Abstract
There are currently no vaccines that provide sterile immunity against malaria. Various proteins from different stages of the Plasmodium falciparum life cycle have been evaluated as vaccine candidates, but none of them have fulfilled expectations. Therefore, combinations of key antigens from different stages of the parasites life cycle may be essential for the development of efficacious malaria vaccines. Following the identification of promising antigens using bioinformatics, proteomics, and/or immunological approaches, it is necessary to express, purify, and characterize these proteins and explore the potential of fusion constructs combining different antigens or antigen domains before committing to expensive and time-consuming clinical development. Here, using malaria vaccine candidates as an example, we describe how Agrobacterium tumefaciens-based transient expression in plants can be combined with a modular and flexible cloning strategy as a robust and versatile tool for the rapid production of candidate antigens during research and development.
Access provided by CONRICYT – Journals CONACYT. Download protocol PDF
Similar content being viewed by others
Keywords
- Agrobacterium tumefaciens
- DsRed
- Gene stacking
- Nicotiana benthamiana
- Modular cloning
- Recombinant protein production
1 Introduction
The limited success of past and current malaria vaccine candidates [1] indicates the need for intensive and accelerated research to identify and characterize new antigens that confer protection against infection, clinical manifestation, and even the transmission of malaria [2]. Furthermore, multi-stage-specific cocktails combining key antigens from the different stages of the Plasmodium falciparum life cycle may be essential for the development of efficacious malaria vaccines [3]. To determine the suitability of novel vaccine candidates as components of vaccine cocktails, the antigens must be rapidly produced, purified, and characterized in terms of their protective efficacy in animal experiments and/or in vitro assays.
Agrobacterium tumefaciens -based transient expression in plants, using either classical T-DNA vectors [4] or amplification systems based on viral replicons [5], is one of the quickest and most versatile strategies for the production of recombinant proteins [6–8]. Although used predominantly for research and development, these systems have also been implemented for the manufacturing of clinical-grade materials, e.g., the experimental antibody cocktail ZMapp, comprising three chimeric monoclonal antibodies against the Ebola virus surface glycoprotein (EBOV-GP) [9], virus-like particles based on human influenza virus hemagglutinin (HA) [10], and the malaria transmission-blocking vaccine candidate Pfs25 [11]. These emerging applications of transient expression are driven by a desire for rapid and inexpensive vaccine development against poverty-related diseases such as malaria [12, 13].
Here we present a well-established and versatile workflow for A.tumefaciens -based transient expression in plants in the context of vaccine development, using the expression of single, multi-domain, and fluorescence-labeled malaria vaccine candidates as case studies. The combination of transient expression with a modular and flexible cloning strategy allows the rapid cloning and expression of multi-domain antigens or DsRedreporter gene fusions by gene stacking , and small to medium scale production without expensive and specialized equipment. We have used this workflow successfully to produce several single and multi-domain malaria vaccine candidates [14–16]. However, the technology is generic and can be applied in any vaccine development scenario where progress is dependent on the rapid production of different candidate antigens for analysis and characterization. Because the downstream purification strategies and functionality assays are highly dependent on the specific antigen, these procedures are not covered in this chapter, but examples of such methods can be found in several reports describing the characterization of plant-derived vaccine candidates [14, 16, 17].
The potential applications of this workflow are illustrated using four examples: (1) the cloning of three individual blood-stage antigens (PfAMA1-DiCo1-3) and their transient expression in Nicotiana benthamiana plants (separately and as a mixture), (2) the subsequent C-terminal fusion of additional blood-stage antigens (PfMSP1_19, PfRH2a, and PfRIPR_EGF7/8) to the PfAMA1-DiCo variants by gene stacking and the generation of stacked fusion antigen constructs comprising proteins and/or domains from the pre-erythrocytic stage (PfCSP_TSR), (3) the blood-stage (PfMSP1_19, PfMSP4, PfMSP8, and PfMSP10) and sexual-stage protein candidates (Pfs25 and Pfs28), and (4) the cloning of a single antigen (PfMSP1_19) as a C-terminal DsRed fusion protein. For additional information on these proteins and domains (seeNotes1–10).
The workflow includes the following procedures:
-
1.
The vectors and cloning strategies to generate expression constructs featuring individual antigens, DsRed–antigen fusion proteins, stacked dual-domain fusion proteins and a panel of stacked multi-domain, multi-stage fusion proteins featuring up to nine different antigens or antigen domains.
-
2.
Techniques for the transformation, screening, and cultivation of recombinant A. tumefaciens.
-
3.
Plant cultivation (N. benthamiana) , syringe and vacuum infiltration as well as incubation.
-
4.
The extraction of recombinant proteins from infiltrated leaf tissue.
2 Materials
2.1 Cloning of the Expression Constructs
-
1.
Bacteria
-
Chemically competent Escherichia coli DH5α (NEB , Frankfurt, Germany).
-
Agrobacterium tumefaciens strain GV3101::pMP90RK [GmR, KmR, RifR] [18].
-
-
2.
Plasmids and synthetic genes
-
pTRAkcERH [19].
-
-
3.
Oligonucleotides
-
4.
Enzymes
-
Restriction enzymes NcoI, NotI, and EagI (NEB, Frankfurt, Germany).
-
DNA-modifying enzymes T4 DNA ligase and Antarctic phosphatase (NEB, Frankfurt, Germany).
-
Please note that the sequences for the construct-specific oligonucleotides (primers) must be added to the target sequence as indicated by dots to allow NcoI/NotI cloning and EagI stacking (Subheading 3.1).
-
Construct-specific forward primer introducing EagI/NcoI restriction sites (5′-AAAAAAAACGGCCGTGGCCATGGCT…-3′).
-
Construct-specific reverse primer introducing NotI restriction site (5′-…GCGGCCGCTTTTTTTT-3′).
-
pTRA-backbone-specific forward primer PS5′ (5′-GACCCCTCCTCTATATAA GG-3′).
-
pTRA-backbone-specific reverse primer PS3′ (5′-GACCCCTCCTCTATATAA GG-3′).
2.2 Buffers and Reagents
-
1.
Lysogeny broth (LB)
-
Tryptone 10 g/L.
-
Yeast extract 5 g/L.
-
NaCl 10 g/L.
-
pH 7.
-
-
2.
Yeast extract broth (YEB)
-
Beef extract 5 g/L.
-
Yeast extract 1 g/L.
-
Tryptone 5 g/L.
-
Sucrose 5 g/L.
-
MgSO4 0.5 g/L.
-
pH 7.
-
-
3.
2× infiltration medium
-
Sucrose 100 g/L.
-
Glucose 3.6 g/L.
-
Ferty®-II-Mega fertilizer (Planta, Regenstauf, Germany) 1 g/L.
-
pH 5.4–5.8.
-
-
4.
Acetosyringone (3′,5′-dimethoxy-4′-hydroxyacetophenone) (Sigma Aldrich, Seelze, Germany).
-
5.
Plant extraction buffer (PBS, pH 7.4)
-
NaCl 8 g/L.
-
KCl 0.2 g/L.
-
Na2HPO4 1.44 g/L.
-
KH2PO4 0.24 g/L.
-
pH 7.4.
-
2.3 Equipment
-
1.
Eppendorf Multiporator (Eppendorf, Hamburg, Germany).
-
2.
Desiccator with connections and tubing (Duran, Wertheim/Main, Germany).
-
3.
Rotary vane vacuum pump RZ 6 (Vacuubrand, Wertheim/Main, Germany).
-
4.
1-mL Ominifx F syringe (Braun, Melsungen, Germany).
-
5.
Blender (Waring, Tampa, FL, USA).
-
6.
Mortar and pestle (Haldenwanger, Waldkraiburg, Germany).
3 Methods
3.1 Cloning the pTRAkc Expression Constructs
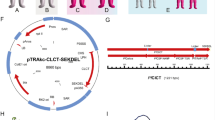
Individual synthetic genes or PCR products encoding selected antigen domains can be inserted into the pTRAkc-ERH vector (Fig. 1) or its variants (seeNote11) by NcoI/NotI cloning (Fig. 2a). Subsequent stacking of additional domains can be achieved by inserting EagI/NotI-fragments into NotI-linearized plasmids (Fig. 2b). When using an appropriate EagI sequence context (Subheading 2.1, item 4, seeNote12), a NotI-site will be reconstituted only at the 5′-end of the fusion gene, allowing the insertion of further domains by repeating the procedure. Similarly, the cloning of DsRed-fusion genes (to generate expression cassettes for tetravalent fluorescence-labeled antigens or antigen domains for different screening approaches), can be achieved by inserting EagI/NotI-fragments into a NotI-linearized plasmid carrying a DsRed cDNA inserted by NcoI/NotI cloning (Fig. 2c). Enzymes should be used according to the manufacturer’s instructions in term of the amounts, buffers, and incubation conditions.
Features of the plant expression vector pTRAkcERH. The plant expression vector pTRAkc is a derivative of the pPAM vector (GenBank AY027531) and contains two origins of replication (ColE1 ori for E.coli and RK2 ori for A.tumefaciens ), and a backbone ampicillin resistance gene (bla) as a bacterial selection marker. Recombinant proteins are expressed under the control of the Cauliflower mosaic virus 35S promoter with duplicated enhancer region, the 5′ untranslated region of the Petrosinella chalcone synthase (CHS) gene and the CaMV polyadenylation signal (pA35SS). Scaffold attachment regions (SAR) were introduced next to the right and left borders (RB and LB) of the T-DNA to improve gene expression following stable transformation (not relevant for transient expression). For the selection of such stable transgenic tobacco lines, the T-DNA contains the kanamycin resistance gene (nptII) under the control of the nopaline synthase promoter (pNos). The cloning cassette in the schematic illustration features the gene of interest (GOI) flanked by NcoI and NotI restriction sites, in-frame between a signal peptide sequence (LPH) and a His6 tag (his6) and an ER-retrieval sequence (SEKDEL)
Modular cloning and gene stacking using pTRAkc-ERH. Single antigen genes can be inserted in the pTRAkc-ERH expression cassette by NcoI/NotI cloning (a), leading to the in-frame insertion of the GOI coding sequence (light gray) between the 5′ signal peptide sequence (black) and the 3′ His6 tag (yellow), and ER-retrieval sequence (blue). The iterative generation of multi-domain fusions (b) can be achieved by subsequent stacking of EagI/NotI-digested GOI fragments into NotI-linearized, de-phosphorylated vectors between a 5′ antigen domain (first step, light gray; second step, dark gray) and the 5′ signal peptide sequence (black) and the 3′ His6 tag (yellow). Fusions to the C-terminus of DsRed (c) can be also realized by inserting EagI/NotI-digested GOI fragments into a NotI-linearized pTRAkc-ERH vector carrying the DsRed gene (red)
3.1.1 Generation of Single Antigen Expression Constructs
-
1.
Digest 2–4 μg of pTRAkc-ERH and target cDNA (insert-specific synthetic gene or PCR product) with NcoI and NotI.
-
2.
Separate the pTRAkc backbone and target cDNA by preparative gel electrophoresis, isolate and purify the DNA fragments.
-
3.
Quantify the purified DNA and use 50–100 ng of pTRAkc backbone for ligation with a five to tenfold molar excess of target cDNA (insert-specific synthetic gene or PCR product).
3.1.2 Generation of Multi-domain or DsRed Expression Constructs
-
1.
Digest 2–4 μg of pTRAkc-ERH containing a single antigen with NotI.
-
2.
Dephosphorylate the linearized vector to avoid re-ligation.
-
3.
Digest target cDNA (insert-specific synthetic gene or PCR product) with EagI.
-
4.
Separate by preparative gel electrophoresis, isolate and purify the DNA fragments.
-
5.
Quantify the purified DNA and use 50–100 ng of pTRAkc backbone for ligation with a five to tenfold molar excess of target cDNA (insert-specific synthetic gene or PCR product).
-
6.
This step can be repeated to fuse additional antigens or antigen domains (stacking).
Transform chemical competent E.coli cells with the ligation reactions, regenerate for 20–60 min at 37 °C and plate on LB agar containing 100 μg/mL ampicillin. Incubate plates overnight at 37 °C.
3.2 Identification of Recombinant E. coli Cells
-
1.
Check recombinant E.coli cells either by restriction digest or PCR using the PS5′ and PS3′ primer pair, or gene-specific primers (seeNote12).
-
2.
Verify all cloning steps by DNA sequencing.
3.3 Preparation of Electrocompetent A.tumefaciens Cells
-
1.
Inoculate 100 mL of YEB containing 25 μg/mL rifampicin and 25 μg/mL kanamycin with an aliquot of cryopreserved A.tumefaciens cells.
-
2.
Grow the culture at 28 °C and 160 rpm for 24–48 h until the OD600 reaches ~5.0 (seeNotes13).
-
3.
Chill cells on ice for 5 min.
-
4.
Transfer cells to two prechilled 50-mL Falcon tubes and centrifuge at 3000 × g for 10 min.
-
5.
Resuspend each cell pellet in 50 mL ice-cold and sterile H2O.
-
6.
Repeat the centrifugation step described above, and resuspend each pellet in 25 mL ice-cold and sterile H2O, and combine both pellets.
-
7.
Repeat the centrifugation step described above, and resuspend the cells in 10 mL ice-cold and sterile 10 % (v/v) glycerol.
-
8.
Repeat step 7, but resuspend the cells in 3 mL ice-cold and sterile 10 % (v/v) glycerol and prepare 50-μL aliquots in sterile 1.5-mL reaction tubes.
-
9.
Store reaction tubes with electrocompetent A.tumefaciens cells immediately at −80 °C.
3.4 Electroporation of A. tumefaciens
-
1.
Thaw an aliquot of electrocompetent A. tumefaciens cells on ice.
-
2.
Add 200–500 ng of pTRAkc plasmid DNA and mix gently with thawed cells.
-
3.
Transfer cells to a prechilled 2-mm electroporation cuvette and apply a pulse of 2.5 kV for 5 ms using an Eppendorf multiporator.
-
4.
Immediately add 950 μL YEB and transfer the cells into sterile 1.5-mL tubes.
-
5.
Incubate the cells for 2–3 h at 28 °C and 160 rpm.
-
6.
Use one spatula to plate, in descending order, 20 μL, 4 μL, and the remaining liquid from the spatula on YEB selection plates containing 50 μg/mL carbenicillin, 25 μg/mL rifampicin, and 25 μg/mL kanamycin (seeNotes14).
-
7.
Incubate the plates for 3–4 days at 28 °C.
3.5 Identification of Recombinant A. tumefaciens Cells
-
1.
Check at least five A. tumefaciens clones growing on selection plates by colony PCR (25 μL final reaction volume).
-
2.
Pick a colony (seeNotes15 and 16) with a standard yellow 200-μL tip, transfer the colony to a YEB master plate containing 50 μg/mL carbenicillin, 25 μg/mL rifampicin, and 25 μg/mL kanamycin and resuspend the colony in 19 μL sterile H2O in a reaction tube.
-
3.
Incubate the master plate for 1–2 days at 28 °C.
-
4.
Prepare a PCR master mix using 0.5 μL of each primer (PS5′ and PS3′) and standard PCR ingredients (10× PCR buffer, dNTPs, and Taq polymerase).
-
5.
Add 6 μL of the PCR master mix to the 19 μL bacterial suspension from step 2.
-
6.
Include pTRAkc plasmid DNA as a positive control and H2O as a negative control, respectively.
-
7.
Run the PCR program shown in Table 1.
Table 1 PCR conditions to identify positive A.tumefaciens colonies -
8.
Analyze PCR products by gel electrophoresis to identify positive recombinant A. tumefaciens clones.
3.6 Cultivation of Recombinant A. tumefaciens Cells
-
1.
Inoculate 3 mL YEB containing 50 μg/mL carbenicillin, 25 μg/mL rifampicin, and 25 μg/mL kanamycin with a recombinant A. tumefaciens colony (seeNote17).
-
2.
Incubate at 28 °C and 160 rpm for 48 h.
-
3.
Enlarge the culture to an appropriate volume (seeNote17) by adding YEB containing 50 μg/mL carbenicillin, 25 μg/mL rifampicin, and 25 μg/mL kanamycin and cultivate the culture at 28 °C and 160 rpm for 24–48 h to achieve an OD600 of 3–6.
-
4.
Prepare glycerol stocks by mixing 500 μL of the culture with 500 μL 100 % (v/v) glycerol and store at −80 °C.
3.7 Preparation of Infiltration Solution
-
1.
Determine the OD600 of the A.tumefaciens culture.
-
2.
Adjust the culture to OD600 = 1 with 2× infiltration medium and an appropriate volume of sterile H2O.
-
3.
Add acetosyringone from 200 mM stock solution in DMSO to a final concentration of 200 μM and incubate the infiltration solution for 30 min at room temperature.
3.8 Cultivation of N. benthamiana Plants
-
1.
Germinate N.benthamiana seeds preferentially on rock wool blocks using a hydroponic culturing system or in standard soil and pots.
-
2.
Irrigate plants with a 0.1 % (w/v) solution of Ferty®-II-Mega in a greenhouse with 25/22 °C day/night temperature, a 16-h photoperiod and 70 % relative humidity.
3.9 Plant Infiltration and Incubation
Two different infiltration and incubation procedures can be used according to the number of expression constructs and the production scale. (a) Syringe-based infiltration of single or multiple leaves using intact plants (Fig. 3, left panel) and (b) vacuum-based infiltration using intact plants (Fig. 3, right panel). The syringe-based infiltration requires much smaller culture volumes (seeNote18) and is more suitable for testing different variants in parallel, whereas vacuum infiltration is typically used for larger-scale production and purification of antigens for detailed characterization (e.g., structural analysis). Note that all work involving genetically modified A.tumefaciens must be carried out in an S1 environment, and all materials should be properly decontaminated according to the applicable regulations (seeNote19).
Infiltration, incubation, and extraction of N. benthamiana. Left panel shows representative images from the syringe infiltration workflow. (a) Syringe infiltration, as the infiltration solution is slowly pushed into the leaf tissue. Darker areas indicate successfully infiltrated regions. (b) The incubation of a N. benthamiana plant (following the syringe infiltration of single leaves) on the shelf of a light cabinet inside a temperature-controlled growth chamber. (c) Small-scale extraction of soluble proteins from leaf tissue using mortar and pestle after syringe infiltration and incubation. Right panel shows representative images from the vacuum infiltration workflow. (A) Vacuum infiltration of a whole plant submerged in infiltration solution, inside a desiccator. (B) After vacuum infiltration, plants are incubated hanging upside down within a light cabinet inside a temperature-controlled growth chamber. (C) Soluble proteins from larger amounts of leaf tissue can be extracted using a commercial homogenizer
3.9.1 Syringe-Based Infiltration
-
1.
Select suitable plants (seeNote20) and prepare them for infiltration by misting (seeNote21).
-
2.
Place the plant on an autoclavable or disposable tray.
-
3.
Wear a laboratory coat and safety glasses.
-
4.
Aspirate 1 mL infiltration solution into a 1-mL syringe without a needle and position the syringe by pressing the tip with moderate pressure against the lower surface of a suitable leaf (seeNote22) close to a main leaf vein. Start infiltrating the solution into the leaf tissue by slowly pushing the solution from the syringe. Infiltrated tissue appears darker and slightly more translucent than non-infiltrated areas (Fig. 3a). Depending on skills and leaf condition, 50–500 μL of solution can be infiltrated into the leaf tissue at one contact point. If the infiltration does not proceed or if the syringe needs to be refilled, change the contact point.
-
5.
Repeat the procedure until the desired number of leaves has been infiltrated.
-
6.
It is possible to use different leaves on the same plant to infiltrate different constructs. In this case, it is important to properly label the leaves and/or contact points and to avoid cross contamination caused by dripping infiltration solution.
-
7.
Transfer plants to a fresh tray and incubate for 3–10 days (Fig. 3b) in a contained growth chamber (16-h photoperiod, 10,000 lx, 22 °C, and 60 % humidity). Check for sufficient watering every 2 days.
3.9.2 Vacuum Infiltration
-
1.
Select suitable plants (seeNote23) and prepare them for infiltration by misting. Make sure that the plants fit into the infiltration vessel (desiccator).
-
2.
Fill a 5-L plastic beaker with 4 L infiltration solution.
-
3.
Carefully invert each plant and lower into the infiltration solution making sure that all leaves are submerged. Use sticks or adhesive tape to prevent the root block from slipping into the solution (Fig. 3A).
-
4.
Place the beaker with the submerged plant into an appropriate 20–20 L desiccator, close the lid and apply an underpressure of <20 mbar using a vacuum pump.
-
5.
Carefully release the vacuum after 5–10 min (seeNote24).
-
6.
Incubate the plants upside down for 3–10 days (Fig. 3B) in a contained growth chamber (16-h photoperiod, 10,000 lx, 22 °C, and 60 % humidity). Check the plants every 2 days for sufficient watering. If the plants appear dry, they should be misted daily.
3.10 Extraction of Total Soluble Proteins
-
1.
Harvest infiltrated leaf material 3–10 days post infiltration (dpi), typically 5 dpi.
-
2.
Weigh the infiltrated leaf material.
-
3.
Grind leaf material to a fine powder in liquid nitrogen using a mortar and pestle for small-scale extraction, and add 2–3 mL of extraction buffer per gram of leaf material (seeNote25).
-
4.
For large-scale extraction, use a blender and mix infiltrated leaves with 3 mL extraction buffer per gram of leaf material.
-
5.
Filter the plant crude extract through a double layer of Miracloth.
-
6.
Centrifuge the extract at 40,000 × g at 4 °C for 15 min to remove insoluble plant compounds.
-
7.
Tobacco crude extract containing total soluble proteins can be used for subsequent analysis, e.g., SDS-PAGE (Fig. 4a) (seeNote26) and immunoblot analysis (Fig. 4b) and/or ELISA . The red fluorescence of DsRed fusion proteins can be observed using a simple red filter with a cold light source and a green excitation filter, in planta (Fig. 4c) or after extraction.
Fig. 4 SDS-PAGE , immunoblot analysis and fluorescence imaging of single and multi-domain malaria vaccine candidates after transient expression in N. benthamiana. SDS-PAGE and immunoblot analysis of plant extracts following the expression of different PfAMA1-DiCo-based single and dual-domain malaria vaccine antigen constructs. (a) The gel was loaded with 6 μL of each sample per lane. M: Page ruler, pre-stained protein marker; 1: Wild-type plant extract; 2: PfAMA1-DiCo1 (61.7 kDa); 3: PfAMA1-DiCo2 (61.7 kDa); 4: PfAMA1-DiCo3 (61.7 kDa); 5: Mixture of PfAMA1-DiCo-1-3 (61.7 kDa); 6: PfAMA1-DiCo1_PfMSP1_19 (72.7 kDa); 7: PfAMA1-DiCo2_PfRH2a (75.3 kDa); 8: PfAMA1-DiCo3_PfRIPR7/8 (70.6 kDa); 9: Mixture of PfAMA1-DiCo1_PfMSP1-19, PfAMA1-DiCo2_PfRH2a and PfAMA1-DiCo3_PfRipr7/8 (70.6–75.3 kDa). Samples were separated under non-reducing conditions on a 4–12 % gradient gel (NuPage, Lifetech, Darmstadt, Germany) and subsequently used for immunoblotting. On the SDS-PAGE gel, proteins were visualized by staining with Coomassie Brilliant Blue (a, left panel), on the blot membrane recombinant proteins were detected using a plant-derived rat–human chimeric version of the PfAMA1-specific monoclonal antibody 4G2 followed by visualization using an alkaline phosphatase-labeled secondary goat anti-human antiserum (a, right panel). (b) Equivalent SDS-PAGE and immunoblot analysis of a series of stacked multi-domain malaria vaccine candidates, with 15 μL of samples loaded per lane. M: Page ruler, pre-stained protein marker; a: Wild-type plant extract; b: PfMSP1-19_EGF1 (6.8 kDa); c: PfMSP1-19_EGF1-PfMSP8_EGF1 (12.3 kDa); d: PfMSP1-19_EGF1-PfMSP8_EGF1/2 (17.0 kDa); e: PfMSP1-19_EGF1-PfMSP8_EGF1/2-PfMSP4_EGF (22.6 kDa); f: PfMSP1-19_EGF1-PfMSP8_EGF1/2-PfMSP4_EGF-PfMSP10_EGF1 (28.1 kDa); g: PfMSP1-19_EGF1-PfMSP8_EGF1/2-PfMSP4_EGF-PfMSP10_EGF1/2 (32.7 kDa); Fig. 4 (continued) h: PfMSP1-19_ EGF1-PfMSP8_EGF1/2-PfMSP4_EGF-PfMSP10_EGF1/2-Pfs25 (51.2 kDa); i: PfMSP1-19_EGF1-PfMSP8_EGF1/2-PfMSP4_EGF-PfMSP10_EGF1/2-Pfs25-PfCSP_TSR (58.8 kDa); j: Pfs28-PfMSP1-19_EGF1-PfMSP8_EGF1/2_PfMSP4_EGF_PfMSP10_EGF1/2 _Pfs25 (69.9 kDa); k: Pfs28_PfMSP1-19_EGF1-PfMSP8_EGF1/2-PfMSP4_EGF-PfMSP10_EGF1/2-Pfs25-PfCSP_TSR. Samples were separated in each lane under non-reducing conditions on a 4–12 % (w/v) gradient gel (NuPage, Lifetech) and subsequently used for immunoblotting. (c) Expression of DsRed-antigen fusions can easily be visualized. 1: Non-infiltrated N.benthamiana leaf under white light (left side) and under green light visualized through a red filter (right side), 2: N. benthamiana leaf infiltrated with DsRed-PfMSP1-19 after 5 days of incubation, under white light (left side) and under green light visualized through a red filter (right side)
-
8.
For purification, adjust the pH of the extract as appropriate and pass the extract through a 0.45-μm filter to avoid clogging the column. The purification strategy for each antigen is highly dependent on its intrinsic properties (seeNote27). However, the crude tobacco extract is compatible with most conventional chromatography resins and strategies such as immobilized metal ion affinity chromatography (IMAC) .
4 Notes
4.1 Cloning Notes
-
1.
Depending on the origin of the vaccine antigens the preparation of synthetic genes adapted to the codon usage of N.benthamiana may be beneficial. AT-rich P.falciparum genes in particular may suffer from low expression levels if native cDNA sequences are used. Transient expression using pTRAkc-ERH expression plasmids will target recombinant proteins for retention in the endoplasmic reticulum (ER) of the plant cell because the vector provides an N-terminal signal peptide sequence and a C-terminal KDEL-ER retrieval sequence. Therefore, surface-exposed recognition sites for N-linked glycosylation will be post-translationally modified with high-mannose type glycans. Many P.falciparum proteins, including many vaccine candidate antigens, contain N-linked glycosylation sites that are not used in the native context because the parasite lacks the corresponding glycosylation machinery [20]. It may therefore be necessary to knock out such N-linked glycosylation sites for the expression of recombinant proteins or protein domains equipped with ER-targeting and retention signals. Depending on the characteristics of the selected antigens, it may be useful to target other subcellular compartments such as the cytosol, vacuole, or plastids. Secreted proteins that require oxidative folding or assembly into homomeric or heteromeric oligomers are suitable for ER-targeting using the pTRAkc-ERH vector, whereas cytosolic proteins lacking disulfide bridges and P.falciparum proteins from which the N-linked glycosylation sites cannot be removed are candidates for cytosolic targeting using another variant of the pTRA vector lacking the signal peptide and ER-retention sequence [21].
-
2.
The gene stacking strategy takes advantage of the compatibility of the single strand 5′ overhangs created by both EagI and NotI. Whereas NotI has an 8-bp recognition site, EagI has a 6-bp recognition site and these features can be used to prevent the reconstitution of a NotI recognition site following the insertion of an EagI-digested fragment. EagI digestion at the 5′ end and NotI digestion at the 3′ end of the stacked insert shifts the NotI site used for stacking to the 3′ end of the proximal gene. Two insert orientations are possible and this can be determined by PCR, using an appropriate combination of PS5′, PS3′, and insert-specific primers that will only generate a product for correct insertion events. Alternatively an NcoI/NotI double digest of the parental and recombinant plasmids will produce identical fragments if the insert is inverted (because the NotI restriction site is reconstituted at the original position), whereas a recombinant plasmid with the correct insert will yield a longer fragment than the parental plasmid.
4.2 Agrobacterium tumefaciens Notes
-
1.
A. tumefaciens regenerates and grows slowly compared to E.coli , especially when inoculated from single colonies. The regeneration times indicated in the protocol should be followed. When inoculating A. tumefaciens cultures from glycerol stocks, at least 50–100 μL should be used to prevent prolonged cultivation times.
-
2.
Strictly avoid incubating A. tumefaciens at temperatures exceeding 30 °C because this will lead to the loss of plasmids and thereby reduce the quality and reproducibility of transient expression experiments.
-
3.
Selection plates containing carbenicillin, rifampicin, and kanamycin should be stored at 4 °C for no longer than 10 days.
-
4.
The transformation of electrocompetent A. tumefaciens cells with pTRA variants is usually highly efficient. Follow the recommendations regarding plasmid amounts and volumes used for plating the transformed cells to avoid overgrown selection plates.
-
5.
After transformation, do not selectively pick the largest or the smallest colonies on the plate.
-
6.
Do not use wooden toothpicks for the inoculation of liquid cultures with A. tumefaciens because phenolic compounds in the stick may inhibit bacterial growth.
-
7.
For troubleshooting, use A. tumefaciens transformed with pTRAkc-DsRedERH as a reporter plasmid.
-
8.
Adjust the size of the A. tumefaciens culture prepared for infiltration according to the amount of leaf tissue that will be infiltrated and the selected infiltration technique. A 20-mL culture usually yields >50 mL infiltration solution sufficient for at least eight leaves using syringe infiltration. When using vacuum infiltration for larger numbers of leaves or whole plants, 4–5 L of infiltration solution is usually required so prepare 500–1000 mL of A. tumefaciens culture. Pre-cultures can be expanded up to 100-fold in one step.
4.3 Plant Infiltration Notes
-
1.
To ensure optimal yields in transient expression experiments, avoid the use of plants that show extensive flowering or clear signs of senescence (crinkled leaves or browning).
-
2.
Spraying plants with water mist 20–30 min before starting the infiltration procedure improves the infiltration efficiency, especially when working with syringe infiltration.
-
3.
When aiming to achieve high expression levels on a small scale (e.g., to produce, purify and compare several construct variants), vital leaves of medium age should be chosen. Using the largest lower leaves will generally not increase the protein yield or integrity.
-
4.
Syringe infiltration is useful for the parallel testing of many construct variants, but proper and efficient infiltration of the leaf tissue does require some practice. Do not apply too much force when contacting the lower leaf surface for injection. Infiltrate the A.tumefaciens suspension using moderate constant pressure, carefully observe the infiltration of the tissue and proceed to a new contact site if necessary. Wear safety glasses and protective clothing because the A. tumefaciens suspension may sputter from the stomata during infiltration.
-
5.
Bubbles will be released from the submerged plant tissues at the beginning of vacuum infiltration. For optimal infiltration, vacuum incubation should continue until bubble formation has ceased. After releasing the vacuum, check the plant for proper infiltration—the infiltrated tissue appears translucent.
-
6.
The whole procedure of A.tumefaciens infiltration must be carried out under containment in an appropriate S1 environment.
4.4 Extraction Notes
-
1.
The efficiency of target protein extraction is strongly dependent on the composition of the extraction buffer. The highest extraction efficiency (as a function of total soluble protein content) is achieved with an extraction buffer at neutral pH (7–8) and declines under more acidic or basic extraction conditions. Under basic conditions (especially pH > 8.0), the extract becomes brown as a result of increased enzymatic oxidation and the formation of polyphenolic compounds. Such compounds can hamper subsequent purification steps by promoting the fouling of chromatography resins thus reducing the resin capacity. These issues can be addressed by including an antioxidant such sodium metabisulfite (final concentration 10 mM) in the extraction buffer. Adding NaCl and increasing the conductivity can further increase the solubility of the target protein.
-
2.
The most abundant protein in the crude extract is ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO), a multi-protein complex comprising eight large subunits (approximately 58 kDa) and eight small subunits (approximately 18 kDa).
-
3.
A heat incubation step can be used to reduce the abundance of plant-derived host cell proteins in the extract. Incubation at 70 °C for 10 min typically removes up to 80 % of plant total soluble proteins and may simplify downstream. The applicability of this step depends on the temperature stability of the recombinant target protein. To avoid unfolding of proteins with internal disulfide bridges, an extraction buffer without reducing agents like sodium metabisulfite is highly recommended when applying the heat incubation step.
4.5 Description of the Proteins Used in the Case Studies
-
1.
The three so-called diversity covering (DiCo) variants of the apical membrane antigen Pf AMA1 have been developed [22] to cover the high allelic diversity of this promising vaccine candidate with a small number of antigens, aiming to elicit cross-strain-specific parasite growth inhibitory immune responses [23].
-
2.
Pf MSP1_19 is an 11-kDa fragment of an abundant blood-stage antigen, the structural motif occurring in many P.falciparum blood-stage antigens. Pf MSP1_19 remains on the surface of the merozoite during erythrocyte invasion underlining the relevance of this antigen as a blood-stage vaccine candidate [24, 25].
-
3.
The RH5 interacting protein (Pf Ripr) is a recently discovered P.falciparum surface protein that forms a complex with PfRH5 and plays a role in the sialic acid-independent erythrocyte invasion pathway [26]. The protein features ten EGF-like domains, and animal studies have indicated that the combination of EFG-7 and EFG-8 is a primary target of protective antibody responses [26].
-
4.
As a member of the P.falciparum reticulocyte binding homologs (PfRH), Pf RH2 is believed to play a role in the sialic acid-independent erythrocyte invasion pathway and studies have shown that IgGs recognizing the full-size as well as various smaller fragments (e.g., Pf RH2a) of the protein correlate with protection from symptomatic malaria and high-density parasitemia [27].
-
5.
The merozoite surface protein 4 (Pf MSP4) is a highly conserved [28], abundant protein found on the surface of P.falciparum merozoites [29]. Immunization with a P. yoelii homolog of MSP4 resulted in protection from lethal parasite challenge [30]. The C-terminal EGF-like domain is recognized by human immune sera [31].
-
6.
Pf MSP8 is another conserved merozoite surface protein that contains EGF-like domains and has been proposed as potential blood-stage vaccine candidate [32].
-
7.
Pf MSP10 is another merozoite surface protein and potential blood-stage vaccine candidate [33, 34] featuring two EGF-like domains.
-
8.
The two closely related P.falciparum ookinete surface proteins Pfs25 [35] and Pfs28 [36] have been shown to elicit transmission-blocking antibodies in animal experiments and are among the leading transmission-blocking vaccine candidates. Both proteins contain four EGF-like domains.
-
9.
The circumsporozoite antigen PfCSP [37, 38] is the major component of the promising malaria vaccine candidate RTS,S and is regarded as an essential pre-erythrocytic antigen. The C-terminal thrombospondin-related domain Pf CSP_TSR [39] represents a defined structural entity that Pf CSP shares with other pre-erythrocytic antigens such as Pf TRAP, and has been selected as pre-erythrocytic component for the multi-stage vaccine candidates used in our case studies.
-
10.
Variants of the red fluorescent protein (RFP) DsRed , initially isolated from the mushroom coral Discosoma sp. [40–43], are commonly used as fluorescent marker proteins. DsRed forms a homomeric tetramer and the mature protein has an excitation optimum of 554 nm and maximum emission at 554 nm. DsRed and its fusion proteins bind copper, and therefore can easily be purified by copper-IMAC . In most cases, DsRed tolerates C-terminal fusions and its strong fluorescence allows the expression screening of antigen domain libraries as well as the sorting of antigen specific B-cell populations taking additional advantage of increased avidity resulting from the multivalent presentation within the context of the DsRed tetramer.
5 Notes
-
1.
The three so-called diversity covering (DiCo) variants of the apical membrane antigen PfAMA1 have been developed [22] to cover the high allelic diversity of this promising vaccine candidate with a small number of antigens, aiming to elicit cross-strain-specific parasite growth inhibitory immune responses [23].
-
2.
Pf MSP1_19 is an 11-kDa fragment of an abundant blood-stage antigen, the structural motif occurring in many P. falciparum blood-stage antigens. PfMSP1_19 remains on the surface of the merozoite during erythrocyte invasion underlining the relevance of this antigen as a blood-stage vaccine candidate [24, 25]
-
3.
The RH5 interacting protein (PfRipr) is a recently-discovered P. falciparum surface protein that forms a complex with PfRH5 and plays a role in the sialic acid-independent erythrocyte invasion pathway [26]. The protein features 10 EGF-like domains, and animal studies have indicated that the combination of EFG-7 and EFG-8 is a primary target of protective antibody responses [26].
-
4.
As a member of the P. falciparum reticulocyte binding homologs (PfRH), PfRH2 is believed to play a role in the sialic acid-independent erythrocyte invasion pathway and studies have shown that IgGs recognizing the full-size as well as various smaller fragments (e.g. PfRH2a) of the protein correlate with protection from symptomatic malaria and high-density parasitemia [27].
-
5.
The merozoite surface protein 4 (PfMSP4) is a highly-conserved [28], abundant protein found on the surface of P. falciparum merozoites [29]. Immunization with a P. yoelii homolog of MSP4 resulted in protection from lethal parasite challenge [30]. The C-terminal EGF-like domain is recognized by human immune sera [31].
-
6.
Pf MSP8 is another conserved merozoite surface protein that contains EGF-like domains and has been proposed as potential blood-stage vaccine candidate [32].
-
7.
Pf MSP10 is another merozoite surface protein and potential blood-stage vaccine candidate [33, 34] featuring two EGF-like domains.
-
8.
The two closely related P. falciparum ookinete surface proteins Pfs25 [35] and Pfs28 [36] have been shown to elicit transmission-blocking antibodies in animal experiments and are among the leading transmission-blocking vaccine candidates. Both proteins contain four EGF-like domains.
-
9.
The circumsporozoite antigen PfCSP [37, 38] is the major component of the promising malaria vaccine candidate RTS,S and is regarded as an essential pre-erythrocytic antigen. The C-terminal thrombospondin-related domain PfCSP_TSR [39] represents a defined structural entity that PfCSP shares with other pre-erythrocytic antigens such as PfTRAP, and has been selected as pre-erythrocytic component for the multi-stage vaccine candidates used in our case studies.
-
10.
Variants of the red fluorescent protein (RFP) DsRed, initially isolated from the mushroom coral Discosoma sp. [40–43], are commonly used as fluorescent marker proteins. DsRed forms a homomeric tetramer and the mature protein has an excitation optimum of 554 nm and maximum emission at 554 nm. DsRed and its fusion proteins bind copper, and therefore can easily be purified by copper-IMAC. In most cases, DsRed tolerates C-terminal fusions and its strong fluorescence allows the expression screening of antigen domain libraries as well as the sorting of antigen specific B-cell populations taking additional advantage of increased avidity resulting from the multivalent presentation within the context of the DsRed tetramer.
-
11.
Depending on the origin of the vaccine antigens the preparation of synthetic genes adapted to the codon usage of N. benthamiana may be beneficial. AT-rich P. falciparum genes in particular may suffer from low expression levels if native cDNA sequences are used. Transient expression using pTRAkc-ERH expression plasmids will target recombinant proteins for retention in the endoplasmic reticulum (ER) of the plant cell because the vector provides an N-terminal signal peptide sequence and a C-terminal KDEL-ER retrieval sequence. Therefore, surface-exposed recognition sites for N-linked glycosylation will be post-translationally modified with high-mannose type glycans. Many P. falciparum proteins, including many vaccine candidate antigens, contain N-linked glycosylation sites that are not used in the native context because the parasite lacks the corresponding glycosylation machinery [20]. It may therefore be necessary to knock out such N-linked glycosylation sites for the expression of recombinant proteins or protein domains equipped with ER-targeting and retention signals. Depending on the characteristics of the selected antigens, it may be useful to target other subcellular compartments such as the cytosol, vacuole or plastids. Secreted proteins that require oxidative folding or assembly into homomeric or heteromeric oligomers are suitable for ER-targeting using the pTRAkc-ERH vector, whereas cytosolic proteins lacking disulfide bridges and P. falciparum proteins from which the N-linked glycosylation sites cannot be removed are candidates for cytosolic targeting using another variant of the pTRA vector lacking the signal peptide and ER-retention sequence [21].
-
12.
The gene stacking strategy takes advantage of the compatibility of the single strand 5’ overhangs created by both EagI and NotI. Whereas NotI has an 8-bp recognition site, EagI has a 6-bp recognition site and these features can be used to prevent the reconstitution of a NotI recognition site following the insertion of an EagI-digested fragment. EagI digestion at the 5’ end and NotI digestion at the 3’ end of the stacked insert shifts the NotI site used for stacking to the 3’ end of the proximal gene. Two insert orientations are possible and this can be determined by PCR, using an appropriate combination of PS5’, PS3’ and insert-specific primers that will only generate a product for correct insertion events. Alternatively an NcoI/NotI double digest of the parental and recombinant plasmids will produce identical fragments if the insert is inverted (because the NotI restriction site is reconstituted at the original position) whereas a recombinant plasmid with the correct insert will yield a longer fragment than the parental plasmid.
-
13.
A. tumefaciens regenerates and grows slowly compared to E. coli, especially when inoculated from single colonies. The regeneration times indicated in the protocol should be followed. When inoculating A. tumefaciens cultures from glycerol stocks, at least 50–100 μL should be used to prevent prolonged cultivation times.
-
14.
Strictly avoid incubating A. tumefaciens at temperatures exceeding 30°C because this will lead to the loss of plasmids and thereby reduce the quality and reproducibility of transient expression experiments.
-
15.
Selection plates containing carbenicillin, rifampicin and kanamycin should be stored at 4°C for no longer than 10 days.
-
16.
After transformation, do not selectively pick the largest or the smallest colonies on the plate.
-
17.
Do not use wooden toothpicks for the inoculation of liquid cultures with A. tumefaciens because phenolic compounds in the stick may inhibit bacterial growth.
-
18.
Adjust the size of the A. tumefaciens culture prepared for infiltration according to the amount of leaf tissue that will be infiltrated and the selected infiltration technique. A 20-mL culture usually yields >50 mL infiltration solution sufficient for at least eight leaves using syringe infiltration. When using vacuum infiltration for larger numbers of leaves or whole plants, 4-5 L of infiltration solution is usually required so prepare 500-1000 mL of A. tumefaciens culture. Pre-cultures can be expanded up to 100-fold in one step.
-
19.
The whole procedure of A. tumefaciens infiltration must be carried out under containment in an appropriate S1 environment.To ensure optimal yields in transient expression experiments, avoid the use of plants that show extensive flowering or clear signs of senescence (crinkled leaves or browning).
-
20.
Spraying plants with water mist 20–30 min before starting the infiltration procedure improves the infiltration efficiency, especially when working with syringe infiltration.
-
21.
Syringe infiltration is useful for the parallel testing of many construct variants, but proper and efficient infiltration of the leaf tissue does require some practice. Do not apply too much force when contacting the lower leaf surface for injection. Infiltrate the A. tumefaciens suspension using moderate constant pressure, carefully observe the infiltration of the tissue and proceed to a new contact site if necessary. Wear safety glasses and protective clothing because the A. tumefaciens suspension may sputter from the stomata during infiltration.
-
22.
When aiming to achieve high expression levels on a small scale (e.g. to produce, purify and compare several construct variants), vital leaves of medium age should be chosen. Using the largest lower leaves will generally not increase the protein yield or integrity.
-
23.
Bubbles will be released from the submerged plant tissues at the beginning of vacuum infiltration. For optimal infiltration, vacuum incubation should continue until bubble formation has ceased. After releasing the vacuum, check the plant for proper infiltration – the infiltrated tissue appears translucent.
-
24.
The efficiency of target protein extraction is strongly dependent on the composition of the extraction buffer. The highest extraction efficiency (as a function of total soluble protein content) is achieved with an extraction buffer at neutral pH (7–8) and declines under more acidic or basic extraction conditions. Under basic conditions (especially pH >8.0), the extract becomes brown as a result of increased enzymatic oxidation and the formation of polyphenolic compounds. Such compounds can hamper subsequent purification steps by promoting the fouling of chromatography resins thus reducing the resin capacity. These issues can be addressed by including an antioxidant such sodium metabisulfite (final concentration 10 mM) in the extraction buffer. Adding NaCl and increasing the conductivity can further increase the solubility of the target protein.
-
25.
The most abundant protein in the crude extract is ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO), a multi-protein complex comprising eight large subunits (approximately 58 kDa) and eight small subunits (approximately 18 kDa).
-
26.
A heat incubation step can be used to reduce the abundance of plant-derived host cell proteins in the extract. Incubation at 70°C for 10 min typically removes up to 80% of plant total soluble proteins and may simplify downstream. The applicability of this step depends on the temperature stability of the recombinant target protein. To avoid unfolding of proteins with internal disulfide bridges, an extraction buffer without reducing agents like sodium metabisulfite is highly recommended when applying the heat incubation step.
References
Crompton PD, Pierce SK, Miller LH (2010) Advances and challenges in malaria vaccine development. J Clin Invest 120:4168–4178
Vaughan AM, Kappe SHI (2012) Malaria vaccine development: persistent challenges. Curr Opin Immunol 24:324–331
Hill AVS (2011) Vaccines against malaria. Philos Trans R Soc Lond B Biol Sci 366:2806–2814
Davey MR, Gartland KM, Mulligan BJ (1986) Transformation of the genomic expression of plant cells. Symp Soc Exp Biol 40:85–120
Gleba Y, Klimyuk V, Marillonnet S (2007) Viral vectors for the expression of proteins in plants. Curr Opin Biotechnol 18:134–141
Pogue GP, Vojdani F, Palmer KE et al (2010) Production of pharmaceutical-grade recombinant aprotinin and a monoclonal antibody product using plant-based transient expression systems. Plant Biotechnol J 8:638–654
Gleba YY, Klimyuk VV, Marillonnet SS (2005) Magnifection—a new platform for expressing recombinant vaccines in plants. Vaccine 23:2042–2048
Santi L, Giritch A, Roy CJ et al (2006) Protection conferred by recombinant Yersinia pestis antigens produced by a rapid and highly scalable plant expression system. Proc Natl Acad Sci U S A 103:861–866
Olinger GG, Pettitt J, KIM D et al (2012) Delayed treatment of Ebola virus infection with plant-derived monoclonal antibodies provides protection in rhesus macaques. Proc Natl Acad Sci U S A 109:18030–18035
D’Aoust M-A, Couture MMJ, Charland N et al (2010) The production of hemagglutinin-based virus-like particles in plants: a rapid, efficient and safe response to pandemic influenza. Plant Biotechnol J 8:607–619
Jones RM, Chichester JA, Manceva S et al (2015) A novel plant-produced Pfs25 fusion subunit vaccine induces long-lasting transmission blocking antibody responses. Hum Vaccin Immunother 11:124–132
Klimyuk V, Pogue G, Herz S et al (2014) Production of recombinant antigens and antibodies in Nicotiana benthamiana using “magnifection” technology: GMP-compliant facilities for small- and large-scale manufacturing. Curr Top Microbiol Immunol 375:127–154
Chichester JA, Manceva SD, Rhee A et al (2013) A plant-produced protective antigen vaccine confers protection in rabbits against a lethal aerosolized challenge with Bacillus anthracis Ames spores. Hum Vaccin Immunother 9:544–552
Voepel N, Boes A, Edgue G et al (2014) Malaria vaccine candidate antigen targeting the pre erythrocytic stage of Plasmodium falciparum produced at high-level in plants. Biotechnol J 9:1435–1445
Boes A, Spiegel H, Edgue G et al (2015) Detailed functional characterization of glycosylated and nonglycosylated variants of malaria vaccine candidate PfAMA1 produced in Nicotiana benthamiana and analysis of growth inhibitory responses in rabbits. Plant Biotechnol J 13:222–234
Beiss V, Spiegel H, Boes A et al (2015) Heat-precipitation allows the efficient purification of a functional plant-derived malaria transmission-blocking vaccine candidate fusion protein. Biotechnol Bioeng 112:1297–1305
Buyel JF, Gruchow HM, Boes A, Fischer R (2014) Rational design of a host cell protein heat precipitation step simplifies the subsequent purification of recombinant proteins from tobacco. Biochem Eng J 88:162–170
Koncz C, Schell J (1986) The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204:383–396
Maclean J, Koekemoer M, Olivier AJ et al (2007) Optimization of human papillomavirus type 16 (HPV-16) L1 expression in plants: comparison of the suitability of different HPV-16 L1 gene variants and different cell-compartment localization. J Gen Virol 88:1460–1469
von Itzstein M, Plebanski M, Cooke BM, Coppel RL (2008) Hot, sweet and sticky: the glycobiology of Plasmodium falciparum. Trends Parasitol 24:210–218
Wandelt CIC, Khan MRM, Craig SS et al (1992) Vicilin with carboxy-terminal KDEL is retained in the endoplasmic reticulum and accumulates to high levels in the leaves of transgenic plants. Plant J 2:181–192
Remarque EJE, Faber BWB, Kocken CHMC, Thomas AWA (2008) A diversity-covering approach to immunization with Plasmodium falciparum apical membrane antigen 1 induces broader allelic recognition and growth inhibition responses in rabbits. Infect Immun 76:2660–2670
Faber BW, Younis S, Remarque EJ et al (2013) Diversity covering AMA1-MSP119 fusion proteins as malaria vaccines. Infect Immun 81:1479–1490
Chappel JA, Egan AF, Riley EM et al (1994) Naturally acquired human antibodies which recognize the first epidermal growth factor-like module in the Plasmodium falciparum merozoite surface protein 1 do not inhibit parasite growth in vitro. Infect Immun 62:4488–4494
Kumar S, Collins W, Egan A et al (2000) Immunogenicity and efficacy in aotus monkeys of four recombinant Plasmodium falciparum vaccines in multiple adjuvant formulations based on the 19-kilodalton C terminus of merozoite surface protein 1. Infect Immun 68:2215–2223
Chen L, Lopaticki S, Riglar DT et al (2011) An EGF-like protein forms a complex with PfRh5 and is required for invasion of human erythrocytes by Plasmodium falciparum. PLoS Pathog 7:e1002199
Reiling L, Richards JS, Fowkes FJI et al (2010) Evidence that the erythrocyte invasion ligand PfRh2 is a target of protective immunity against Plasmodium falciparum malaria. J Immunol 185:6157–6167
Wang LL, Richie TLT, Stowers AA et al (2001) Naturally acquired antibody responses to Plasmodium falciparum merozoite surface protein 4 in a population living in an area of endemicity in Vietnam. Infect Immun 69:4390–4397
Gilson PR, Nebl T, Vukcevic D et al (2006) Identification and stoichiometry of glycosylphosphatidylinositol-anchored membrane proteins of the human malaria parasite Plasmodium falciparum. Mol Cell Proteomics 5:1286–1299
Goschnick MW, Black CG, Kedzierski L (2004) Merozoite surface protein 4/5 provides protection against lethal challenge with a heterologous malaria parasite strain. Infect Immun 72:5840–5849
de Silva HD, Saleh S, Kovacevic S et al (2011) The antibody response to Plasmodium falciparum Merozoite Surface Protein 4: comparative assessment of specificity and growth inhibitory antibody activity to infection-acquired and immunization-induced epitopes. Malar J 10:266
Black CG, Wu T, Wang L et al (2001) Merozoite surface protein 8 of Plasmodium falciparum contains two epidermal growth factor-like domains. Mol Biochem Parasitol 114:217–226
Black CG, Wang L, Wu T, Coppel RL (2003) Apical location of a novel EGF-like domain-containing protein of Plasmodium falciparum. Mol Biochem Parasitol 127:59–68
Puentes A, Ocampo M, Rodríguez LE, Vera R (2005) Identifying Plasmodium falciparum merozoite surface protein-10 human erythrocyte specific binding regions. Biochimie 87:461–472
Vermeulen AN (1985) Sequential expression of antigens on sexual stages of Plasmodium falciparum accessible to transmission-blocking antibodies in the mosquito. J Exp Med 162:1460–1476
Duffy PE, Kaslow DC (1997) A novel malaria protein, Pfs28, and Pfs25 are genetically linked and synergistic as falciparum malaria transmission-blocking vaccines. Infect Immun 65:1109–1113
Yoshida N, Nussenzweig RS, Potocnjak P et al (1980) Hybridoma produces protective antibodies directed against the sporozoite stage of malaria parasite. Science 207:71–73
Nussenzweig V, Nussenzweig RS (1985) Circumsporozoite proteins of malaria parasites. Cell 42:401–403
Plassmeyer ML, Reiter K, Shimp RL et al (2009) Structure of the Plasmodium falciparum circumsporozoite protein, a leading malaria vaccine candidate. J Biol Chem 284:26951–26963
Campbell RE, Tour O, Palmer AE (2002) A monomeric red fluorescent protein. Proc Natl Acad Sci U S A 99:7877–7882
Wall MA, Socolich M, Ranganathan R (2000) The structural basis for red fluorescence in the tetrameric GFP homolog DsRed. Nat Struct Biol 7:1133–1138
Shaner NC, Campbell RE, Steinbach PA et al (2004) Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 22:1567–1572
Bevis BJ, Glick BS (2002) Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed). Nat Biotechnol 20:83–87
Acknowledgements
We thank Dr. Thomas Rademacher for cloning the pTRAkc vector series. The chimeric antibody 4G2 was kindly provided by Stefan Menzel. The work described in this chapter was partly supported by the Fraunhofer Future Foundation via the malaria vaccine project “Innovative technologies to manufacture ground-breaking biopharmaceutical products in microbes and plants”.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer Science+Business Media New York
About this protocol
Cite this protocol
Boes, A., Reimann, A., Twyman, R.M., Fischer, R., Schillberg, S., Spiegel, H. (2016). A Plant-Based Transient Expression System for the Rapid Production of Malaria Vaccine Candidates. In: Thomas, S. (eds) Vaccine Design. Methods in Molecular Biology, vol 1404. Humana, New York, NY. https://doi.org/10.1007/978-1-4939-3389-1_39
Download citation
DOI: https://doi.org/10.1007/978-1-4939-3389-1_39
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-4939-3388-4
Online ISBN: 978-1-4939-3389-1
eBook Packages: Springer Protocols