Abstract
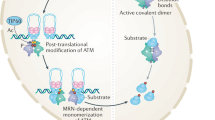
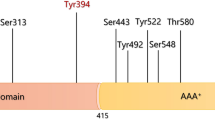
ATM (ataxia-telangiectasia mutated) protein kinase is a key regulator of cellular responses to DNA damage and oxidative stress. DNA damage triggers complex cascade of signaling events leading to numerous posttranslational modification on multitude of proteins. Understanding the regulation of ATM kinase is therefore critical not only for understanding the human genetic disorder ataxia-telangiectasia and potential treatment strategies, but essential for deciphering physiological responses of cells to stress. These responses play an important role in carcinogenesis, neurodegeneration, and aging. We focus here on the identification of DNA damage inducible ATM phosphorylation sites to understand the importance of autophosphorylation in the mechanism of ATM kinase activation. We demonstrate the utility of using immunoprecipitated ATM in quantitative LC-MS/MS workflow with stable isotope dimethyl labeling of ATM peptides for identification of phosphorylation sites.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Johnson LN, Noble ME, Owen DJ (1996) Active and inactive protein kinases: structural basis for regulation. Cell 85(2):149–158
Durocher D, Jackson SP (2001) DNA-PK, ATM and ATR as sensors of DNA damage: variations on a theme? Curr Opin Cell Biol 13(2):225–231
Kim ST, Lim DS, Canman CE, Kastan MB (1999) Substrate specificities and identification of putative substrates of ATM kinase family members. J Biol Chem 274(53):37538–37543
O'Neill T, Dwyer AJ, Ziv Y, Chan DW, Lees-Miller SP, Abraham RH, Lai JH, Hill D, Shiloh Y, Cantley LC, Rathbun GA (2000) Utilization of oriented peptide libraries to identify substrate motifs selected by ATM. J Biol Chem 275(30):22719–22727. doi:10.1074/jbc.M001002200
Kozlov S, Gueven N, Keating K, Ramsay J, Lavin MF (2003) ATP activates ataxia-telangiectasia mutated (ATM) in vitro. Importance of autophosphorylation. J Biol Chem 278(11):9309–9317
Bakkenist CJ, Kastan MB (2003) DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421(6922):499–506. doi:10.1038/nature01368
Sun Y, Jiang X, Chen S, Fernandes N, Price BD (2005) A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci U S A 102(37):13182–13187. doi:10.1073/pnas.0504211102
Gupta A, Sharma GG, Young CS, Agarwal M, Smith ER, Paull TT, Lucchesi JC, Khanna KK, Ludwig T, Pandita TK (2005) Involvement of human MOF in ATM function. Mol Cell Biol 25(12):5292–5305 . doi:10.1128/MCB.25.12.5292-5305.2005doi:25/12/5292 [pii]
Ali A, Zhang J, Bao S, Liu I, Otterness D, Dean NM, Abraham RT, Wang XF (2004) Requirement of protein phosphatase 5 in DNA-damage-induced ATM activation. Genes Dev 18(3):249–254. doi:10.1101/gad.1176004
Goodarzi AA, Jonnalagadda JC, Douglas P, Young D, Ye R, Moorhead GB, Lees-Miller SP, Khanna KK (2004) Autophosphorylation of ataxia-telangiectasia mutated is regulated by protein phosphatase 2A. EMBO J 23(22):4451–4461. doi:10.1038/sj.emboj.7600455
Shreeram S, Demidov ON, Hee WK, Yamaguchi H, Onishi N, Kek C, Timofeev ON, Dudgeon C, Fornace AJ, Anderson CW, Minami Y, Appella E, Bulavin DV (2006) Wip1 phosphatase modulates ATM-dependent signaling pathways. Mol Cell 23(5):757–764. doi:10.1016/j.molcel.2006.07.010
Uziel T, Lerenthal Y, Moyal L, Andegeko Y, Mittelman L, Shiloh Y (2003) Requirement of the MRN complex for ATM activation by DNA damage. EMBO J 22(20):5612–5621. doi:10.1093/emboj/cdg541
Lee JH, Paull TT (2005) ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science 308(5721):551–554. doi:10.1126/science.1108297
Pellegrini M, Celeste A, Difilippantonio S, Guo R, Wang W, Feigenbaum L, Nussenzweig A (2006) Autophosphorylation at serine 1987 is dispensable for murine Atm activation in vivo. Nature 443(7108):222–225. doi:10.1038/nature05112
Lavin MF, Kozlov S (2007) ATM activation and DNA damage response. Cell Cycle 6(8):931–942
Guo Z, Kozlov S, Lavin MF, Person MD, Paull TT (2010) ATM activation by oxidative stress. Science 330(6003):517–521. doi:10.1126/science.1192912
Paull TT (2015) Mechanisms of ATM activation. Annu Rev Biochem 84:711–738. doi:10.1146/annurev-biochem-060614-034335
Nagahara H, Latek RR, Ezhevsky SA, Dowdy SF (1999) 2-D phosphopeptide mapping. Methods Mol Biol 112:271–279
Boyle WJ, van der Geer P, Hunter T (1991) Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol 201:110–149
Kozlov SV, Graham ME, Peng C, Chen P, Robinson PJ, Lavin MF (2006) Involvement of novel autophosphorylation sites in ATM activation. EMBO J 25(15):3504–3514. doi:10.1038/sj.emboj.7601231
Kozlov SV, Graham ME, Jakob B, Tobias F, Kijas AW, Tanuji M, Chen P, Robinson PJ, Taucher-Scholz G, Suzuki K, So S, Chen D, Lavin MF (2011) Autophosphorylation and ATM activation: additional sites add to the complexity. J Biol Chem 286(11):9107–9119. doi:10.1074/jbc.M110.204065
Jenkins CW, Xiong Y (1996) Immunoprecipitation and immunoblotting in cell cycle studies. In: Pagano M (ed) Cell cycle-materials and methods. Springer lab manuals. Springer, New York, pp 250–264. doi:10.1007/978-3-642-57783-3
Sarkaria JN, Busby EC, Tibbetts RS, Roos P, Taya Y, Karnitz LM, Abraham RT (1999) Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res 59(17):4375–4382
Neuhoff V, Stamm R, Eibl H (1985) Clear background and highly sensitive protein staining with Coomassie Blue dyes in polyacrylamide gels: a systematic analysis. Electrophoresis 6(9):427–448. doi:10.1002/elps.1150060905
Chiang GG, Abraham RT (2004) Determination of the catalytic activities of mTOR and other members of the phosphoinositide-3-kinase-related kinase family. Methods Mol Biol 281:125–141. doi:10.1385/1-59259-811-0:125
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Stone KL, Williams KR (1996) Enzymatic digestion of proteins in solution and in SDS polyacrylamide gels. In: Walker JM (ed) The protein protocols handbook. Humana Press, New York, pp 415–425. doi:10.1007/978-1-60327-259-9_71
Hellman U (2000) Sample preparation by SDS/PAGE and in-gel digestion. In: Jollès P, Jörnvall H (eds) Proteomics in functional genomics: protein structure analysis. Birkhäuser Basel, Basel, pp 43–54. doi:10.1007/978-3-0348-8458-7_3
Boersema PJ, Raijmakers R, Lemeer S, Mohammed S, Heck AJ (2009) Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat Protoc 4(4):484–494. doi:10.1038/nprot.2009.21
Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B (2007) Quantitative mass spectrometry in proteomics: a critical review. Anal Bioanal Chem 389(4):1017–1031. doi:10.1007/s00216-007-1486-6
Bantscheff M, Lemeer S, Savitski MM, Kuster B (2012) Quantitative mass spectrometry in proteomics: critical review update from 2007 to the present. Anal Bioanal Chem 404(4):939–965. doi:10.1007/s00216-012-6203-4
Engholm-Keller K, Larsen MR (2016) Improving the phosphoproteome coverage for limited sample amounts using TiO2-SIMAC-HILIC (TiSH) phosphopeptide enrichment and fractionation. Methods Mol Biol 1355:161–177. doi:10.1007/978-1-4939-3049-4_11
Kozlov SV, Waardenberg AJ, Engholm-Keller K, Arthur JW, Graham ME, Lavin M (2016) Reactive Oxygen Species (ROS)-activated ATM-dependent phosphorylation of cytoplasmic substrates identified by large-scale phosphoproteomics screen. Mol Cell Proteomics 15(3):1032–1047. doi:10.1074/mcp.M115.055723
Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26(12):1367–1372. doi:10.1038/nbt.1511
Nielsen ML, Vermeulen M, Bonaldi T, Cox J, Moroder L, Mann M (2008) Iodoacetamide-induced artifact mimics ubiquitination in mass spectrometry. Nat Methods 5(6):459–460. doi:10.1038/nmeth0608-459
McDonald WH, Ohi R, Miyamoto DT, Mitchison TJ, Yates Iii JR (2002) Comparison of three directly coupled HPLC MS/MS strategies for identification of proteins from complex mixtures: single-dimension LC-MS/MS, 2-phase MudPIT, and 3-phase MudPIT. Int J Mass Spectrom 219(1):245–251. doi:http://dx.doi.org/10.1016/S1387-3806(02)00563-8
Bunkenborg J, Espadas G, Molina H (2013) Cutting edge proteomics: benchmarking of six commercial trypsins. J Proteome Res 12(8):3631–3641. doi:10.1021/pr4001465
Chan DW, Son SC, Block W, Ye R, Khanna KK, Wold MS, Douglas P, Goodarzi AA, Pelley J, Taya Y, Lavin MF, Lees-Miller SP (2000) Purification and characterization of ATM from human placenta. A manganese-dependent, wortmannin-sensitive serine/threonine protein kinase. J Biol Chem 275(11):7803–7810
Dephoure N, Gould KL, Gygi SP, Kellogg DR (2013) Mapping and analysis of phosphorylation sites: a quick guide for cell biologists. Mol Biol Cell 24(5):535–542. doi:10.1091/mbc.E12-09-0677
Sechi S, Chait BT (1998) Modification of cysteine residues by alkylation. A tool in peptide mapping and protein identification. Anal Chem 70(24):5150–5158
Wall MJ, Crowell AM, Simms GA, Carey GH, Liu F, Doucette AA (2011) Implications of partial tryptic digestion in organic-aqueous solvent systems for bottom-up proteome analysis. Anal Chim Acta 703(2):194–203. doi:10.1016/j.aca.2011.07.025
Acknowledgments
M.E.G. and M.F.L. were funded by the National Health and Medical Research Council. M.E.G. was funded by the Brain Foundation and equipment grants from the Cancer Institute New South Wales, the Australian Cancer Research Foundation, and Rebecca L Cooper Medical Research Foundation.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer Science+Business Media LLC
About this protocol
Cite this protocol
Graham, M.E., Lavin, M.F., Kozlov, S.V. (2017). Identification of ATM Protein Kinase Phosphorylation Sites by Mass Spectrometry. In: Kozlov, S. (eds) ATM Kinase. Methods in Molecular Biology, vol 1599. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-6955-5_10
Download citation
DOI: https://doi.org/10.1007/978-1-4939-6955-5_10
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-6953-1
Online ISBN: 978-1-4939-6955-5
eBook Packages: Springer Protocols