Abstract
The ubiquitin-proteasome system (UPS) plays a key role in maintaining proteostasis by degrading most of the cellular proteins. Traditionally, UPS activity is studied in vitro, in yeast, or in mammalian cell cultures by using short-lived GFP-based UPS reporters. Here, we present protocols for two fluorescent tools facilitating real-time imaging of UPS activity in living animals. We have generated transgenic Caenorhabditis elegans (C. elegans) expressing a photoconvertible UbG76V-Dendra2 UPS reporter, which permits measurement of reporter degradation by the proteasome independently of reporter protein synthesis, and a fluorescent polyubiquitin-binding reporter for detection of the endogenous pool of Lys48-linked polyubiquitinated proteasomal substrates. These reporter systems facilitate cell- and tissue-specific analysis of UPS activity especially in young adult animals, but can also be used for studies during development, aging, and for example stress conditions.
Access provided by CONRICYT – Journals CONACYT. Download protocol PDF
Similar content being viewed by others
Key words
- Ubiquitin-proteasome system (UPS)
- UbG76V-Dendra2
- Polyubiquitin-binding reporter
- Photoconversion
- Live imaging
- Proteostasis
- C. elegans
1 Introduction
The ubiquitin-proteasome system (UPS) is one of the main safeguards of protein homeostasis by degrading cellular proteins including short-lived regulators, unfolded and damaged proteins. In UPS, the substrate is polyubiquitinated through the actions of ubiquitin-activating (E1 ), -conjugating (E2 ), and -ligating (E3 ) enzymes. The polyubiquitinated substrate is then recognized and degraded by the proteasome. The proteasome is a large (over 2.5 megadaltons) multisubunit protein complex consisting of a barrel-shaped 20S core particle capped from one or both ends with 19S regulatory particles or alternative activators [1]. Dysfunctions of the UPS are associated with severe proteotoxic conditions such as age-related neurodegenerative diseases and some cancers [2]. In addition, changes in proteasomal degradation have been detected in aging organisms including humans and C. elegans [3–5].
Development of short-lived GFP-based UPS reporters for human cell culture studies has provided key tools for investigations on UPS-mediated proteolysis [6, 7]. More recently, the nematode C. elegans has started to be utilized as a multicellular model system to unravel functions of the UPS [5, 8, 9]. C. elegans is a widely used model organism in biomedical research and, due to its short life-span and conserved signaling pathways, it is also a popular model for aging studies. Moreover, the transparent body of C. elegans makes it highly suitable for live imaging . We have created two fluorescent tools to study UPS-mediated protein degradation in C. elegans: a photoconvertible UPS reporter and a fluorescent polyubiquitin-binding reporter [5, 10].
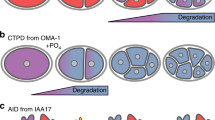
Our photoconvertible fluorescent UPS reporter (Fig. 1a) is based on the coral fluorescent Dendra2 protein, which can be irreversibly photoconverted from green-to-red fluorescent form by using 405 nm or 488 nm wavelength [11]. We have tagged Dendra2 with the uncleavable ubiquitin molecule UbG76V turning it into an ubiquitin fusion degradation (UFD) substrate [12, 13]. This N-terminal ubiquitin form cannot be removed by deubiquitinases and functions as an anchor for polyubiquitin chains, thereby targeting the complete fusion protein for degradation by the proteasome. UbG76V-Dendra2 is expressed under tissue-specific promoters in C. elegans , which enables UPS activity studies in different cell types and tissues (Fig. 1b). Photoconversion of UbG76V-Dendra2 from a green-to-red state enables quantification of proteasomal turnover rate for the subset of photoconverted reporter proteins, thus avoiding the effect of newly synthesized reporter proteins on the experimental outcome. In comparison, constitutively fluorescent GFP-based UPS reporters require expression of a second fluorescent protein (e.g., mRFP, mCherry), preferably in the same tissue of the animal, to distinguish whether the observed effect is due to changes in degradation rate or rate of protein synthesis.
In vivo UPS reporter systems. (a) UPS reporter for direct measurement of proteasome-mediated protein degradation. The UbG76V-Dendra2 UPS reporter can be irreversibly photoconverted from a green to a red fluorescent state by exposure to intense light (e.g., 405 nm). The UbG76V-Dendra2 is degraded by the proteasome in a polyubiquitin-dependent manner. (b) Animal model reporting on cell or tissue-specific UPS activity. Transgenic C. elegans expressing UbG76V-Dendra2 in for example intestinal cells can be exposed to single-cell photoconversion . Normal, enhanced or impaired proteasomal degradation of the photoconverted reporter can be measured independently of new reporter synthesis in a living animal. (c) Animal model for detection of the cellular pool of endogenous polyubiquitinated proteins. The polyubiquitin-binding reporter is stabilized upon binding to Lys48-linked polyubiquitinated substrates in the cell. The polyubiquitin-binding reporter is targeted for ubiquitin-independent proteasomal degradation via the mouse ornithine decarboxylase degradation domain (MODC domain) fused to the fluorescent ZsGreen protein. The reporter binds polyubiquitinated endogenous substrates via the ubiquitin-interacting motif (UIM) domains of the C. elegans proteasome subunit RPN-10. Impaired proteasomal degradation results in accumulation of polyubiquitinated proteins and increased reporter fluorescence
The stability of ubiquitin-tagged UPS reporters is not only affected by proteasome activity, but also by the activity of upstream UPS components such as E3 ligase(s). We therefore used an alternative approach to design another UPS reporter for live imaging of the cellular pool of endogenous Lys48-linked polyubiquitinated proteins in C. elegans . This polyubiquitin-binding reporter has two ubiquitin-interacting motif (UIM) domains derived from the C. elegans proteasome subunit RPN-10 fused to the N-terminus of the commercially available ZsProSensor-1 fluorescent reporter (Fig. 1c) [10]. ZsProSensor-1 composes of the fluorescent ZsGreen protein couple in the C-terminus to the mouse ornithine decarboxylase (MODC) domain, leading to a short-lived fusion protein targeted for ubiquitin-independent degradation by the proteasome. Accordingly, expression of ZsProSensor-1 in C. elegans intestinal cells did not result in detectable reporter fluorescence. The UIM-domains capture Lys48-linked polyubiquitinated endogenous substrates in the cell, thus stabilizing the reporter and leading to fluorescent worms. As a readout, increased fluorescence can be interpreted as an accumulation of polyubiquitinated proteins due to impaired proteasome-mediated degradation. Polyubiquitin-binding domains have previously been used as tools for capturing the cellular pool of polyubiquitin chains from mammalian cell lysates [14] and for visualizing polyubiquitinated proteins in mammalian cells [15].
The experimental procedures and equipment requirements differ between the above described reporters, as UbG76V-Dendra2 reporter worms should preferably be individually imaged with confocal microscope, whereas the polyubiquitin reporter worms can be imaged with standard fluorescence microscope. This protocol description focuses on providing key points in the UbG76V-Dendra2 reporter animal analysis, as well as a brief description of imaging of the polyubiquitin reporter worms. By using these complementing in vivo UPS reporter systems, we have been able to start unravelling cell-type- and aging -specific changes in UPS activities in C. elegans , as well as identified tissue-specific regulatory mechanisms of the UPS [5, 10, 16].
2 Materials
2.1 Transgenic C. elegans
Transgenic C. elegans strains expressing UbG76V-Dendra2 or the polyubiquitin-binding reporter in body wall muscle cells, neurons, or intestinal cells [5, 10, 16].
2.2 Agarose (Fischer Scientific)
3–5 % melted agarose in H2O.
2.3 Glass Slides and Cover Slips (Thermo Scientific)
1 mm × 26 mm × 76 mm (thickness, lenght, width) glass slides and 0.13 – 0.16 mm × 20 mm × 20 mm (thickness, lenght, width) cover slips.
2.4 Levamisole Hydrochloride (Sigma-Aldrich)
0.5–1 mM in M9 buffer (22 mM KH2PO4, 41 mM Na2HPO4, 8.5 mM NaCl, and 19 mM NH4Cl ). If worms are not paralyzed, the concentration can be carefully increased.
2.5 Microscopes
For UbG76V-Dendra2 C. elegans imaging, we preferably like to use motorized Zeiss Axio Observer Z1 inverted confocal microscope with LSM 5 Live line scanner and LSM AIM software Rel. 4.2. 518F immersion oil (Zeiss) is used with 63× objective. However, by optimizing photoconversion , also other confocal microscopes can be used. Live imaging of polyubiquitin-binding reporter strains can be performed with Zeiss Axioplan microscope or any other equivalent fluorescent microscope.
3 Methods
3.1 Agarose Pads
Use 3–5 % melted agarose to prepare thick agarose pads for worm mounting. Thick agarose pads can be prepared by placing spacers, e.g., two glass slides of 1 mm on top of each other, on each side of the sample slide to which ~400 μl melted agarose is added. A glass slide is then temporarily placed on the melted agarose and the spacers to generate a flattened 1 mm thick agarose pad. The top glass slide is then removed and the pad is placed in a humidified box for immediate use.
3.2 Transgenic C. elegans Maintenance
-
1.
Transgenic C. elegans strains are grown under standard conditions [17] at 20 °C (see Note 1 ). Worms can be imaged at any time during their life. However, UbG76V-Dendra2 strains tend to loose fluorescence upon aging , and therefore it is recommended to image them as young adult worms when possible.
3.3 Live Imaging
-
1.
For imaging, worms are mounted on agarose pads with a drop of 1 mM levamisole and topped with a cover slip. It is recommendable to use a 1 mm thick agarose pad, as it functions as a cushion, and the worm does not flatten when the cover slip is placed on top of it. The mounted worms should be used for immediate imaging.
-
2.
At its unconverted state, Dendra2 has excitation and emission maxima at 490 and 507 nm, respectively, and at its photoconverted state, the excitation and emission maxima are at 553 and 573 nm, respectively. After the worm has been localized under the microscope, fast scanning with the GFP channel should be used to identify and focus the cell for photoconversion . Images are acquired with 63× 1.4 NA plan-apochromat objective and 518F immersion oil. Before the photoconversion, one scan should be taken and saved with both green and red channels to serve as a “before photoconversion” time point. For photoconversion we use 405 nm diode laser. Dendra2 can also be photoconverted, but less efficiently, with 488 nm laser. After selecting the target cell, photoconversions are done by using 25–50 iterations (scanning speed 1.6 μs per pixel) with 100 % laser output. In addition to single cells, whole tissues and worms can also be photoconverted and images acquired with 10× 0.45 NA plan-apochromat objective (see Notes 2 and 3 ).
-
3.
After acquiring the image “after photoconversion, ” the worm should be removed from the pad to recover on NGM-agar plate before later time-point imaging. If the worm will be used in time-lapse imaging with short intervals, it can be kept on the agarose pad for 2–3 h. In this case, it is good to check that the worm does not dry out. This can be prevented by sealing the cover slip with agarose. For measurements of UbG76V-Dendra2 proteasomal degradation in the dorsorectal ganglion, we have imaged the worm every 10 min for up to 3 h, while imaging of the UPS reporter in dopaminergic neurons, body wall muscle cells, and intestinal cells has been performed at 3, 6, 12, and 24 h after photoconversion due to slower degradation rates. For time-lapse imaging manual focusing is required at each time-point, as the worm may slightly twist or subside in the agarose during microscopy (see Notes 4 and 5 ).
-
4.
We have used LSM AIM software Rel. 4.2 to analyze signal intensities. For analysis, we have quantified similar pixel amount at each image from different time points. To calculate the relative intensities, the absolute values of fluorescence before and right after photoconversion are set as 100 % for the green and red fluorescence signals, respectively.
-
5.
For analysis of polyubiquitin-binding reporter worms, the worms are immobilized with levamisole on similar agarose pads as UbG76V-Dendra2 worms, and imaged with Zeiss Axioplan microscope, or any other equivalent fluorescent microscope. Alternatively, worms can be immobilized with levamisole on a foodless agar growth plate, and imaged by using standard fluorescent stereomicroscope used for normal fluorescence worm maintenance. Fluorescent intensities can be quantified by using ImageJ.
4 Notes
-
1.
Detailed information on worm maintenance and generation of transgenic animals are described in the online published WormBook at www.wormbook.org.
-
2.
Depending on the cell-type of expression, the short-lived UbG76V-Dendra2 may not be visible in all cells of the same tissue in the worm. Especially extrachromosomal arrays give mosaic expression, which can be avoided by transgene integration. For example, an extrachromosomal array with intestinal expression does not usually show UbG76V-Dendra2 fluorescence in every intestinal cell. In addition, in worms with stronger UPS activity than in the wild-type (N2) background, such as the long-lived daf-2(e1370) mutants, it may be difficult to find young adults with fluorescent intestinal cells. Worms exhibiting mosaic reporter expression can also be taken advantage of by investigating if variation in reporter expression levels, as reflected by differences in fluorescence intensity, affects its degradation rate. For photoconversion , it is recommended to pick worms with fluorescence in at least two cells, because depending on the orientation of the worm, the other cell may not come into clear focus. This is affected by the position of the gonad: if the gonad is above the cell being photoconverted, it hampers focusing. It is very important to keep the same lateral orientation of the worm during photoconversion and imaging at the later timepoint(s), i.e., when the worm previously exposed to photoconverison is placed back on the agarose pad after a recovery period in the normal growth plate. If the photoconverted cell resides on the other side of the worm compared to it its position during photoconversion, it will reduce the fluorescence intensity due to the lack of focus. The lateral orientation is convenient to check, e.g., from the position of the vulva (left vs. right) in relation to head of the worm.
-
3.
At the start of photoconversion , when searching for the worm on the pad under the confocal microscope, it is recommended to avoid using the fluorescence light source and use only transmitted light. Especially with high magnification objectives (40× and 63×) the fluorescent light easily photoconverts the whole worm. Scanning speed and number of iterations for the photoconversion step should be determined separately for each microscope. In our Dendra2 experiments, we have used Zeiss LSM 5 Duo confocal microscope and its fast line-scanner LSM 5 Live. We have noticed that confocal microscopes with slow scanning speed are not suitable for the photoconversion experiments described here.
-
4.
It is important to notice that lasers on confocal microscopes can heat up when they are kept on for several hours. Laser heating increases the signal intensity and can affect the results. For example, it is common that the imaging lasers produce a stronger signal in the afternoon compared to the signal right after the photoconversion in the morning, if the lasers have been on the whole day. This problem can be avoided by keeping the lasers on for a while before starting the photoconversion.
-
5.
It is always important to choose healthy looking age-synchronized worms for experiments.
References
Finley D (2009) Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem 78:477–513
Dahlmann B (2007) Role of proteasomes in disease. BMC Biochem 8(Suppl 1):S3
Chondrogianni N, Gonos ES (2005) Proteasome dysfunction in mammalian aging: steps and factors involved. Exp Gerontol 40:931–938
Chondrogianni N, Voutetakis K, Kapetanou M, Delitsikou V, Papaevgeniou N, Sakellari M, Lefaki M, Filippopoulou K, Gonos ES (2014) Proteasome activation: an innovative promising approach for delaying aging and retarding age-related diseases. Ageing Res Rev 23:37–55
Hamer G, Matilainen O, Holmberg CI (2010) A photoconvertible reporter of the ubiquitin-proteasome system in vivo. Nat Methods 7:473–478
Dantuma NP, Lindsten K, Glas R, Jellne M, Masucci MG (2000) Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nat Biotechnol 18:538–543
Bence NF, Sampat RM, Kopito RR (2001) Impairment of the ubiquitin-proteasome system by protein aggregation. Science 292:1552–1555
Liu G, Rogers J, Murphy CT, Rongo C (2011) EGF signalling activates the ubiquitin proteasome system to modulate C. elegans lifespan. EMBO J 30:2990–3003
Segref A, Torres S, Hoppe T (2011) A screenable in vivo assay to study proteostasis networks in caenorhabditis elegans. Genetics 187:1235–1240
Matilainen O, Arpalahti L, Rantanen V, Hautaniemi S, Holmberg CI (2013) Insulin/IGF-1 signaling regulates proteasome activity through the deubiquitinating enzyme UBH-4. Cell Rep 3:1980–1995
Chudakov DM, Lukyanov S, Lukyanov KA (2007) Tracking intracellular protein movements using photoswitchable fluorescent proteins PS-CFP2 and Dendra2. Nat Protoc 2:2024–2032
Johnson ES, Bartel B, Seufert W, Varshavsky A (1992) Ubiquitin as a degradation signal. EMBO J 11:497–505
Johnson ES, Ma PC, Ota IM, Varshavsky A (1995) A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem 270:17442–17456
Bennett EJ, Shaler TA, Woodman B, Ryu KY, Zaitseva TS, Becker CH, Bates GP, Schulman H, Kopito RR (2007) Global changes to the ubiquitin system in huntington’s disease. Nature 448:704–708
Sims JJ, Scavone F, Cooper EM, Kane LA, Youle RJ, Boeke JD, Cohen RE (2012) Polyubiquitin-sensor proteins reveal localization and linkage-type dependence of cellular ubiquitin signaling. Nat Methods 9:303–309
Li X, Matilainen O, Jin C, Glover-Cutter KM, Holmberg CI, Blackwell TK (2011) Specific SKN-1/nrf stress responses to perturbations in translation elongation and proteasome activity. PLoS Genet 7:e1002119
Brenner S (1974) The genetics of caenorhabditis elegans. Genetics 77:71–94
Acknowledgement
We thank the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440), for strains and the Biomedicum Imaging Unit, University of Helsinki, for imaging services. This work was supported by grants to C.I.H. from the Academy of Finland (131408 and 259797), University of Helsinki Funds, International Human Frontier Science Program Foundation, and the Sigrid Juselius Foundation. The authors would like to acknowledge networking support by the Proteostasis COST Action (BM1307).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer Science+Business Media New York
About this protocol
Cite this protocol
Matilainen, O., Jha, S., Holmberg, C.I. (2016). Fluorescent Tools for In Vivo Studies on the Ubiquitin-Proteasome System. In: Matthiesen, R. (eds) Proteostasis. Methods in Molecular Biology, vol 1449. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-3756-1_12
Download citation
DOI: https://doi.org/10.1007/978-1-4939-3756-1_12
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-3754-7
Online ISBN: 978-1-4939-3756-1
eBook Packages: Springer Protocols