN and YONBAN) and the TT virus-like mini virus. Intervirology 43:119–123
Takasawa N, Munakata Y, Ishii KK, Takahashi Y, Takahashi M, Fu Y, Ishii T, Fujii H, Saito T, Takano H, Noda T, Suzuki M, Nose M, Zolla-Pazner S, Sasaki T (2004) Human parvovirus B19 transgenic mice become susceptible to polyarthritis. J Immunol 173:4675–4683
Thacker TC, Johnson FB (1998) Binding of bovine parvovirus to erythrocyte membrane sialylglycoproteins. J Gen Virol 79:2163–2169
Todd D, McNulty MS, Adair BM, Allan GM (2001) Animal circoviruses. Adv Virus Res 57:1–70
Truyen U, Gruenberg A, Chang SF, Obermaier B, Veijalainen P, Parrish CR (1995) Evolution of the feline-subgroup parvoviruses and the control of canine host range in vivo. J Virol 69:4702–4710
Truyen U, Everman JF, Vieler E, Parrish CR (1996) Evolution of canine parvovirus involved loss and gain of feline host range. Virology 215:186–189
Tsao J, Chapman MS, Agbandja M, Keller W, Smith K, Wu H, Luo M, Smith TM, Rossman M, Compans RW, Parrish CR (1991) The three-dimensional structure of canine parvovirus and its functional implications. Science 251:1456–1464
Tzang BS, Lee YJ, Yang TP, Tsay GJ, Shi JY, Tsai CC, Hsu TC (2007) Induction of antiphospholipid antibodies and antiphospholipid syndrome-like autoimmunity in naive mice with antibody against human parvovirus B19 VP1 unique region protein. Clin Chim Acta 382:31–36
Verschoor EJ, Langenhuijzen S, Heeney JL (1999) TT viruses (TTV) of non-human primates and their relationship to the human TTV genotypes. J Gen Virol 80:2491–2499
von Landenberg P, Lehmann HW, Knöll A, Dorsch S, Modrow S (2003) Antiphospholipid antibodies in pediatric and adult patients with rheumatic disease are associated with parvovirus B19 infection. Arthritis Rheum 48:1939–1947
von Poblotzki A, Hemauer A, Gigler A, Puchhammer-Stöcke E, Heinz F-X, Pont J, Laczika K, Wolf H, Modrow S (1995) Antibodies to the nonstructural protein of parvovirus B19 in persistently infected patients: implications for pathogenesis. J Infect Dis 172:1356–1359
Walters RA, Yi SMP, Keshavjee S, Brown KE, Welsh MJ, Chorioni JA, Zabner J (2001) Binding of adeno-associated virus type 5 to 2,3-linked sialic acid is required for gene transfer. J Biol Chem 276:20610–20616
Weger S, Wendland M, Kleinschmidt JA, Heilbronn R (1999) The adeno-associated virus type 2 regulatory proteins rep78 and rep68 interact with the transcriptional coactivator PC4. J Virol 73:260–269
Wonderling RS, Kyostio SR, Owens RA (1995) A maltose-binding protein/adeno-associated virus rep68 fusion protein has DNA-RNA helicase and ATPase activity. J Virol 69:3542–3548
Young SM Jr, McCarty DM, Degtyareva N, Samulski RJ (2000) Roles of adeno-associated virus Rep protein and human chromosome 19 in site-specific recombination. J Virol 74:3953–3966
Zhi N, Mills IP, Lu J, Wong S, Filippone C, Brown KE (2006) Molecular and functional analyses of a human parvovirus B19 infectious clone demonstrates essential roles for NS1, VP1, and the 11-kilodalton protein in virus replication and infectivity. J Virol 80:5941–5950
zur Hausen H, de Villiers EM (2009) TT viruses: oncogenic or tumorsuppressive properties? Curr Top Microbiol Immunol 331:109–116
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Copyright information
© 2013 Springer-Verlag Berlin Heidelberg
About this entry
Cite this entry
Modrow, S., Falke, D., Truyen, U., Schätzl, H. (2013). Viruses with a Single-Stranded DNA Genome. In: Molecular Virology. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-20718-1_20
Download citation
DOI: https://doi.org/10.1007/978-3-642-20718-1_20
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-20717-4
Online ISBN: 978-3-642-20718-1
eBook Packages: Biomedical and Life SciencesReference Module Biomedical and Life Sciences