Abstract
Posttranslational modifications are crucial in determining the functions of proteins in the cell. Modification of the NLRP3 inflammasome by the ubiquitin system has recently emerged as a new level of regulation of the inflammasome complex. Here, we describe a method to detect polyubiquitination of NRLP3 using two different approaches: (1) detection with an ubiquin antibody or (2) using TUBE (Tandem Ubiquitin Binding entities). This approach can be used to detect ubiquitination of other NLR or other components of the inflammasome.
Access provided by CONRICYT – Journals CONACYT. Download protocol PDF
Similar content being viewed by others
Key words
1 Introduction
Ubiquitination is a posttranslational protein modification crucial to maintain cellular homeostasis. The addition of ubiquitin to a protein is mediated by an E1-ubiquitin activating enzyme, an E2-ubiquitin-conjugating enzyme, and an E3 ubiquitin ligase [1]. This is a reversible process and this is controlled by a family of enzymes called deubiquitinases [2]. Disturbing this balanced ubiquitination state can have detrimental consequences for the cell impairing important processes that maintain the normal functioning of the cells such as protein degradation, trafficking, or gene expression [3] and hence the body.
The ubiquitin system also plays a crucial role in the regulation of inflammation. This is especially relevant in innate immune responses where key signalling pathways are finely tuned by ubiquitin [4]. Pathways such as the NFkB rely on this posttranslational modification to successfully initiate the transcription of important proinflammatory mediators [5, 6]. Increasing evidencing, however, is showing that ubiquitin is not only important in the regulation of transcriptional pathways but also in the assembly of the NLRP3 inflammasome [7]. This is a molecular complex that mediates the release of the proinflammatory cytokine IL-18 and IL-1β [8]. Although it has been shown that the NLRP3 is ubiquitinated, how the ubiquitination state of this receptor regulates the inflammasome still remains a mystery. In order to further study these mechanisms, it is necessary to determine the level of ubiquitination of the NLRP3. In this chapter, we described a method to detect endogenous NLRP3 ubiquitination in NLRP3-expressing HEK293 cells by two different methods. Here, NLRP3 is immunoprecipitated and then the ubiquitination state detected by western blot using either a ubiquitin antibody or Tandem Ubiquitin Binding Entities (TUBE) . This method could also be applied to detect ubiquitination in cells that endogenously express NLRP3 or for other components of the inflammasome complex.
2 Materials
2.1 Cell Lysis
-
1.
SDS-lysis buffer: 20 mM Tris–base (pH 8.0), 250 mM NaCl, 3 mM EDTA, 10 % glycerol, 1 % SDS, 0.5 % NP-40, 20 mM N-Ethylmaleimide (NEM), and 5 mM 1,10-phenanthroline monohydrate, protease inhibitors [9].
-
2.
Nondenaturing lysis buffer: 50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1 % Triton X-100, 10 % glycerol, 20 mM NEM, and 5 mM 1,10-phenanthroline monohydrate, protease inhibitors (see Note 1 ).
2.2 Transfection
-
1.
Complete cell culture media: Dulbecco’s Modified Eagle’s Medium (DMEM) containing glucose, l-glutamine, sodium pyruvate, and sodium bicarbonate, supplemented with 10 % of decomplemented Fetal Bovine Serum (FBS).
-
2.
Microcentrifuge tubes.
-
3.
12-well cell culture plates.
-
4.
Transfection media: DMEM without FBS supplementation.
-
5.
Lipofectamine 2000® transfection reagent (Life Technologies). Other transfection reagents might be also suitable if transfection efficiency is high.
-
6.
Plasmid coding for NLRP3-Flag (see Note 2 ).
2.3 Immunoprecipitation
-
1.
SDS-immunoprecipitation (SDS-IP) buffer: 20 mM Tris–Base (pH 8.0), 250 mM NaCl, 3 mM EDTA, 10 % glycerol, 0.1 % SDS, 0.5 % NP-40, 20 mM NEM, and 5 mM 1,10-phenanthroline monohydrate, protease inhibitors (see Note 3 ).
-
2.
Nondenaturing lysis buffer: 50 mM Tris, 150 mM NaCl, 1 mM EDTA, 1 % Triton X-100, 10 % glycerol, 20 mM NEM, and 5 mM 1,10-phenanthroline monohydrate, protease inhibitors (see Note 4 ).
-
3.
Bicinchoninic acid (BCA) protein assay kit.
-
4.
Anti-Flag M2 affinity gel.
-
5.
Protein G Sepharose.
-
6.
Hamilton syringe or equivalent device, such a narrow-end Pasteur pipette .
2.4 Western Blot
-
1.
Tris-glycine gels (resolving gel 6 % and stacking gel 5 %).
-
2.
Tris-Glycine-SDS (TGS) running buffer: 25 mM Tris, 192 mM Glycine, 0.1 % (w/v) SDS, pH 8.3.
-
3.
Laemmli Sample Buffer 4× supplemented with 10 % β-mercaptoethanol.
-
4.
Protein electrophoresis system. We use the Mini-PROTEAN Tetra Cell electrophoresis system (Bio-Rad).
-
5.
Semidry blot transfer system. We use the Trans-Blot Turbo transfer system (Bio-Rad).
-
6.
Nitrocellulose membrane with pore size 0.45 μm.
-
7.
Transfer buffer, for 1 L: 200 ml of 5× Transfer buffer (Bio-Rad), 600 ml of nanopure water, and 200 ml of ethanol 100 %.
-
8.
Powdered skim milk.
-
9.
Bovine serum albumin (BSA).
-
10.
Tris buffered saline (TBS) 10×: 1.5 M NaCl, 0.1 M Tris–HCl, pH 7.4.
-
11.
Phosphate buffered saline (PBS) 10×: 1.2 M Sodium chloride, 90 mM disodium hydrogen orthophosphate, 37 mM sodium dihydrogen orthophosphate, and 26 mM potassium chloride (pH 7.4).
-
12.
PBST solution: PBS containing 0.1 % Tween-20.
-
13.
TTBS solution: TBS containing 0.1 % Tween-20.
-
14.
Blocking solution: 5 % milk in PBST (BS1) or TBST (BS2). Store at 4 °C.
-
15.
Diluent solution: 5 % milk in TBST (TBST1) or PBST (PBST2). Store at 4 °C.
-
16.
Antibodies: Mouse anti-ubiquitin (SantaCruz, sc-8017), mouse anti-ubiquitin M1-specific (Lifesensor, AB130), mouse anti-NLRP3 (Enzo, ALX-804-818), and rabbit anti-Flag (Cell Signaling, 2368).
-
17.
Secondary Polyclonal rabbit antimouse or goat antirabbit immunoglobulins conjugated with HRP.
-
18.
TUBE2-Biotin (Binds to K6-, M1-, K48-, and K63-linked polyubiquitin) (Lifesensors).
-
19.
Streptavidin-HRP.
-
20.
Enhanced chemoluminescence (ECL) substrate.
-
21.
Imaging System, such as the ChemiDoc MP (Bio-Rad).
3 Methods
3.1 Transfection
-
1.
Plate HEK293 cells (0.15 × 106 cells per well in a 12 well plate) in 1 ml of complete DMEM cell culture media (see Note 5 ).
-
2.
The following day transfect cells with 1 μg of NLRP3-Flag plasmid. Control cells were transfected with an empty plasmid. Mix 50 μl of transfection media with 1 μl of NLRP3-Flag-tag vector or empty vector (1 μg/ul) (Tube 1). Mix 50 μl of transfection media with 2.5 μl of Lipofectamine2000 (Tube 2). Incubate for 5 min a room temperature. Add contents from tube 1 into tube 2 and mix gently. Incubate at room temperature for 15 min.
-
3.
Add this mix to the well and incubate the cells for 48 h (see Note 6 ).
3.2 Cell Lysis for Detection with Ubiquitin Antibody
-
1.
Wash cells in PBS.
-
2.
Add 200 μl of SDS-lysis buffer per well.
-
3.
Boil samples at 90–95 °C by placing the cell culture plate on the heat block for 20–30 min. To help lysis, mix and pipette up and down using a pipette (10–1000 μl range) for 5–10 s every 3 min during the boiling step. Continue procedure until viscosity is eliminated.
-
4.
Transfer samples to a microcentrifuge tube and centrifuge the samples at 12,000 × g, 10 min at room temperature in a standard table-top microfuge and transfer supernatants to new tubes. Cell lysates can now be used or stored at −20 °C before further analysis.
3.3 Cell Lysis for Detection with TUBE-Biotin
-
1.
Perform lysis at 4 °C.
-
2.
Lyse cells by adding 200 μl of nondenaturing lysis buffer per well.
-
3.
Incubate on ice for 10 min.
-
4.
Transfer samples to an microcentrifuge tube and centrifuge the samples at 12,000 × g, 10 min at 4 °C in a standard table-top microfuge and transfer supernatants to new tubes. Cell lysates can now be used or stored at −20 °C before further analysis.
3.4 Immunoprecipitation of NLRP3
-
1.
Subject equal amounts of protein lysates (around 100–150 μg) to immunoprecipitation (IP) assay. Measure protein concentration using a BCA assay. Save 5–10 μg of cell lysates for use as the pre-IP samples.
-
2.
Dilute lysates into IP buffer to a final 800 μl of and perform all subsequent steps at 4 °C.
-
3.
To prepare the Sepharose and/or anti-Flag M2 affinity gel, thoroughly suspend them to make a uniform suspension of the resin. Immediately transfer 40 μl of the suspension per reaction to a fresh test tube (see Note 7 ).
-
4.
Wash the resin with IP buffer, 6 times with 1 ml of the corresponding IP buffer. Centrifuge at 8000 × g for 30 s and discard buffer after each wash. In order to let the resin settle in the tube, wait for 1–2 min before removing the buffer (see Notes 8 and 9 ).
-
5.
Preclearing step: Add the 800 μl of cell lysate to the washed resin (Protein G sepharose).
-
6.
Incubate at 4 °C for 1 h with rotation (a roller shaker is recommended). Centrifuge at 8000 × g for 30 s, collect supernatant, and d iscard agarose (see Notes 10 and 11 ).
-
7.
Add this supernatant to the anti-Flag M2 affinity gel previously washed (see Note 12 ).
-
8.
Agitate or shake all samples and controls gently (a roller shaker is recommended) for 4 h at 4 °C.
-
9.
Centrifuge the resin for 30 s at 8000 × g. Remove the supernatants with a narrow-end pipette tip.
-
10.
Wash the resin 5 times with 0.5 ml of the appropriate IP buffer depending on protocol (as in step 4). Make sure all the supernatant is removed by using a Hamilton syringe or equivalent device.
-
11.
Dilute the SDS-PAGE sample buffer 4× to 1× with water.
-
12.
Add 40 μl of 1× sample buffer to each sample and control.
-
13.
Boil the samples and controls for 3 min.
-
14.
Centrifuge the samples and controls at 8000 × g for 30 s to pellet undissolved agarose. Transfer the supernatants to fresh test tubes with a Hamilton syringe or a narrow-end Pasteur pipette. These samples are ready to be loaded in a SDS-pag e gel.
3.5 Western Blot Analysis
-
1.
Load samples obtained from IP in two different 6 % Tris-glycine gels (a) to detect NLRP3 ubiquitination (load 30 μl of IP-sample) and (b) to confirm that IP has worked (load 10 μl of IP-sample).
-
2.
Optional: Load a third gel with Pre-IP samples. This will inform us of the success rate of transfection (see Note 13 ).
-
3.
Run the gel at 150 V in TGS buffer for 1 h and transfer into a nitrocellulose membrane using the High Molecular weight settings in the Trans-Blot Turbo transfer system.
3.6 Detection of Polyubiquitination by Using Ubiquitin Antibodies
-
1.
Transfer membrane into BS1 and block for 2 h at room temperature.
-
2.
Add the desired primary antibodies: anti-Ub (1:200), anti-M1 Ub (1:1000), anti-Flag (1:800), or anti-NLRP3 (1:1000), and incubate overnight at 4 °C.
-
3.
Wash membrane 10×, 2 min each with PBST.
-
4.
Add secondary antibody in BS1 and incubate for 1 h at room temperature.
-
5.
Wash membrane 10×, 2 min each with PBST.
-
6.
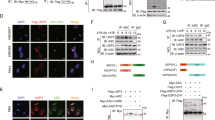
Develop using ECL detection reagents according to manufacturer instructions and a luminescence compatible Imaging System (Fig. 1a, b).
Fig. 1 Western blotting detection of NLRP3 ubiquitination in HEK293 cells. The blots at the top show a smear above 120 kDa typical of protein polyubiquitination. This is detected using ubiquitin general antibody (Ub) (a), specific linear M1-ubiquitin antibody (M1-Ub) (b), or TUBE-Biotin (c). The blots at the bottom show that the NLRP3 receptor was successfully immunoprecipitated, validating the assay
3.7 Detection of Polyubiquitination by Using TUBE-Biotin
-
1.
Transfer membrane into BS2 and block for 2 h at room temperature.
-
2.
Add 1 μg/ml of TUBE-Biotin or primary antibodies: anti-Flag (1:800) or anti-NLRP3 (1:1000), and incubate overnight at 4 °C.
-
3.
Wash membrane 10×, 2 min each with PBST.
-
4.
To detect TUBE-Biotin add streptavidin-HRP (1:10,000) in BS2. For other antibodies, add appropriate secondary antibodies in BS1. Incubate for 1 h at room temperature.
-
5.
Wash membrane 10×, 2 min each with PBST.
-
6.
Develop using ECL detection reagents according to manufacturer instructions and a luminescence compatible Imaging System (Fig. 1c).
4 Notes
-
1.
These buffers can be made ahead of time with the exception that the 10 μl/ml of 100× protease inhibitor, NEM, and 1,10-phenanthroline monohydrate should be added immediately before use. The buffers can be stored at room temperature.
-
2.
Plasmid coding for NLRP3 was a gift of J. Tschopp.
-
3.
For detection using ubiquitin antibodies.
-
4.
For detection using TUBE-Biotin.
-
5.
Cells should be 75–80 % confluent when transfecting.
-
6.
Scale the transfection reagents up or down depending on the samples required for your experiment.
-
7.
For resin transfer, use a clean, plastic pipette tip with the end enlarged to allow the resin to be transferred.
-
8.
SDS-IP buffer for ubiquitin and nondenaturing buffer for TUBE protocol.
-
9.
In case of numerous immunoprecipitation samples, wash the resin needed for all samples together and after washing, divide the resin according to the number of samples tested.
-
10.
This step is to remove nonspecific binding to agarose.
-
11.
Remove the supernatant with a narrow-end pipette tip or a Hamilton syringe, being careful not to transfer any resin. Narrow-end pipette tips can be made using forceps to pinch the opening of a plastic pipette tip until it is partially closed.
-
12.
If not using anti-Flag M2 affinity gel, combine the appropriate antibody (e.g., anti-NLRP3 if using endogenous expression) with the cell lysates for 1–4 h at 4 °C to form the immune complexes. Depending on the antibody, different amounts of antibody could be used. Typically, 3–5 μg of antibody is used for each IP. This could be increased to 10 μg if no signal is observed.
-
13.
Dilute them in SDS-load buffer to 1× and boil for 3 min previous to loading.
References
Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67:425–479
Clague MJ, Barsukov I, Coulson JM, Liu H, Rigden DJ, Urbe S (2013) Deubiquitylases from genes to organism. Physiol Rev 93:1289–1315
Aguilar RC, Wendland B (2003) Ubiquitin: not just for proteasomes anymore. Curr Opin Cell Biol 15:184–190
Malynn BA, Ma A (2010) Ubiquitin makes its mark on immune regulation. Immunity 33:843–852
Harhaj EW, Dixit VM (2012) Regulation of NF-kappaB by deubiquitinases. Immunol Rev 246:107–124
Skaug B, Jiang X, Chen ZJ (2009) The role of ubiquitin in NF-kappaB regulatory pathways. Annu Rev Biochem 78:769–796
Lopez-Castejon G (2013) Regulation of NLRP3 activation by the ubiquitin system. Inflammasome 1:15–19
Latz E, Xiao TS, Stutz A (2013) Activation and regulation of the inflammasomes. Nat Rev Immunol 13:397–411
Hwang J, Kalejta RF (2011) In vivo analysis of protein sumoylation induced by a viral protein: detection of HCMV pp 71-induced Daxx sumoylation. Methods 55:160–165
Acknowledgements
This work was supported by funds from the Manchester Collaborative Centre for Inflammation Research and a Sir Henry Dale fellowship from the Wellcome Trust (UK).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer Science+Business Media New York
About this protocol
Cite this protocol
Palazón-Riquelme, P., López-Castejón, G. (2016). Method to Measure Ubiquitination of NLRs. In: Di Virgilio, F., Pelegrín, P. (eds) NLR Proteins. Methods in Molecular Biology, vol 1417. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-3566-6_16
Download citation
DOI: https://doi.org/10.1007/978-1-4939-3566-6_16
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-3564-2
Online ISBN: 978-1-4939-3566-6
eBook Packages: Springer Protocols