Abstract
Polioviruses are enteric viruses that cause paralytic poliomyelitis in less than 0.5 % of infections and are asymptomatic in >90 % infections of naïve hosts. Environmental surveillance monitors polio in populations rather than in individuals. When this very low morbidity to infection ratio, drops drastically in highly vaccinated populations, environmental surveillance employing manual or automatic sampling coupled with molecular analysis carried out in well-equipped central laboratories becomes the surveillance method of choice since polioviruses are excreted by infected individuals regardless of whether or not the infection is symptomatic. This chapter describes a high throughput rapid turn-around time method for molecular characterization of polioviruses from sewage. It is presented in five modules: (1) Sewage collection and concentration of the viruses in the sewage; (2) Cell cultures for identification of virus in the concentrated sewage; (3) Nucleic acid extractions directly from sewage and from tissue cultures infected with aliquots of concentrated sewage; (4) Nucleic Acid Amplification for poliovirus serotype identification and intratypic differentiation (discriminating wild and vaccine derived polioviruses form vaccine strains); and (5) Molecular characterization of viral RNA by qRT-PCR, TR-PCR, and Sequence analysis. Monitoring silent or symptomatic transmission of vaccine-derived polioviruses or wild polioviruses is critical for the endgame of poliovirus eradication. We present methods for adapting standard kits and validating the changes for this purpose based on experience gained during the recent introduction and sustained transmission of a wild type 1 poliovirus in Israel in 2013 in a population with an initial IPV vaccine coverage >90 %.
Access provided by CONRICYT – Journals CONACYT. Download protocol PDF
Similar content being viewed by others
Key words
- Environmental surveillance
- RNA purification
- RT-PCR
- qRT-PCR
- Sequence analysis
- Intratypic differentiation
- Poliovirus
- Wild poliovirus
- Vaccine-derived poliovirus (VDPV)
- Oral polio vaccine (OPV)
- Tissue culture
- Plaque assay
1 Introduction
Polioviruses like other member of the Picornaviridae are encapsidated in an icosahedral structure formed from 60 capsomeres containing one copy each of viral capsid proteins 1 through 4, (VP1, VP2, VP3, and VP4 [1]). There are three serotypes of poliovirus [1, 2]. Poliovirus can cause irreversible paralysis of the infected host and even death, however, most poliovirus infections are asymptomatic [1]. The most common and most efficient route of host-to-host transmission is fecal–oral but poliovirus can also be transmitted by oral–oral transmission [3]. Excretion, duration of excretion, and the amount excreted are dependent on host factors and on vaccination history of the infected individual [2, 3]. A poliovirus is designated as polio vaccine-like, vaccine derived poliovirus (VDPV), or wild poliovirus based on whether the molecular sequence of its VP1 differs from that of the homologous Sabin serotype in live oral polio vaccine (OPV) by ≤1 % (or <0.6 % for serotype 2), from 1 to 15–18 %, or >20 %, respectively [4].
The Global Poliovirus Eradication Initiative (GPEI), launched at the World Health Assembly (WHA, resolution WHA41.28) in 1988, is the single largest, internationally coordinated public health project the world has ever known. Early detection of highly pathogenic infectious organisms such as polio is essential for containing the spread of these organisms. Early detection and measurement of efficacy of response requires high quality surveillance over extended periods of time. Acute flaccid paralysis (AFP) surveillance, is based on monitoring infections in individuals and is the Gold Standard for poliovirus surveillance. AFP surveillance is based on investigating all cases of AFP in children under 15 to rule-in or rule-out poliovirus etiology. Two trivalent vaccines (trivalent since they contain vaccine strains for each or the three poliovirus serotypes) have been used for eradication; IPV formulated from inactivated neurovirulent polioviruses and OPV a vaccine formulated from attenuated live polioviruses. A region is considered to be poliovirus free when over a period of 3 years, the number of AFP cases is equivalent to the expected incidence due to non-poliovirus causes (1 per 100,000 children) and none of the investigated cases were due to poliovirus.
In contrast to AFP surveillance, environmental surveillance for poliovirus is based on the monitoring of poliovirus transmission in human populations by examining environmental samples that contain human feces. The rationale for environmental surveillance is based on the fact that all poliovirus-infected individuals shed large amounts of poliovirus in their feces for several weeks whether or not they have symptoms [5]. The protein capsid is relatively stable enabling poliovirus viruses to remain viable in the environment at ambient temperatures [3, 5]. The length of time when viruses remain viable is extended when the virus adsorbs to solids in sewage [3, 5]. The probability of detecting poliovirus in environmental samples [6] depends on the duration and amount of poliovirus excreted by one or more infected individuals, the effect of physical and mechanical factors on the dilution and survival of poliovirus in the sewage system (reviewed by Dowdle [3]), the frequency of collection and laboratory processing of the environmental samples [7] and the location of the excretor relative to the sample site [5].
Poliovirus may be recovered quantitatively from the environment [8–10]. Two complementary assays for quantifying poliovirus are presented here, plaque assay and quantitative (or semiquantitative) reverse transcription polymerase chain reaction (qRT-PCR). Decreasing the distance between the excretor or excretors and the sample site is usually more effective in increasing the probability of detection, the amount of virus detected and is less labor intensive and more cost efficient than increasing the frequency of sampling [11].
When the very low morbidity to infection ratio (<1:200) of poliovirus infections in naïve populations drops drastically in highly vaccinated populations, environmental surveillance employing manual or automatic sampling coupled with molecular analysis carried out in well-equipped central laboratories becomes the surveillance method of choice. In fact environmental surveillance has been used [5]: to determine extent of a poliovirus outbreak in a population; pinpoint the putative reservoirs and/or epicenters of wild or vaccine-derived polioviruses; to calculate the risk for emergence and transmission of vaccine-derived viruses after immunization campaigns switch from inclusion of live vaccines to exclusive use of inactivated poliovirus vaccine; to screen for unidentified persistently infected asymptomatic individuals in a given population; to monitor for introduction of wild or vaccine-derived viruses into poliomyelitis-free regions; and to determine whether there is sustained person-to-person transmission after introduction of a non-vaccine poliovirus. Environmental surveillance will play an increasingly important role in quality assurance as the amount of wastewater reclamation for agriculture, recreation and drinking purposes grows [5]. Finally, environmental surveillance will also play a critical role in post-eradication surveillance in demonstrating the absence of wild and vaccine strains and for detecting reemergence of poliovirus (see Note 1 ).
Standard protocols and algorithms for determining the serotype of a poliovirus, e.g., typic differentiation (TD) and whether the isolate is vaccine-like, VDPV, or wild, e.g., intratypic differentiation (ITD) of poliovirus from tissue culture by molecular means are provided in detail in the WHO Polio Laboratory Manual 4th edition, 2004 WHO /IVB/04.10 [12]. The different steps must be performed in appropriate designated Molecular Areas that are defined in Note 2 . The molecular assays are based on qRT-PCR using specific primers and labeled TaqMan probes that recognize the 5′ untranslated region (5′ UTR) of enteroviruses and the VP1 genes of polioviruses in RNA extracted from supernatants of infected tissue cultures (see Note 3 ). These standard algorithms and reagents (see Note 4 ) are in the process of revision and late draft versions of these revisions were discussed at the 20th Informal Consultation of the Global Polio Laboratory Network held at WHO Headquarters in Geneva Switzerland, on June 26–27, 2014. Thus it would not be productive to provide step-by-step instructions for molecular characterization of the poliovirus RNA extracted in Subheading 3.4 at this time. For the latest routine procedures and algorithms we recommend that the reader contact the Polio Laboratory at the Centers for Disease Control and Prevention in Atlanta, GA, USA for information on what latest kits, reagents, and methods are available; and the WHO Global Polio Laboratory Network (GPLN) Regional Laboratory Coordinator for the region in which the laboratory is situated to obtain the latest SOP including algorithms for molecular characterization. A degenerate primer is a mixture of oligonucleotide primers where the bases at one or more nucleotide positions differ or a primer that contains a universal nucleotide at one or more position, such as inosine, that can complement all four nucleotide bases. Sequence variations among wild poliovirus still in circulation have made it necessary to use degenerate primers especially for wild polioviruses for some of the reactions in these kits.
What we will present is a protocol based on these general protocols that can be used to build a qRT-PCR assay with high sensitivity and specificity for a poliovirus of interest once a wild polio or VDPV has been identified based on experience gained during a silent outbreak (persistent transmission without cases) of type 1 wild poliovirus (WPV1) in Israel in 2013–2014 [9, 13]. After its viral capsid protein 1 (VP1) has been sequenced by the protocol in Subheading 3.4 below, it is possible to develop and validate specific nondegenerate primers and probe for qRT-PCR for that strain [9]. Nondegenerate primers such as those used to identify vaccine strains, usually have 100- to 1000-fold lower limits of quantitation (LOQs) and limits of detection (LODs) than degenerate primers. The qRT-PCRs that will be described are multiplex, e.g., each reaction tube includes primers and probes for more than one target sequence. In PCR, the amount of target is doubled during each cycle. The amount of nucleic acid in the solution being assayed is inversely proportional to the Ct or cycle at which specific signal from the probe is first detected above the threshold of background noise. It is important to correlate the Ct for the specific qRT-PCR results with the number of plaque forming units (PFU) of virus [9]. This will enable the lab to provide quantitative information about the load of the virus of interest even when it is present a mixture of heterotypic or homotypic polioviruses (see Note 5 ). Finally the nondegenerate qRT-PCR primers that have been designed to specifically identify the virus of interest can also be used to specifically sequence the VP1 of the virus of interest in RNA extracted from mixtures of heterotypic and homotypic polioviruses without the need for first isolating the virus of interest from these mixtures. Next-generation sequencing (NGS) procedures for RNA extracted from concentrated sewage with or without enrichment for polioviruses by affinity precipitation using anti-poliovirus antibodies or poliovirus receptors are being developed as this chapter goes to press and may supplement or replace some of the procedures described above at specialized molecular laboratories equipped with the necessary machinery and expertise.
2 Materials and Equipment
All solutions use autoclave-sterilized ultrapure water (prepared either by purifying deionized water to attain a sensitivity of 18 MΩ at 25 °C or by distillation) and analytical (AR), molecular biology, or tissue culture grade reagents. Stocks and working solutions were aliquoted before freezing to limit the number of freeze–thaw cycles and preserve component reactivity and uniformity of results. The solutions should be clearly labeled and the label should contain the date of preparation and the expiration date. Prepare and store all reagents at room temperature (unless indicated otherwise). Sewage and all materials derived from sewage must be treated as if contaminated with pathogens. Therefore, appropriate personal protective equipment (PPE) must be worn at all times; lab coats, protective sleeves, splashguard for face or safety glasses, gloves that must be changed frequently, and laboratory approved footwear (no open shoes or sandals) and used biological reagents must be decontaminated before disposal. Appropriate materials to contain and decontaminate spills must be near at hand in accordance with your institutions safety regulations. All stocks of cells should be tested periodically to ensure that they are mycoplasma-free and sensitive to infection by poliovirus. Do not eat, drink, or smoke in laboratory work areas and never pipette by mouth.
2.1 Sewage Processing Components
-
1.
Sterile 0.5, 1, and 2 L screw cap bottles for storage of unprocessed sewage at 4 and at −20 °C.
-
2.
Wide-mouthed screw cap 2 L plastic bottles: 12 cm diameter × 22.5 cm tall, mouth >6 cm in diameter.
-
3.
Magnetic stirrers and magnetic stirring bars (see Note 6 ).
-
4.
Vortex mixers.
-
5.
Rotary shakers for containers ≤500 and ≤50 mL.
-
6.
Test tubes: 1.5, 2.0, 15, 50, 250, and 500 mL tubes suitable for centrifugation and for freezing.
-
7.
Centrifuge with head suitable for spinning 500 mL or 250 mL centrifuge bottles at 10,000 × g.
-
8.
Centrifuge suitable for spinning tubes ≤50 mL at 1400 × g.
-
9.
Microcentrifuges for 1.5 and 2.0 mL microcentrifuge test at up to 14,000 × g.
-
10.
Chemical fume hood or biological–chemical biosafety level 2 (BSL-2) laminar flow hood for initial stages of processing.
-
11.
Biosafety level 2 (BSL-2) laminar flow hood.
-
12.
+4 °C refrigerator(s) for temporary storage of large bottles containing volumes up to 2 L.
-
13.
−20 °C freezers.
-
14.
−70 °C freezers.
-
15.
Polyethylene glycol 6000 (Sigma or Merck) (see Note 7 ).
-
16.
AR grade NaCl.
-
17.
AR grade chloroform .
-
18.
Chlorine for disinfection: Chlorine tablets (Actichlor Plus (EchoLabs, Northwich, Cheshire CW8 4DX, UK) containing 1.7 g sodium dichloroisocyanurate or the equivalent to prepare fresh solutions containing 1000 ppm of chlorine solution weekly (one tablet per liter of tap water); and/or 3 % 30,000 ppm) or 12 % (120,000 ppm) concentrated chlorine stock solutions which can be diluted to 1000 ppm for decontamination (see Note 8 ).
-
19.
PBS: Dulbecco’s calcium- and magnesium-free phosphate-buffered saline (NaCl 8 g, KCl 0.2 g, Na2HPO4 1.15 g, KH2PO4 0.2 g, 1 L water; pH = 7.3–7.4).
-
20.
PBS–Tween 80: PBS containing 0.1 % Tween 80.
-
21.
PSF stock: Penicillin 50 mg/mL, Streptomycin 50,000 U/mL, and Fungizone 0.5 mg/mL.
-
22.
Mycostatin stock [6250 U/mL].
-
23.
PSMY stock: Penicillin G 50,000 U/mL, Dihydro-Streptomycin 50 mg/mL, and Mycostatin 6250 U/mL.
-
24.
3 % DIFCO (Becton, Dickinson and Company, NJ, USA) beef extract in water (pH 7.2).
2.2 Cell Cultures Components
-
1.
World Health Organization (WHO) certified cell lines: human rhabdomyosarcoma (RD) currently available form the National Institute of Standards and Controls (NIBSC UK; Blanche Lane, South Mimms, Potters Bar, Hertfordshire, EN6 3QG) and transgenic mouse cells expressing the human CD155 receptor for polioviruses (L20B) and Buffalo Green Monkey Cells (BGM: ATCC CCL-161) and human epithelial cancer cells (HEp2C: ATCC CCL-23) from the ATCC collection (American Type Culture Collection (ATCC), P.O. Box 1549, 10801 University Boulevard, Manassas, VA 20110, USA) (see Note 9 ).
-
2.
PSMY stock: Penicillin G 50,000 U/mL, Dihydro-Streptomycin 50 mg/mL, and Mycostatin 6250 U/mL.
-
3.
PSF: Penicillin G 50,000 U/mL, Dihydro-Streptomycin 50 mg/mL, and Fungizone 0.5 mg/mL.
-
4.
Fetal bovine serum (FBS) that has NOT been heat inactivated.
-
5.
L-glutamine 3 % (w/v) in water.
-
6.
Complete M199 medium: M199 medium, 10 % FBS (v/v), 2 mL per 500 mL PSMY antibiotics (v/v). Store at 4 °C.
-
7.
2× concentrated M199 Medium: 2× concentrated M199 medium, 2 % FBS (v/v), 1 mL per 200 mL PSMY antibiotic (v/v). Store at 4 °C.
-
8.
Complete Eagle’s MEM-NAA medium: Eagle’s MEM-NAA medium, 10 % fetal bovine serum (v/v), 2 mL per 500 mL PSMY antibiotics (v/v), 4 mL per 500 mL L-glutamine (v/v). Store at 4 °C.
-
9.
Complete Viral Growth Medium: Eagle’s MEM-NAA medium, 2 % FBS (v/v), 4 mL per 500 mL PSMY antibiotics (v/v), 4 mL per 500 mL l-glutamine (v/v). Store at 4 °C.
-
10.
Plaque plating medium: M199 medium, 2 mL per 500 mL PMSY (v/v) without FBS.
-
11.
Plaque suspension medium: M199, 4 mL per 500 mL PMSY (v/v) without FBS.
-
12.
Trypsin-Versene solution: 0.25 % trypsin, 0.05 % EDTA.
-
13.
1.8 % DIFCO (Becton, Dickinson and Company, NJ, USA) Bacto agar in sterile deionized water.
-
14.
Agar overlay: a solution of 1.8 % DIFCO Bacto Agar in water melted in a microwave (approximately 1 min at the maximum setting).—CAUTION: the solution is very hot and may boil vigorously and spill out of the bottle when the bottle is removed from the microwave—Keep melted agar at 50 °C in a water bath until just before use when an equal volume of 2× concentrated M199 medium (brought to 50 °C) is added.
-
15.
Plaque staining solution: M199 medium, 1 mL per 100 mL 1 % neutral red in H2O (v/v).
-
16.
Tissue culture plates, tubes, and flasks: 10 cm style disposable, tissue culture grade petri dishes; 25, 75 of 150 cm2 style tissue culture flasks; Greiner Bio-One CELLSTAR 12 mL Cell Culture Tubes or their equivalent (Greiner Bio-One International AG, 4550 Kremsmünster Austria).
-
17.
Separate incubators for uninfected and infected cell cultures (see Note 10 ).
-
18.
Sterile pipettes and/or transfer pipettes suitable for volumes of 1, 2, 5, 10, and 25 mL.
-
19.
Micropipettes with adjustable volumes with the following ranges up to 0.5 to 10, ≤2 to 20, 10 to 100, 20–200, and 200–1000 μL.
-
20.
Sterile micropipette tips for volumes up to 10, 20, 100, 200, 1000 μL with aerosol barriers.
-
21.
Disposable test tubes: sterile 1.5, 2.0, 15, and 50 mL tubes suitable for centrifugation and frozen storage with appropriate test tube racks.
-
22.
Reusable centrifuge tubes for 250 and 500 mL.
-
23.
Biosafety level 2 laminar flow hood.
-
24.
+4 °C refrigerator(s).
-
25.
−20 °C freezers.
-
26.
−70 °C freezers.
-
27.
Liquid nitrogen freezing and storage facilities for maintenance of frozen cell stocks.
-
28.
Regular or inverted (phase contrast optional) light microscopes.
-
29.
Decontaminating solutions for wiping down work areas before and after work.
-
30.
Optional but preferred—Lab mat (absorbent paper with a nonpermeable bottom layer) to place on working surfaces before starting and discarded in a container for biological decontamination afterwards.
-
31.
Vacuum line, tubing, and flasks for removing tissue culture medium from tissue cultures (see Note 11 ).
2.3 RNA and cDNA Extraction
-
1.
QIAamp Viral RNA Mini Kit (QIAGEN Inc, Valencia, CA, USA) or its equivalent.
-
2.
KingFisher Purification System (Thermo Scientific, Waltham, MA, USA) using the viro_totRNA_KFmini extraction protocol with NucleoMag 96 RNA extraction Kits (Macherey-Nagel GmbH Duren, Germany) or Thermo Fisher Total RNA Kits (Thermo Scientific, Waltham, MA, USA) or their equivalents.
-
3.
NucliSENS easyMag semiautomatic extractor (bioMérieux, Marcy l’Etoile, France) using the Specific B extraction protocol with easyMag extraction kits or their equivalent.
-
4.
High Pure PCR Product Purification Kit (Roche Diagnostics, Indianapolis, IN, USA) or its equivalent.
-
5.
QIAquick Gel Extraction Kit (QIAGEN Inc, Valencia, CA, USA) or its equivalent.
-
6.
Big Dye Terminator Purification Kit (Life Technologies, Foster City, CA, USA).
-
7.
Test tubes: 1.5, 2.0, 15, 50 mL suitable for centrifugation and frozen storage with appropriate test tube racks.
-
8.
Storage boxes (9×9) for storing tubes ≤2 mL.
-
9.
+4 °C refrigerator(s).
-
10.
−20 °C freezers.
-
11.
−70 °C freezers.
-
12.
Centrifuge for spinning tubes ≤50 mL at ≤1500 × g.
-
13.
Microcentrifuge for spinning 1.5 and 2.0 tubes up to 15,000 × g.
-
14.
Spin-down centrifuges for 0.2 mL tubes or strips.
-
15.
BSL-2 laminar flow hood.
-
16.
External RNA control: shielded RNA or MS2 coliphage as an external control for the presence of inhibitors of PCR or RT-PCR that might be co-extracted with the RNA.
2.4 Nucleic Acid Amplification
-
1.
QIAGEN one-step RT-PCR kit (QIAGEN Inc, Valencia, CA, USA) or its equivalent.
-
2.
ABI Prism 7500 sequence detection system (Life Technologies, Foster City, CA, USA) for qRT-PCR using AgPath-ID™ One-Step RT-PCR Kit (Life Technologies/Rhenium LTD).
-
3.
PCR machines for 0.2 mL tubes or 8-tube strips.
-
4.
Test tubes: 0.2, 0.5, 1.5, 2.0, 15, and 50 mL suitable for centrifugation and frozen storage with appropriate test tube racks.
-
5.
Axygen 0.2 mL 8-test tube strips compatible with ABI 7500 instruments or their equivalent.
-
6.
Centrifuge for spinning tubes ≤50 mL at ≤1500 × g.
-
7.
Microcentrifuge for spinning 1.5 and 2.0 tubes up to 15,000 × g.
-
8.
Centrifuges for spin down for 0.2 mL tubes or strips.
-
9.
Metal cooling blocks for holding twelve, 8-tube strips (0.2 mL volume) and maintaining the tubes at cold temperatures.
-
10.
Test tube strip transfer holders for transporting filled test tube strips (can be sterilized, nucleic acid-free (chlorine treated), empty aerosol barrier tip boxes).
-
11.
Micropipettes with adjustable volumes with the following ranges up to 0.5 to 10, ≤2 to 20, 10–100, 20–200, and 200–1000 μL.
-
12.
Sterile micropipette tips for volumes up to 10, 20, 100, 200, 1000 μL with aerosol barriers.
-
13.
Aliquots of stock solutions of primers and probes. Sequences and stock concentrations are listed in Table 1.
Table 1 Sequences and stock concentrations -
14.
E buffer (for 10× concentrated stock: 48.44 g Trizma Base, 16.4 g Na-acetate, and 362 mg EDTA per liter water brought to pH = 7.2 with acetic acid).
-
15.
Ethidium bromide (1 mg/mL in water).
-
16.
2 % agar for gels: (1 % agarose; 1 % NuSieve agarose in E buffer).
2.5 Sequencing and Preliminary Sequence Analysis
-
1.
PCR machines for 0.2 mL tubes or 8-tube strips.
-
2.
Automatic Sanger Sequence Detection System: 3100 Genetic Analyzer; 3500 Genetic Analyzer (Life Technologies, 850 Lincoln Centre Drive, Foster City, CA 94404, USA) or their equivalent.
-
3.
Test tubes: 0.2, 0.5, 1.5, 2.0, 15, and 50 mL suitable for centrifugation and frozen storage with appropriate test tube racks.
-
4.
Corning Axygen 0.2 mL 8-test tube strips compatible with ABI 7500 instruments.
-
5.
Centrifuge for spinning tubes ≤50 mL at ≤1500 × g.
-
6.
Microcentrifuge for spinning 1.5 and 2.0 tubes up to 15,000 × g.
-
7.
Centrifuges for spin down for 0.2 mL tubes or strips.
-
8.
Metal blocks for holding twelve 8-tube strips and maintaining the tubes at cold temperatures.
-
9.
Spectrophotometer for measuring the amount of nucleic: NanoDrop ND-1000 Spectrophotometer (Thermo Fisher Scientific, 3411 Silverside Road, Bancroft Building, Suite 100, Wilmington, DE 19810, USA) or equivalent.
-
10.
Micropipettes with adjustable volumes with the following ranges up to 0.5 to 10, ≤2 to 20, 10 to 100, 20–200, and 200–1000 μL.
-
11.
Sterile micropipette tips for volumes up to 10, 20, 100, 200, 1000 μL with aerosol barriers.
-
12.
Computer with connection to the internet and loaded with computer programs for analysis of nucleic acid sequences. Highly recommended: Sequencher version 5 or later (GeneCodes, Ann Arbor, MI, USA).
3 Methods
3.1 Sewage Collection and Concentration with High Throughput with Minimal Turnaround Time
Composite sewage samples (1–2 L) are collected by pooling aliquots of sewage collected at timed intervals over a 24 h period by in-line automatic collectors at the inlet to sewage treatment facilities. Composite sewage samples are collected at upstream sites and at sewage treatment facilities lacking in-line collectors by using automatic portable, computerized composite sewage collectors (Sigma SD900 portable samplers, HACH, Loveland, CO, USA) (see Note 12 ). Samples must be transported under cold chain conditions (kept below 4 °C) to the processing laboratory to avoid loss of viability of any poliovirus in the samples. Surveillance sites are chosen according to WHO recommended Guidelines for environmental surveillance of poliovirus circulation [7]. A revised version of these WHO guidelines is available on-line from the WHO (http://www.polioeradication.org/Portals/0/Document/Resources/GPLN_publications/GPLN_GuidelinesES_April2015.pdf last accessed 17-05-2015).
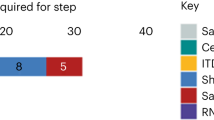
Method for concentrating virus in sewage by 30- to 40-fold (see Fig. 1 for timing and time course for performing each step):
Poliovirus isolation and identification working algorithms employed during the routine environmental surveillance (a) and after discovery of the WPV1-SOAS (b). The working algorithm shown in A allows isolation of any poliovirus type by sequence analysis within 16–23 days and allows processing of 6–12 samples/week. The quantitative evaluation of plaque number is done after the final identification. The working algorithm shown in b incorporating additional steps (blue shapes) allows primary detection and quantification of WPV1-SOAS by qRT-PCR within 3–4 days, confirmation by ITD and quantitative evaluation of plaque number within 7–9 days and full sequence analysis within 10–16 days. It also allows processing of up to 50 samples per week
-
1.
The starting volume of sewage for concentration is 500 mL (±50 mL). Volume in excess of 1 L is discarded after decontaminating with chlorine solution. Pour approximately 500 mL of the sewage plus sediment into a 2 L wide mouth plastic bottle. The remaining half liter is poured into a 500 mL disposable plastic bottle and stored at 4 °C for backup if needed. Sewage samples may be stored at 4 °C for up to 2 weeks until processing is begun without significant loss of virus viability and for up to a month for recovery of genomic RNA for molecular assay but this should be validated for conditions relevant for your laboratory.
-
2.
Add 40 g of polyethylene glycol 6000 and 8.8 g of AR grade NaCl (see Note 13 ).
-
3.
Add a sterile magnetic bar (see Note 6 ) and stir for ≥60 min at room temperature in a chemical hood.
-
4.
Precipitate the virus by letting the sample sit overnight at 4 °C.
-
5.
Gently spill off some of the uppermost liquid, mix the sediment and remaining liquid, and add this suspension to one 500 mL centrifuge tube or two 250 mL centrifuge tubes (see Note 14 ). Decontaminate any excess liquid with chlorine solution.
-
6.
Centrifuge the samples at 10,000 × g for 1 h at 4 °C (see Note 15 ). Caution: Use a transfer pipette to balance tubes before centrifugation and decontaminate any excess liquid with chlorine solution.
-
7.
The virus is in the pellet. Therefore, carefully pour off the supernatant into a new bottle into which you add chlorine solution (for decontamination) and dispose the chlorine-treated liquid after 30 min.
-
8.
Resuspend the pellet in 7 mL of PBS–Tween 80. Pipette and wash the sides of the centrifuge tube to resuspend all pelleted material.
-
9.
Transfer this suspension to a 50 mL centrifuge tube and add ½ volume of chloroform .
-
10.
Agitate vigorously for 20 min on a rotary shaker at room temperature to break apart small clumps and inactivate bacteria (see Note 16 ).
-
11.
Centrifuge the suspension at 1400 × g for 20 min at 4 °C.
-
12.
Transfer the supernatant that contains most of the virus to a fresh 50 mL test tube.
-
13.
Reextract any virus trapped in the pellet by adding 2–3 mL of 3 % beef extract (pH 7.2) to the pellet and mixing vigorously for 5 min (see Note 16 ).
-
14.
Centrifuge at 1400 × g for 20 min at 4 °C.
-
15.
Add the supernatant to the liquid from step 12. Decontaminate (with chlorine solution) and discard the pellet and any chloroform that was carried over.
-
16.
Add antibiotics for concentrated sewage to the combined supernatants (for each 50–60 mL of supernatant, add 0.5 mL of PSF, 0.5 mL of Mycostatin stock, and 0.8 mL of PSMY stock). The final volume should be approximately 15 mL. If it is less, bring the volume to 15 mL by adding medium containing PSF and PSMY. Important: Record the volume of the concentrated sewage for calculating the fold concentration (see Note 17 ).
-
17.
Incubate the processed sewage from step 16 for 20 min at room temperature before proceeding or freezing the sample at −20 °C (see Note 18 ).
3.2 Cell Cultures for Identification of Virus in Concentrated Sewage
3.2.1 Preparation and Maintenance of Stocks of Uninfected L20B, RD, HEp2C and BGM Cells
Cell cultures are essential for testing for the presence of viable virus in sewage surveillance samples. Cell lines are passed once a week on the fifth workday into flasks for maintaining the cell lines and into tube cultures for viral challenge on the second workday of the following week. Each week there must be enough flasks of cell cultures on hand to maintain continuous passage of each cell line and provide enough extra for preparing the necessary number of uninfected tube cultures and plate cultures anticipated for characterization of the virus in the samples during the coming week. Cell cultures are renewed periodically from frozen stocks. Confirm the sensitivity of L20B and RD to polioviruses midway through the expected use of 15 passages or every 3 months. The other cell lines should also be checked but the interval between sensitivity testing can be longer (see Note 19 ). Uninfected and infected cell culture tubes should be maintained in separate incubators.
-
1.
Growth medium and passage ratios for maintaining uninfected cell cultures: Grow cultures at 37 °C in closed 75 cm2 flasks (see Note 10 ).
-
L20B cells stock cultures are passage at a ratio of 1:3 or 1:4.
-
RD at a ratio of 1:5; HEp2C at a ratio of 1:6 or 1:7.
-
BGM at a ration of 1:4. Complete M199 medium is used for L20B, RD, and BGM.
-
HEp2C cells are cultured in complete Eagle’s MEM-NAA. L20B can also be cultured in complete Eagle’s MEM-NAA.
-
-
2.
Preparations of cell suspensions for maintenance and for tube and plate cultures.
-
Pour off medium.
-
Add 20–30 mL of Trypsin-Versene solution (see Note 20 ).
-
Pour off all but a few milliliters of the Trypsin-Versene solution.
-
Incubate the culture at 37 °C for >5 min.
-
Tap the bottle to release cells.
-
Add 5 mL of complete tissue culture medium (with FBS).
-
Pipette up and down a few times to break apart any clumps.
-
Either aliquot the cell suspensions in the ratios indicated above for continuing passage or add complete medium in the amounts indicated in the next paragraph for plating the cells in tube cultures or petri dishes for viral challenge and isolation (see Note 21 ).
-
-
3.
Tube cultures per sample to be tested: Four L20B and four RD tube cultures are prepared 1 week before use. You will also need to prepare at least one L20B culture tube for repassage of supernatant from RD cells that develop cytopathic effects (CPE) during the previous week (see Note 22 ). Prepare at least one additional tube for each cell line to serve as uninfected control.
-
Dilute cell suspensions from one 75 cm2 flask diluted into a final volume of 85 mL of complete M199 medium.
-
Add 2 mL of this is suspension to each 12 mL cell culture tubes (see Notes 23 and 24 ).
-
Close the cap tightly and incubate at 37 °C in test tube racks that keep the tubes at a 20° slant (see Note 10 ).
-
-
4.
Tube cultures for HEp2C cells. Prepare one HEp2C culture tube for each plaque isolate to be amplified. Prepare at least one additional tube to serve as uninfected control (see Note 24 ).
-
Resuspend cells from one 75 cm2 flask in a final volume of 85 mL of complete MEM-NAA medium and
-
Add 2 mL of this is suspension to 12 mL cell culture tube.
-
Close the cap tightly and incubate at 37 °C in test tube racks that keep the tubes at a 20° slant (see Note 10 ).
-
-
5.
L20B monolayers for plaque assay and plaque isolation. Prepare three 10 cm petri dishes of L20B cells for each sewage sample and one for negative challenge control 4 days before intended use.
-
Dilute suspended cells from one 150 cm2 flask in 100 mL of complete M199 medium.
-
Plate 12.5 mL of this suspension into each 10 cm style tissue culture petri dishes.
-
Incubate plates in a 5 % CO2 incubator at 37 °C.
-
-
6.
BGM and RD monolayers for plaque assay and plaque isolation (see Note 25 ). Prepare one 10 cm petri dishes of RD cells for each sewage sample and one for negative challenge control 4 days prior to viral challenge.
-
Dilute suspended cells from one 150 cm2 flask in 100 mL of complete Eagle’s MEM-NAA medium—Plate 12.5 mL of this suspension into each 10 cm style tissue culture petri dishes 4 days before intended use.
-
Incubate plates in a 5 % CO2 incubator at 37 °C.
-
3.2.2 Viral Challenge of Uninfected Cell Cultures
Viral challenge of tube cultures
-
1.
Thaw concentrated sewage in room temperature water and gently vortex to ensure homogeneous resuspension of the virus.
-
2.
Replace complete medium with viral growth medium in the tube cultures.
-
Pour off or remove complete medium by vacuum.
-
Replace with 2 mL of virus growth medium.
-
Tightly close the cap tubes and incubate tubes at 37 °C.
-
-
3.
Add concentrated sewage (200 μL) to each of four replicate tube cultures of L20B cells and four replicate tube cultures of RD cells.
-
4.
Maintain the tube cultures at 37 °C for 5–7 days (see Note 10 ).
-
5.
Visually inspect for CPE.
-
CPE-positive L20B cultures most likely contain polioviruses. The presence of poliovirus is confirmed and the serotype and type determined by molecular TD and ITD assays described in Subheading 3.4 below.
-
RD CPE-positive cultures may contain human enteroviruses including poliovirus.
-
-
6.
Confirm whether CPE is caused by poliovirus by characterizing the virus present in CPE-positive cultures (see Notes 26 and 27 , and especially Note 28 ).
-
Perform molecular ITD (Subheading 3.4) on RNA extracted (Subheading 3.3) from CPE-positive supernatants of L20B cells.
-
Perform molecular ITD (Subheading 3.4) on RNA extracted (Subheading 3.3) from CPE-positive supernatants of L20B cells . Significant amounts of work and molecular reagents can be saved by repassing 200 μL of supernatant from CPE-positive RD tube cultures (or pooled supernatant from replicate RD tube cultures) on L20B tube cultures. These cultures are called RL cultures to indicate that the L20B tube culture was infected with supernatant from and RD tube cultures (see Note 27 ).
-
If CPE develops on RL tube cultures, the presence of poliovirus is confirmed by molecular ITD assay.
-
Plaque assays and plaque purification
-
1.
Infect the cell monolayer.
-
Dilute 4 mL of concentrated sewage with 12 mL of plaque plating medium.
-
Remove medium by vacuum from three petri dishes of L20B and one petri dish of BGM plated 4 days earlier
-
Add 4 mL of diluted sample from step 1 to each plate (see Note 29 ).
-
Rock the plates gently to ensure that the liquid is evenly distributed over the entire surface.
-
Incubate for 1 h at 37 °C. Rock the plates gently every 15 min to keep all of the surface area of the monolayer moist and all cells alive.
-
Prepare the agar overlay and keep it at 50 °C in a water bath during this incubation period in the previous step.
-
Remove the 4 mL of diluted sample by vacuum
-
Gently overlay the monolayer with 12.5 mL of the agar overlay. Add the agar to the lower sidewall of the plate that is held at a shallow angle rather than directly onto the cell monolayer to avoid tearing the monolayer. Make an effort not to create bubbles.
-
Let the overlay solidify at room temperature (approximately 5–10 min).
-
Invert the petri dishes.
-
Incubate them at 37 °C for a minimum of 2 days.
-
-
2.
Inspect the monolayers for plaques and count the number of plaques.
-
Invert the petri dishes so the cells are on the bottom.
-
Add 8.5 mL of plaque staining solution.
-
Incubate at 37 °C for 1 h.
-
Remove the plaque staining solution with vacuum.
-
Incubate the petri dishes in an upright position at 37 °C overnight.
-
Count the number of plaques on each 10 cm-style petri dish. CAUTION: never hold the plates above your face when counting plaques to reduce the chance of bio-contamination by liquid falling from the plates onto your face or body (see Note 30 ).
-
Calculate the number of plaques in 1 mL of unconcentrated sewage (PFU/mL) by multiplying the average of the number of plaques on the three plates by the dilution factor (starting volume approximately 500 mL divided by the volume of concentrated sewage recorded) (recorded in Subheading 3.1).
-
-
3.
Pick individual plaques of virus.
-
Pick virus in a plaque by stabbing through the agar overlay using a disposable plastic volume transfer pipette (see Note 31 ).
-
Transfer the agar plug from the stab into a 2 mL microcentrifuge test tube containing 200 μL of plaque suspension medium (see Note 32 ). Caution: Take precaution so that the transfer of the agar plug does not cause the liquid to splash.
-
The tube with the agar plug can be store at 4 °C for up to 4 days or at −20 °C until primary stocks of the virus are prepared in tube cultures (HEp2C or RD preferred).
-
-
4.
Primary amplification of virus in the plaque—preparation of a primary stock.
-
Vortex the plaque suspension.
-
Spin down the liquid by a brief centrifugation.
-
Add 150 μL of the supernatant HEp2C or RD tube culture containing viral growth medium.
-
Incubate cultures for up to 5 days at 37 °C.
-
Inspect the cultures for CPE daily starting on the third day.
-
Transfer tubes with full CPE to 4 °C or freeze them at −20 °C or at −70 °C (preferred) (see Note 33 ).
-
-
5.
Prepare high-titer secondary stocks (see Note 34 ).
High-titer secondary stocks better suited for some types of molecular analysis such as sequencing can be prepared as follows:
-
Add 150–200 μL of primary stock to a fresh HEp2C or RD tube culture.
-
Incubate for 3–5 days at 37 °C.
-
Transfer tubes with full CPE to 4 °C or freeze them at −20 °C or at −70 °C (preferred) (see Note 34 ).
-
3.3 RNA Extraction from Tissue Culture Supernatants and Concentrated Sewage
RNA is extracted from supernatants of individual CPE-positive tube cultures, from pools supernatants of tube cultures (see Notes 27 and 35 ) and from concentrated sewage. The volume used for extraction varies between 50 and 1000 μL depending on the extraction procedure used. Most commercial extraction systems provide RNA of more or less equivalent quality (for example see [14]). Two alternatives are provided here: (1) total nucleic acid (NA) extraction using the EasyMag system (bioMérieux, Marcy l’Etoile, France); and (2) RNA extraction using a KingFisher System (Waltham, MA, USA). We recommend adding protected non-polio RNA such as shielded RNA or MS2 coliphage (ATCC 15597-B1) either to the lysis buffer to serve as an external control for the presence of inhibitors of PCR or RT-PCR chemistry that might be co-extracted with the RNA or adding them to each sample before extraction to serve as a control for extraction and inhibitors. We add MS-2 to the lysis buffer at a concentration sufficient to produce 28–32 Cts in a multiplex qRT-PCR. Each extraction run should also include extraction of (1) a negative control that also serves as positive for MS2 in the absence of inhibitors; and (2) an extract positive control (see Note 36 ), a sample containing an amount of enterovirus that will give a consistent Ct in the range of 28–32 after extraction for quality control for extraction.
-
1.
Thaw stored samples for samples that have been frozen.
-
Thaw in room temperature water.
-
Vortex to prepare a uniform suspension.
-
-
2.
Extract the RNA in the sample (see Note 28 ):
EasyMag extraction system
-
Add up to 1 mL of tissue culture supernatant or concentrated sewage to 2.0 mL of lysis buffer containing MS2 coliphage at a concentration of 10,000 PFU/mL.
-
Vortex.
-
Extract according to the manufacturer’s instructions on the NucliSENS EasyMAG system.
-
Elute the RNA in 55 μL elution buffer (supplied with the kit) or sterile nuclease-free water.
KingFisher Extraction System
-
Add 50 μL of tissue culture supernatant or concentrated sewage to the lysis well on the 8-well strip of the RNA extraction kit.
-
Extract using the recommended viro_totRNA_KFmini protocol that can be downloaded and installed on the KingFisher extractor using the Thermo Scientific BindIt Software program (Waltham, MA, USA).
-
RNA is eluted in 55 μL elution buffer (supplied with the kit) or sterile nuclease-free water.
-
-
3.
Store the extracted RNA.
-
Store RNA at −70 °C pending analysis.
-
3.4 Nucleic Acid Amplification for Poliovirus Molecular Serotype Identification (TD) and Intratypic Differentiation (ITD)
The latest WHO recommended standard kits for poliovirus TD and determining whether the poliovirus is vaccine-like, vaccine-derived, or wild, e.g., ITD differentiation (and the SOPs to perform them can be obtained from the Polio laboratory at the CDC, Atlanta, Georgia). These kits can directly identify all wild and vaccine polioviruses from sewage and indirectly identify VDPVs by a process of elimination when all of the reactions are run (pan-entero; pan-polio; generic type 1, 2, and 3; Sabin 1, 2, and 3; and optionally VDPV 1 and 2 and SOAS wild type 1 and 3 and WEAF wild type 1 and 3). Since the kits are currently under revision, we will not include their protocols here. Instead we will provide detailed instructions on how to provide critical rapid, high throughput information necessary for making evidence-based decisions during outbreaks [9, 15, 16] by modifying the kits, primers, and/or probes and/or procedures for a virus of interest once that virus has been detected and identified. [Caution: the procedures outlined here are for detecting the virus of interest. Unless the other standard assays are also applied, other viruses of interest may be overlooked.] All specific assay modifications should conform to guidance for assay design and optimization, analytical validation, and environmental field validation in Saunders et al. [17]. For these procedures, plaque purified poliovirus stocks need to be prepared for designing specific primers and probes to identify the virus of interest. The steps that follow were designed for amplification and detection by TaqMan technology using an Life Technologies 7500 sequence detection systems (Life Technologies, NY, USA) (see Note 37 ).
-
1.
Prepare primary and secondary virus stocks of the virus of interest for preparing specific reagents for detection and characterization.
-
Plaque-purify the virus of interest as described in Subheading 3.2 above (see Note 38 ).
-
Prepare a primary stock, from one plaque.
-
Determine the PFU/mL of the primary stock.
-
Prepare a secondary stock at a multiplicity of infection (MOI) of one PFU per cell (see Note 38 ).
-
Incubate at 35 °C for 48 h.
-
Freeze-thaw the tube at −70 °C to increase viral yield (see Note 33 ).
-
Clarify the cell debris by centrifugation at 3000 × g for 10 min.
-
Determine the PFU/mL of the secondary stock as above.
-
Adjust the virus concentration of the secondary stocks to 2 × 108 PFU/mL.
-
Aliquot the secondary stock and store at −70 °C.
-
-
2.
Design primers and probes specific for the virus of interest.
-
Sequence the complete viral capsid protein 1 (VP1) of the twice plaque-purified secondary stock or stocks using generic primers Y7 and Q8 (Table 1) and the procedure outlined in Subheading 3.5 below (see Note 38 ).
-
Align VP1 sequences from more than one related virus isolate of interest (see Note 38 ).
-
Use a computer program (see Note 39 ) to design at least two different nondegenerate pairs of primers and probe suitable for qRT-PCR that will not react with the corresponding OPV strain [9] (see Note 40 ).
-
-
3.
Optimize reagents for a standard qRT-PCR assay protocol.
It is assumed that the operator is familiar with the use of the qRT-PCR machine. Remember to set the qRT-PCR machine to read the dyes used on the probe. We optimize all qRT-PCR reactions to the same run conditions so that we can test different viruses or combinations of viruses in the same run (see Note 41 ). The procedures here are for qRT-PCR on an ABI 7500 instrument (Life Technologies, Thermo Fisher Scientific, Waltham, MA, USA) using 8-well microtiter strips for maximum flexibility (see Note 42 ). Modifications must be validated if different qRT-PCR machines, enzymes, and buffers are used.
Molecular Area 1
-
Prepare a work sheet listing all RNAs to be tested and the position they will appear on each strip before starting any qRT-PCR run. Include positive and negative controls for amplified target and the extraction positive control (see Note 36 ).
-
Take cold metal blocks kept at 4 °C in the refrigerator to keep reagents and mixtures cold throughout preparation.
-
Remove an aliquot of Mastermix from the AgPath-ID™ One-Step RT-PCR Kit (Life Technologies/Rhenium LTD) kit that had been kept at −20 °C, thaw it by spinning the tube in a microcentrifuge, vortex it gently for 5 s, and then respinning it for a few seconds in a centrifuge.
-
Place the Master mix tube in the cold block and transfer the calculated amount into a 1.5 mL test tube. Add a black dot on the top of the Master mix tube each time it is thawed and immediately return the Master mix to the freezer. Do not use after five freeze–thaw cycles.
-
Once the amount of Mastermix has been transferred to the 1.5 test tube, remove the Reverse Transcriptase/Polymerase enzymes tube from the −20 °C freezer, briefly centrifuge it for 5 s, and transfer the required amount of enzyme to the tube with the Master mix. Return the enzyme immediately to the −20 °C freezer to maintain activity of the enzyme.
-
Place 0.2 mL tube strips on the cold block. Label the top end of each strip with strip numbers corresponding to those on the work sheet.
-
Vortex the tube containing Master mix plus enzyme for 5 s, spin-down for 5 s, and then aliquot 20 μL to each reaction tube.
-
Gently place a cover on top of the 0.2 mL strip. Hold the strip cap in place by closing only the tubes at both ends of the strip, but do not close them completely (see Note 43 ).
-
Do NOT remove the cold block from Molecular Area 1. Instead, transfer the strips to an empty chemically and biologically sterile pipette tip box kept in Molecular Area 1 and bring the transfer box to a 4 °C refrigerator in Molecular Area 2.
Molecular Area 2
-
Transfer the strips to a metal cold block in a dead-air box in Molecular Area 2.
-
Remove the extracted RNA from the −20 °C freezer.
-
Thaw the unknowns by centrifuging, vortexing, and respinning the extracted RNA samples in a microcentrifuge.
-
Load 8-μL of RNA from the samples using a separate aerosol barrier tip for each RNA sample.
-
Place the caps back on the strip and close firmly using a strip cap-closing tool kept in the dead-air box.
-
Include additional reactions for control RNAs (positive poliovirus RNA, MS2 RNA, negative control, and extract positive and negative controls) stored in the −20 °C freezer.
-
Spin-down the contents of the tubes in a small strip centrifuge.
-
-
4.
Optimization of qRT-PCR specific for the virus of interest.
Standard qRT-PCR conditions are followed for a reaction mix of 25 μL; 5 μL of RNA is added to a 20 μL reaction mixture containing AgPath-ID One-Step RT PCR reagents (Life Technologies, NY, USA), primers and probe specific for the poliovirus of interest, and MS-2 primers and probes.
-
RT at 48 °C for 30 min.
-
Taq polymerase activation at 95 °C for 10 min.
-
50 cycles of strand separation at 95 °C for 15 s, and elongation at 60 °C for 1 min. Data collection is ON only for the 60 °C elongation step.
-
Interpretation of the assay results is according to Table 2 and the number of reactions for any given RNA is dependant on these initial results.
-
Evaluate different concentrations of the newly designed VP-1 nondegenerate primers (300, 600, and 900 nM) and probe (200 and 300 nM) to optimize the multiplex qRT-PCR in a multiplex using 150 nM each of MS2 external control primer and 50 nM MS-2 probe.
-
Choose the concentrations that provide the lowest limit of quantitation and detection as described in the next two subsections.
Table 2 Assay results -
-
5.
Determine the Analytical sensitivity, the limit of detection (LOD), and precision of the qRT-PCR specific for the virus of interest.
-
Determine the analytical sensitivity, the limit of detection (LOD), and precision of the qRT-PCR by testing in parallel 4 tenfold serial dilutions in water of RNA extracted from the secondary virus stock starting with 2 × 107 PFU/mL as in Dreier et al. [18].
-
Inspect the multicomponent curve for each qRT-PCR reaction to confirm that a positive Ct is actually the result of a true event.
-
-
6.
Determine the analytical specificity of the qRT-PCR specific for the virus of interest.
-
Determine the analytical specificity of the specific primers and probe in silico using Basic Local Alignment Search Tool (BLAST) search (available at: http://www.ncbi.nlm.nih.gov, last accessed July 25, 2014)
-
Determine the analytical specificity of the specific primers and probe in vitro by testing them on RNA and DNA from polio and non-polio enteroviruses and as wide a range of reference strains of non-enteroviruses as the your laboratory possesses.
-
-
7.
Validation of the primers and probe for the qRT-PCR specific for the virus of interest.
-
Validate the qRT-PCR by extracting 200 μL of pools of poliovirus-negative concentrated sewage collected before first appearance of the poliovirus of interest spiked with tenfold serial dilutions of the secondary stock (starting from 2 × 106 PFU/mL).
-
In parallel, confirm the actual titer in each dilution by plaque assay of 200 μL after bring the volume to 4 mL and using the plaque assay conditions as described above.
-
Finally validate the assay by testing 20–50 concentrated sewage samples known to contain the poliovirus of interest. Reconfirm that they contain polio by retesting on L20B tube cultures in parallel with the qRT-PCR.
-
-
8.
Validate your qRT-PCR for Sabin strains.
-
Validate a multiplex qRT-PCR as above for a qRT-PCR multiplex using your Master mix, buffers and enzymes with CDC-designed [19] primers and probes for Sabin 1, Sabin 2, and Sabin 3 (Table 1).
-
Inspect the multicomponent curve for each qRT-PCR reaction to confirm that a positive Ct is actually the result of a true event.
-
-
9.
Important note added in proof: The CDC has started to distribute new ITD v4.0 and VDPV v4.0 Kits in 2015.
The reaction protocol of the ITD Kit v4.0 has been simplified and differs from previous versions of ITD kits. There are five not six reaction mixtures: (1) a quadriplex for pan-enterovirus, Sabin 1, Sabin 2, and Sabin 3; (2) a singleplex for pan-poliovirus; (3) a duplex for both currently circulating lineages of wild type 1 polio virus (SOAS and WEAF); and separate singleplex reactions for (4) wild type 3 SOAS; and (5) wild type 3 WEAF polioviruses. The three serotype-specific reactions from the previous ITD kit have been discontinued. In addition, changes have been made in the composition of some of the primers and probes and/or the dyes and quenchers. The sequences are provided in the Kit insert.
The VDPV v4.0 kits are still based identification of a VDPV by ruling out the presence of Sabin strains of the same serotype. The reaction protocol of the ITD Kit v4.0 also has been simplified and differs from previous versions of ITD kits.
As before when using both kits to characterize polioviruses in sewage in areas where OPV is still administered and sewage contains Sabin-like isolates, VDPVs especially highly diverged VDPVs may be missed in environmental samples containing homotypic mixtures of highly diverged VDPVs and Sabin strains of the same serotype.
The CDC has tested the new reaction mixtures and amplification protocols from the v4.0 kits using many available commercial enzymes. The efficiency of the reactions varied widely. For example for one of the Kit reaction components, Cts for the same RNA sample ranged from 12 to 36 and the quality of the amplification plots ranges from poor to excellent. The CDC recommends two commercial enzyme preparations that provided the best results for all of v4.0 kit reaction components. It is possible to purchase the primers and probes separately, formulate mixes with different buffers than these in the CDC kits, and use different enzymes. However given the extensive validation needed and the requirement for uniformity of analysis conditions for all the laboratories of the Global Poliovirus Laboratory Network, we now strongly recommend using the kits supplied by the CDC and the enzymes they recommend. The kits also contain positive controls that can be used to evaluate differences between labs when standard protocols are used. Furthermore we recommend validating the assay in your laboratory for both recommended enzymes in case there arise problems in availability for one of them.
3.5 Molecular Characterization of Poliovirus RNA by RT-PCR and Sequence Analysis
The complete VP1 sequence must be determined to confirm the presence of a poliovirus of interest once it has been identified by the qRT-PCR described in Subheading 3.4 above. For this, RNA is amplified by RT-PCR and both strands of the amplicon are sequenced. This section describes how to amplify the poliovirus VP1 gene, purify the amplicon, and sequence both strands on a Sanger-based [20] automatic sequencer. It is assumed that the reader either knows how to use a Sanger-based automatic sequencer or can send the processed cDNA to a sequencing laboratory. In the future it will be possible to obtain full poliovirus genomes using next-generation sequencing. Subheading 3.5 concludes with instructions on how to correct the raw sequence data generated by the automatic sequencer and how to easily and rapidly perform TD and ITD on the sequence.
As throughout this section, the work flow is unidirectional from Molecular Area 1 to Molecular Area 4 for both RT-PCR and Sequencing. The RT-PCR reaction mix (50 μL final volume with nucleic acid) is prepared in Molecular Area 1; the RNA is added to the RT-PCR mix in Molecular Area 2; the RT-PCR is performed in Molecular Area 3; RT-PCR results are analyzed in Molecular Area 4; and the amplicon is purified and labeled in Molecular Area 4. Sequencing reaction mix is prepared in Molecular Area 1; the amplicon is added and labeled in Molecular Area 4, the labeled amplicon is purified in Molecular Area 4; and the labeled amplicon is sequenced in Molecular Area 3 or 4. RT-PCR conditions are described for RT-PCR using the QIAgen one-step RT-PCR Kit (QIAGEN GmbH, Chatsworth, CA, USA). Conditions need to be validated and optimized when equivalent RT-PCR kits are used.
-
1.
Preparing specific amplicons for sequencing.
-
Prepare the reaction mix in Area 1: 28 μL sterile distilled water, 10 μL of 5× buffer, 2 μL of 10 mM dNTP mix, 2 μL of RT-PCR enzyme mix, 0.5 μL of RNase inhibitor, 0.5 μL of 100 mM dDTT per reaction and 1 μL each of forward and reverse primers chosen by the following three algorithms based on whether or not the RNA contains mixtures of polioviruses and the serotypes of the viruses in the mixtures. Stock concentrations of the primers are listed in Table 1 and were calculated to provide the correct final concentration when 1 μL is added to the mix.
-
Add 5 μL of RNA in Area 2 (see Note 44 ).
-
Run the RT-PCR using the following conditions: 42 °C for 45 min; 94 °C for 10-min; 35 cycles of 94 °C for 30 s, 42 °C for 45 s, ramp to 60 °C T 0.4 °C/s, and 60 °C for 2 min; 60 °C for 5 min; and 4 °C hold.
-
Algorithm 1: the RNA contains only the virus of interest or mixtures of sequence variants of this virus.
-
Generate an amplicon containing the complete VP1 sequence by RT-PCR using generic forward primer Y7 or Y7R with generic reverse primer Q8.
-
Purify the amplicon.
-
Confirm that it is of the correct size (as described below).
-
Label the amplicons for sequencing in separate reactions with the same primers used to prepare the VP1 amplicons.
-
Sequence both strands of the amplicon in separate reactions using primers Y7 and Q8.
-
-
Algorithm 2: Heterotypic mixes - the RNA contains the virus of interest or mixtures of sequence variants of this virus together with polioviruses of different serotypes.
-
Generate two overlapping amplicons specific for the virus of interest. Specificity is obtained by running two hemi-nested RT-PCRs. One hemi-nested RT-PCR is with generic forward upstream primer Y7 (or Y7R) and the reverse primer from the specific primer pair designed for qRT-PCR in step 2 of Subheading 3.4 above. The second hemi-nested RT-PCR is with the forward primer from the specific primer pair designed for qRT-PCR in step 2 of Subheading 3.4 above and generic primer Q8 (see Note 45 ).
-
Purify the amplicon as described below.
-
Confirm that it is of the correct size by gel electrophoresis.
-
Label the amplicons for sequencing in separate reactions with the same primers used to prepare each amplicon.
-
-
Alternative for Algorithm 2:
-
Use generic primers Y7 or Y7R with Q8 to amplify the VP1 from all viruses in the mix.
-
Purify the amplicon as described below.
-
Confirm that it is of the correct size by gel electrophoresis.
-
Specifically label the amplicons for the virus of interest in separate reactions using the specific qRT-PCR primers for the virus of interest as sequencing primers (see Note 46 ).
-
When Sabin 2-like viruses may need to be sequenced from heterotypic mixtures of polio to rule out emergence or silent circulation of type 2 VDPVs, first try RT-PCR with primer Q8 and 247 s (1061 nt) and then Y7 with 253A (989 nt), and use the same primers for sequencing (see Note 47 ).
-
-
Algorithm 3 Homotypic mixes: The RNA contains the virus of interest or mixtures of sequence variants of this virus together with polioviruses of the same serotype, for example the vaccine strain.
The most difficult situation is to specifically sequence the genome of interest from a mixture of viruses of the same serotype.
-
Produce Amplicons using any of the methods in Algorithm 2.
-
Purify the amplicon.
-
Confirm that they are of the correct size by gel electrophoresis.
-
Selectively label the amplicons using qRT-PCR specific primers or any serotype specific primers that will not cross hybridize with the other viruses in the homotypic mix (see Notes 46 and 47 ).
-
-
-
2.
Confirm that the amplicons generated from algorithms 1, 2, or 3 above are of the correct size by gel electrophoresis.
-
Load one tenth of the reaction mix onto a lane on a 2 % agarose gel containing 1 μL of a 1 mg/mL aqueous Ethidium bromide solution per 10 mL of agarose solution in a bath containing 1× E buffer.
-
Visualize the results with UV light, and record the results with a camera or other permanent recording device.
-
Estimate the size by comparison with fragments in a 100 base pair ladder DNA size marker loaded onto an adjacent lane and run at the same time.
-
-
3.
Purify the specific amplicon from the mixture of buffers, enzymes and primers according to the whether the electrophoresis pattern in the previous step indicated a single band of expected size or more than one band, including one of the anticipated size.
-
Algorithm 1: a single band of anticipated size.
Purify the amplicon directly from the mix remaining in the RT-PCR tube using a High Pure PCR product Purification Kit (Roche Diagnostics, Indianapolis, IN, USA).
-
Add binding buffer (green cap) in the amount of five times the volume of the reaction mix not including the volume of the mineral oil (if any). It is not necessary to remove the mineral oil.
-
Mix by pipetting gently to avoid spilling.
-
Transfer all of the liquid to the High Pure filter column (see Note 48 ).
-
Centrifuge for 1 min at 8000 × g at room temperature.
-
Transfer the column to a clean 2 mL test tube. Discard the flow through.
-
Wash the column by adding 500 μL of wash buffer (blue cap) and centrifuging for 1 min at 8000 × g.
-
Transfer the column to a clean 2 mL test tube. Discard the flow through.
-
Wash the column by adding 200 μL of wash buffer (blue cap) and centrifuging for 1 min at 8000 × g.
-
Transfer the column to a clean 1.5 mL test tube. Discard the flow through.
-
Centrifuge for 1 min at 8000 × g.
-
Transfer the column to a new clean 1.5 mL test tube labeled on the lid with the name of the sample.
-
Add 50 μL of elution buffer (vial #3), or sterile distilled water. If the band is faint elution can be in a minimum of 25 μL.
-
Let stand for 1 min at room temperature.
-
Centrifuge for 1 min at 8000 × g. Discard column.
-
Save the flow through liquid containing the amplicon at −20 or −70 °C.
-
-
Algorithm 2: Multiple bands, one of which is a band of anticipated size.
-
Load the remaining volume of reaction mix onto a 2 % agarose gel (see Note 49 ).
-
Load a 100 base pair size marker in a separate lane.
-
Electrophorese the sample and marker.
-
After electrophoresis, excise the band of the correct size for the amplicon from the agarose gel with a clean sharp disposable scalpel blade and place it in a pre-weighed 15 mL test tube. Re-weigh the tube to determine the net weight of the agarose gel slice by subtracting the initial weight from the final weight (see Note 50 ).
-
Add 3 volumes of Buffer QG to 1 volume of 1 % agarose gel or 6 volumes of Buffer QG to 1 volume of 2 % agarose gel using the approximation that 100 μL is equivalent to 100 mg (see Note 50 ).
-
Incubate at 50 °C for 10 min with flicking every 2–3 min. Ensure that pH remains OK (solution yellow) otherwise correct with 3 M sodium acetate (pH = 5.0).
-
Add 1 volume of isopropanol and mix.
-
Load sample on column (≤800 μL per run) and centrifuge at 10,000 × g for 1 min on a microcentrifuge.
-
Place the QIAquick column into new 2 mL eppendorf centrifuge tube, not into same tube as specified in manufacturer’s protocol. Discard tube and flow through (see Note 51 ).
-
Add 500 μL of Buffer QG and centrifuge at 10,000 × g for 1 min on a microcentrifuge centrifuge.
-
Transfer the QIAquick column into new 2 mL eppendorf centrifuge tube, not into same tube as specified in manufacturer’s protocol (see Note 51 ). Discard tube and flowthrough.
-
Wash the column by adding 750 μL of Buffer PE, incubate at RT for 5 min, and centrifuging at 10,000 × g for 1 min.
-
Transfer the column into new 2 mL tube and centrifuge at 10,000 × g for 1 min rather than discarding flowthrough and reusing same tube as in manufacturer’s protocol (see Note 51 ). Discard the original tube with flowthrough.
-
Transfer the spin column into 1.5 mL microcentrifuge tube and elute the DNA by applying 30–50 μL of water directly to the center of the QIAquick spin column membrane, letting the spin column stand for 2 min at RT, and centrifuging at 10,000 × g for 1 min.
-
Discard the spin column.
-
Store DNA in the flow through at −20 or −70 °C.
-
-
-
4.
Determine the concentrations of the amplicon DNA using a NanoDrop ND1000 spectrophotometer (Thermo Scientific, Waltham, MA, USA).
-
Turn the machine on, and then open the computer program for the NanoDrop.
-
Clean the spectrophotometer by lifting the sample arm and adding sterile distilled water to the measurement pedestal, closing and opening the sample arm and wiping both surfaces with a soft clean absorbent tissue (see Note 52 ).
-
First blank the NanoDrop machine by adding 1.8 μL of sterile water to the pedestal of the sample arm, close the sample arm, and then press the blank (F3) button on the computer program.
-
Open the sample arm and wipe both surfaces with a soft clean absorbent tissue.
-
Add an aliquot of sterile water to the lower measurement pedestal.
-
Close the sample arm and initiate a spectral measurement by pressing the measure button (F1) on the operating software on the computer.
-
After the measurement wipe both surfaces of the sample arm with a soft clean absorbent tissue.
-
Clean and repeat until the water blank gives a flat spectrum.
-
Finally, add samples and measure by pressing the measure button (F1) on the operating software on the computer (see Note 53 ).
-
Enter the name for each sample on the computer program and finished, print out a report.
-
Clean the spectrophotometer at the end of use.
-
-
5.
Label the dsDNA amplicon for sequencing with the Z reaction.
-
Prepare the Z reaction mix for n + 0.5 reactions in Molecular Area 1: (2 μL Big Dye Terminator Ready Reaction Premix (Life Technologies, Foster City, CA, USA), 1 μL of 5× buffer Tris–HCl, pH = 9.0 and MgCl2), 5 μL of primer diluted to give 3 pmol/reaction, and 5 μL of sterile deionized water: the final volume with cDNA is 10 μL.
-
Transfer the tube with the Z mix to Molecular Area 3.
-
In Molecular Area 3, add 1 μL the cDNA from step 11 above to the Z mix after first diluting the cDNA to a concentration of 3–10 ng per 100 nucleotides per 1 μL water and add.
-
Incubate the mix in a PCR machine using the following settings: 25 cycles of 94 °C for 20 s, 42 °C for 15 s, ramping to 60 °C at 0.4 °C/s, and 60 °C for 4 min, followed by a hold at 4 °C.—Purify the end-labeled cDNA using a BigDye X Terminator Purification Kit (Life Technologies, Foster City, CA, USA).
-
Transfer the 10 μL reaction mix into a well of a 96-well microtiter plate.
-
Add 45 μL of SAM Solution and 10 μL of BigDye X Terminator solution to the well. Use a wide bore tip for the BigDye X Terminator solution.
-
Cover the palate with adhesive film.
-
Thoroughly mix plate on a vortex or rotary shaker for 30 min.
-
Centrifuge the plate at 1000 × g for 2 min in a centrifuge with buckets suitable for 96-well microtiter plates.
-
-
6.
Sequencing the labeled VP1 amplicons
-
Transfer 15 μL of the Z reaction supernatant to the 96 well microtiter plate sample holder of the AB1 3100 or 3500 Genetic Analyzer (see Note 54 for suggestions on how to name the sample).
-
Sequence the amplicons.
-
Save the graphic sequence output files (they end in .AB1) from the automatic sequencing machine on a CD, DVD, disc-on-key, or equivalent.
-
-
7.
ITD and sequence analysis of VP1.
The protocol provided here is for the commercial Sequencher program v5.0 or later (GeneCodes, Madison, WI, USA) because we feel offers a very user-friendly platform for correcting primary sequence data and for immediately resolution of ITD. It can also highlight amino acid substitutions or differences and aligned sequences can be easily exported in a number of different formats including FASTA NEXUS and Phylip for more advanced phylogenetic analysis using other computer programs. Bold italicized text refers to the menu commands and a forward slant “/” indicate that the command following the forward slant is in a submenu. There are other commercial, share-ware, and free-ware programs for sequence correction and preliminary analysis.
-
1.
Prepare a Generic Sequencher project file for correction and preliminary ITD analysis of all polioviruses.
-
This file should contain text files for the complete genome of Sabin 1, Sabin 2, and Sabin 3 (DDBJ/GenBank/EMBL database access numbers V01150, AY184220, and X00925, respectively)
-
The file should contain text files of the complete VP1 sequence of all three Sabin strains extracted from the complete sequence files (906, 903, and 900 nt, respectively, for Sabin 1, Sabin 2, and Sabin 3).
-
This file should contain three copies of Y7 and Q8 primer sequences (see Note 55 ).
-
The file may contain graphic sequence files (files ending in .AB1) for amplicons of all three Sabin strains amplified using Y7 and Q8 primers.
-
Select all sequences (with the exception of the primers) and assemble automatically using the default values for Maximum Match Percentage (85 %) and minimum overlap (20 bp) in the Assembly Parameters to produce three separate contigs, one for each vaccine serotype. A contig is a sequence generated in silico by de novo assembly of sequences that partially or fully overlap.
-
Select each contig and a single copy of Y7 and Q8 primers and assemble automatically using the default values for Maximum Match Percentage (85 %) and minimum overlap (20 bp) in the Assembly Parameters.
-
Save this file with the name “proj polio”.
-
Duplicate this “proj polio” file and use the duplicate COPY for ITD analysis of all poliovirus sequences.
-
-
2.
Prepare a separate folder for each virus that will be sequenced and name it after the isolate. This folder should contain all raw (uncorrected) graphic sequence files from the given isolate even if from different regions of the genome and a copy of the “proj polio” file. The folder and project file should be renamed to reflect the name of the isolate (see Note 56 ).
-
3.
Transfer the .AB1 raw sequence files to the computer with the Sequencher program.
Add the name of the isolate to the beginning of the .AB1 file name (see Note 57 ).
-
4.
Load the raw data .AB1 file into the project file by using the File/import and one of the options in the next submenu commands or by dragging and dropping the icons of the .AB1 sequence files onto the icon of Sequencher or the open Sequencher window.
-
5.
Activate all newly loaded sequences (place the curser over the file name and for Mac: Command click; PC: control click) and trim the ends of the sequence (Sequences/Trim ends without preview).
-
6.
Preliminary correction of individual sequences in the individual sequence windows.
-
Open all of the newly trimmed sequences, e.g., double-click on the name or icon. This will open separate windows for each of the trimmed sequences.
-
Click on the button Show Chromatogram. The computer generated base calls are located below the line above the graphic peak for each nucleotide. These computer generated base calls cannot be edited, however there is an editable copy of each base call that appears above the line. It is these above-line base calls that are used by the Sequencher program for comparisons.
-
Correct the bases where the program cannot unambiguously call a base and returns a degenerate IUB base code (examples: “R” for a puRine, an “A” or a “G”, “N” for a Ny base) by placing the curser on the base call.
-
Visually inspect the beginning and end of the chromatogram. Trim ends without preview removes bases above the line at the 5′ and 3′ ends. You can restore as many trimmed bases that are readable by placing the cursor on the computer generated bases below the line and choosing Sequence/Revert to Experimental Data.
-
Correct ambiguous base calls of the restored sequences after visual inspection of the graphic data (see Note 58 ).
-
Use the Backspace key NOT the Delete key to remove extra bases.
-
-
7.
Prepare a contig of the corrected sequences from the previous step:
-
Activate the all the sequences form the same isolate (place the curser on the file name and Mac: Command click; PC: control click).
-
Press the Assembly Automatically button.
-
Place the cursor on the temporary text name of the contig, double-click to open the name for editing.
-
Rename the contig with the name of the viral isolate.
-
-
8.
Correct the sequences in the contig.
-
Open the contig by double-clicking on the icon to visualize the graphic results (change from Overview to Bases). While in overview, drag window to the region you wish to correct before using bases.
-
Correct the mismatches between overlapping sequences based on the graphics (see Note 58 ).
Use degenerate bases if there are more than one graphic trace at a given nucleotide position and the graphic traces are approximately equal on all overlapping strands at that nucleotide position. Otherwise choose the major peak if it is the same on all overlapping strands.
-
Save file at frequent intervals.
-
-
9.
Prepare a text file containing the consensus sequence of the corrected contig.
-
Select all bases in the consensus sequence (place cursor on consensus line and press Mac: Command A; PC: control A).
-
Copy the consensus (Mac: Command C; PC: control C).
-
Open a new sequence Sequence/Create a new sequence.
-
Type in the name of the new sequence in the small window that opens (isolate name; space or underline; VP1).
-
Click enter.
-
Paste the consensus sequence into the new window that opens (Mac: Command V: PC: Control V); closing the window and selecting “Record as Experimental data” from the new window that opens (see Note 59 ).
-
Close all open windows.
-
Add the newly created consensus sequence to the contig by activating the contig and the newly created consensus text sequence and clicking on “Assemble automatically”.
-
Open the contig file and switch to Bases.
-
Place the cursor on the name of consensus sequence and drag the newly created text sequence to the top of the list of sequences.
-
Activate all sequence names in the Sequence Name window and click on Sequence/compare bases to/“top sequence”. The position of all mismatches will be indicated in the window that opens. Go to each nucleotide position and correct any mismatches.
-
Save the file. This is critical so that if there is a problem in the next steps before the next save, work will not be lost. if there is a problem it allows the option of closing the file without saving and then restarting from the saved version
-
-
10.
Final correction and Automatic ITD determination. Determine the serotype and whether the virus is vaccine, a minimally diverged VDPV, a highly diverged VDPV, or wild by performing the following steps in order.
-
Activate the contig file containing the corrected consensus file and all three Sabin serotype files and form a new contig containing graphic and text files of the new sequence and one of the three Sabin prototypes by clicking on “Assemble automatically” (see Note 60 ).
-
Bring the consensus file of the sequence of interest to top of the list of sequences and the Sabin VP1 sequence immediately below it. If all has gone well, the consensus sequence of interest will be longer than the Sabin VP1 file.
Trim the consensus text file to the length of the Sabin VP1 sequence. If the sequence is shorter, it is possible to restore more of the trimmed sequences by placing the cursor on the position of the consensus sequence that includes the last base in the consensus sequence of the sequence of the virus of interest in Base view mode (bottom line), activating Show Chromatogram, placing the cursor over the lower window and clicking to activate the chromatogram window, going to the chromatogram of the graphic file of the sequence of interest in the lower window, moving the cursor to bases below the line on the graphic display of the graphic file of the sequence of interest and restoring trimmed bases as in step 6. Once again review and correct the restored segment (see Note 61 ).
-
-
11.
Save the proj file. Copy/paste or export the corrected and trimmed consensus sequence for the virus of interest to your sequence data base.
-
12.
If the unknown does not form a contig with one of the Sabin contigs using the default value for Assembly parameters, open the Assembly Parameters and successively increase the Minimum Match Percentage in steps of 2 % until a match occurs or you reach 81 %, since at 80 % all Sabin strains will form a single contig. If no match occurs at the 81 % setting, BLAST search the text consensus sequence of the unknown against the nt library at DDBJ/GenBank/EMBL either directly from Sequencher or by copy pasting the sequence into the BLAST window at NCBI (http://www.ncbi.nlm.nih.gov/ last accessed July 2014) to determine whether it is a poliovirus and which polioviruses serotype of wild or highly diverged VDPV it most closely resembles. Return to the Sequencher project file, activate the contig of the unknown and the Sabin serotype corresponding to the serotype of the closest matches in the BLAST search, and continue to increase the Minimum Match Percentage in steps of 2 % until a match occurs. Correct the file by comparison with the corresponding Sabin VP1 after removing paired gaps (see Note 60 ). Save the proj file. Copy or export the consensus sequence of your unknown isolate into your data base file. Change the “n” in “pvn” for the name of the folder and proj file to the serotype of the unknown virus.
-
1.
4 Notes
-
1.
In practice, most poliovirus-positive environmental samples contain one or at best a few polioviruses of interest [2, 5]. Under these conditions environmental surveillance protocols operate at the lower limits of detection. Thus negative findings cannot rule out the presence of polioviruses at levels below detection. However negative findings gain significance when they are part of a long sequence of negative results from frequent routine surveillance at the given site. It also requires that the limits of quantitation (LOQ) and limits of detection (LOD) of your molecular assays are well documented.
-
2.
The procedures of reagent preparation; nucleic acid extraction; nucleic acid amplification; detection of amplified products, and other post-amplification manipulation should be carried out in separate rooms designates as Molecular Areas 1 through 4 using equipment and even personal protective equipment such as color coded lab coats dedicated for exclusive use in each area. (Use clearly separated work areas if it is not possible to use separate rooms.) The work schedule must be planned in advance so that there is unidirectional movement from Area 1 to Area 4 on the same workday in order to avoid molecular or biochemical contamination. Molecular Area 1 is the area dedicated to the preparation of reagents for amplification by PCR. Use gloves and color-coded disposable lab coats at all times when working in this room. Do not use lab coats that have been outside of the room. Use a biological cabinet or dead-air box equipped for UV irradiation for handling all reagents using pipettors, pipettes tips, etc. that are kept in this room at all times. The only nucleic acids allowed in the room are primers and probes. It is prohibited to bring RNA or DNA, and especially amplified DNA into this room. After finishing work, the pipettors and any test tube racks should be decontaminated using chemical means or a UV Cross linker. Molecular Area 2 is for work with unamplified nucleic acids for example nucleic acid extraction from sewage and for the addition of extracted nucleic acids to the master mix. The room should contain a BSL-2 laminar flow hood for working with the samples and a dead air box for addition of the nucleic acids to the reaction mix. The biological safety hood and the pipettes should be cleaned with bleach/PCR decontamination spray All pipettors, pipettes, tips, etc. used in Molecular area 2 should be kept in this room at all times. These pieces of equipment and this area should not be used for pipetting or processing amplified DNA. Molecular Area 3 is an area used for molecular amplification and detection of amplified DNA. Infected tissue cultures that have reached full CPE, contain very high titers of virus and probably should be manipulated in Area 3 rather than Area 2. If Area 2 is used, work with the tissue cultures should be done last and the area thoroughly disinfected using agents that inactivate the virus and destroy any residual nucleic acids. Dedicated pipettors, pipettes and equipment should be kept in this area at all times. Use a color-coded lab coat in this area. Do not bring amplified DNA or any supplies or equipment from Area 3 into Areas 1 or 2. Molecular Area 4 is the area where amplified nucleic acids are manipulated. Manipulation includes but is not limited to gel electrophoresis, purification and labeling of amplified nucleic acids, labeling for sequencing and cloning,
-
3.
Initial test results and downstream molecular tests that are used depend on the types of poliovirus that may be present in sewage and in some cases on which WHO region of the Global Polio Laboratory Network a specific laboratory is located. For example in OPV-free areas (vaccinated exclusively with IPV) all viruses recovered from sewage are viruses of interest, whereas in OPV areas where viruses of interest may be a minority among OPV strains, selective tests must be used to pick out these viruses of interest. Histories of VDPVs from persistently infected individuals and population movements from areas with circulating VDPVs and wild polioviruses also play some role. The full battery of positive rule in tests recommended by the WHO /CDC will cover all these possibilities. The negative or rule out molecular tests for vaccine derived viruses may fail to pick up VDPVs if there are other viruses of the same serotype present in the sample. Rule-in tests are currently under development. See also Note 4 .
-
4.
The primers and probes for semiquantitative or quantitative reverse transcript polymerase chain reaction (qRT-PCR) that have been developed and distributed by the CDC for the GPLN are based on positive identification of poliovirus in general, and wild poliovirus lineages still in circulation and for vaccine strains in particular [19, 21–23]. There are three CDC/WHO molecular kits for characterization of polioviruses based on positive results (rule in). (Kit 1) The CDC/WHO poliovirus diagnostic rRT-PCR Kit contains reagents for six qRT-PCR tests for each RNA preparation: pan enterovirus, pan-poliovirus, type 1, type 2, and type 3 poliovirus, and a multiplex for Sabin 1, 2, and 3 (Kits 2 and 3). There are two separate kits, one for identifying wild type one or wild type three polioviruses for lineages circulating in South Asia and the other for lineages circulation western Africa. Additional qRT-PCR Kits test for the presence of vaccine derived polioviruses (VDPVs); however, they are based on ruling out that a poliovirus is vaccine or wild, and thus it is possible to obtain false-negative results when there are mixtures of VDPVs with vaccine or wild poliovirus strains. Protocols reagents and kits are currently under development by the CDC for positive identification of VDPVs.
-
5.
When the virus of interest is the only virus in the environmental sample, for example in sewage from a population vaccinated exclusively with IPV, plaque assay for correlating the number of viable viruses with RNA can be performed on concentrated sewage. Once the correlation has been made, it is possible to continue to follow the amount of virus of interest in a sample even after the introduction of the corresponding OPV strain in Supplementary Immunization Activities (SIAs) when plaque-based assays lose meaning because environmental samples contain homotypic (if monovalent OPV is used) or homotypic and heterotypic polio viruses (when bivalent or trivalent OPV is used) ([9] and Shulman unpublished). When the poliovirus of interest is already in homotypic or heterotypic mixtures, such as in sewage from a population vaccinated with OPV or combined program using both OPV and IPV, the correlation must be done using purified virus of interest and or by spiking poliovirus-negative environmental samples with the virus of interest.
-
6.
It is preferably that the magnetic bars be oval or egg shaped to minimize mechanical failure by entrapment in sediment in the sewage.
-
7.
Polyethylene glycol is hygroscopic and will adsorb moisture—therefore store it in closed containers.
-
8.
Chlorine solutions are corrosive. Wash surfaces after 15 min to remove chlorine residues with water or 70 % ethanol followed by water.
-
9.
Obtain stocks with the lowest passage numbers and freeze aliquots of early passages for secondary laboratory stocks. The WHO recommends that fresh cultures should be prepared from frozen secondary stocks after 15–20 passages. Sensitivity of cells must be demonstrated at frequent intervals for cultures that have been passed more than 20 times after restarting from the secondary stock. Cell sensitivity results should br reported to the WHO Regional Polio Laboratory Coordinator within 48 h of test completion.
-
10.
It is better if the incubators are 5 % CO2 incubators. However non-CO2 incubators can be used when plating tube cultures or after adding small volumes of virus to tube cultures as long as the tubes are tightly closed. The high number of cells that adhere to the wall and their metabolism in complete medium will keep the culture form turning alkaline. Tube cultures that are alkaline should be discarded. It is better to use CO2 incubators after viral challenge. Cells in viral growth medium do not metabolize as fast as in complete medium and are much more sensitive to pH changes. Cells in alkaline medium will undergo cytotoxic CPE that is difficult to distinguish from CPE caused by virus infection
-
11.
With the exception of plaque assays , liquids from uninfected cell cultures can be carefully poured off into collecting bottles that are disinfected afterwards.
-
12.
When it is not possible to use automatic composite samplers, collect 4–6 grab samples (about 400–200 mL each) during 2–3 h of the peak flow rate of the sewage. Mix the grab samples and keep 1 L for the concentration procedure.
-
13.
It is possible to pre-weigh 40 g portions of polyethylene glycol 60000 and add 8.8 g of NaCl as long as the portions are stored under conditions that prevent hydration of the hygroscopic polyethylene glycol.
-
14.
Most of the virus is in the sediment so pouring off a does not result in significant losses. The liquid is poured off primarily to avoid spillage during centrifugation using fixed angle centrifuge heads when centrifuge tubes are completely filled. When the head spins, the top of the liquid will turn perpendicular to the radius of the centrifuge head. Make sure that either before or during centrifugation no liquid will enter the capped region of the centrifuge tube.
-
15.
We use one 500 mL reusable centrifuge bottles per sample, a Sorval SLA 3000 Superlite rotor, and a refrigerated Sorval RC5C Plus centrifuge. Smaller rotors can be used for 250 mL centrifuge tubes
-
16.
This is important to minimize loss of virus during processing in general and for quantitation by decreasing micro-clumping of viruses (viruses trapped together by organic matter).
-
17.
In some cases the volume may >15 mL. Record the volume and proceed. This is important for determining the quantity of PFU per mL in unconcentrated sewage.
-
18.
This delay is very important to ensure the uptake and effectiveness of the antibiotics and antimycotics.
-
19.
Cell Sensitivity Testing of L20B and RD cells must be performed routinely (at least midway through their expected use of 15 passages or every 3 months) according to written WHO Standard Operating Procedures using Laboratory Quality Control (LQC) standards prepared according to the WHO recommended procedure. Sufficient quantities of appropriately labeled aliquots of the standards are stored at –20 °C. Permanent records of results must be kept in a format that allows analysis of trends. Corrective action must be taken (e.g., replacement of cell lines, retesting of samples) in response to evidence of reduced cell sensitivity.
-
20.
Some people prefer to wash the cell monolayer with PBS or a few mL of the Trypsin–Versene solution, pour it off and then add the rest of the 20–30 mL of Trypsin–Versene as indicated. This removes FBS and increases the efficiency of the Trypsin–Versene and may yield more single cells and fewer clumps.
-
21.
In cases when it is important to know the number of cells in the tube culture, place a drop of the suspension on a Neubauer hemocytometer counting chamber (1/400 mm2 and 1/10 mm deep). The counting chamber contains a grid that forms 9 large squares containing 16 smaller squares. The smaller squares in the center and middle large outer squares on all sides are further subdivided into 16 even smaller squares. Count the number of cells in any of the nine largest squares (or average the number of cells in two or more of the large squares) and multiply this by 104 to get the number of cells per mL. When counting choose two adjacent sides of the large cell and include all cells that overlap these lines. Ignore any cells that overlap the lines forming the remaining two adjacent sides.
-
22.
The WHO protocol currently requires that CPE-negative cultures be repassed once as well to increase the chance of isolating poliovirus.
-
23.
Pipette the cell suspensions up and down a few times before plating in order to break up any clumps. Do not let the cells sit too long before distribution or else they will start reattaching to the sides or bottom of the plastic flask. Reattachment may take longer if the cells are transferred to a sterile glass bottle or non-tissue culture plastic 50 mL centrifuge tube.
-
24.
To save on medium and serum, only prepare as many tubes needed. Remember, 5 mL of suspension diluted in medium is enough for twenty 2 mL-tube cultures, so 1 mL of this suspension diluted to 8 mL is sufficient for four test tubes.
For tube culture placed on test tube racks with a 20° slant, it is also important to place a dot or equivalent indicator on the upper side of the tube cultures after adding cell suspensions so that it is easy to locate the lower surface of the tube where the cells have settled and adhered to the surface. The dot should be on the bottom when using a regular microscope and on the top when visualizing the cells with an inverted microscope. We recommend adding a string of linked rubber bands through the holes on the slanted test tube rack to keep the tubes from turning once the cells have adhered since this will dry and kill the cells. More cells are needed if a roller apparatus is used instead of a slanted rack since more surface area of the tube will need to be covered by the monolayer. To economize, round culture tubes can be used instead of tubes with a plat culture surface.. Cells on the inner surface of the round tubes can be viewed on standard or inverted microscopes if tubes are kept from rolling with a simple angled stand made from two parallel rods with a leg at one end.
-
25.
BGM and RD plaque assay plates are used to determine the number of enteroviruses in the sewage as a quality control for viral recovery from concentrated sewage. According to WHO guidelines is enough to challenge tube cultures or small flask cultures of these cell lines and score the culture as CPE positive or negative. The WHO recommends a minimum positivity rate of 30 % for sewage as an indication that conditions have been maintained that would enable isolation of polio from the sample if it were present. We count plaques to obtain information on the level of non-polio enterovirus infections in a community that could potentially compete with and reduce the efficiency of vaccination with OPV. The number of non-polio enterovirus sewage samples will vary for each country. In Israel, 80 % of the sewage samples normally contain enteroviruses.
-
26.
When there are very low numbers of poliovirus particles and/or viable particles in a sewage sample it is not unusual to receive positive results for either tissue culture or molecular assay of RNA extracted from concentrated sewage, but not both. It is also possible to find different viruses or combinations of viruses in replicate tube cultures.
-
27.
The WHO recommends a blind passage of CPE negative L20B cultures on L20B and a second culture on L20B for each RD culture without regard to CPE. During outbreaks and especially after outbreak response with OPV, many RD cultures are positive. For rapid results, any poliovirus in the RD CPE-positive cultures can be characterized directly by molecular ITD assay (Subheading 3.4) from RNA extracted from the supernatant (Subheading 3.3) when molecular methods are sensitive enough to detect 0.2 PFU per RT-PCR reaction for RNA extracted from concentrated sewage [9]. This saves 1 week.
It is possible to pool aliquots from all replicate CPE-positive RD cultures from a single sewage sample (or to pool supernatants from all replicate RD cultures regardless of CPE when at least one tube culture is positive). It is critical to change micropipette aerosol barrier tips between tubes when pooling to prevent cross-contamination of the RD tubes. Unpooled RD tubes should be stored at −20 °C. To save on costly reagents it is possible to characterize the poliovirus directly from pooled aliquots by molecular ITD assay (Subheading 3.4) using RNA extracted from the pooled supernatants (Subheading 3.3) and/or to challenge a single L20B tube culture with the pooled supernatant. The reason for storing unpooled RD samples is that each of the tubes may contain different mixtures of poliovirus and sometimes this makes downstream processing involving sequencing and characterization of a virus of interest from mixture easier to perform (see Subheading 3.4). Characterizing the poliovirus in individual RD tubes may also provide a crude indication of viral load.
-
28.
During an outbreak and especially as the outbreak winds down and samples become negative it is crucial to be as careful as possible to avoid cross contamination of samples with intact virus and/or purified nucleic acids. It is also crucial when the laboratory also performs tests on clinical samples. Molecular assays and sequencing works best with RNA extracted from tissue cultures with full CPE. However these cultures contain very high titers of virus. To prevent cross contamination from intact virus and/or subsequently purified nucleic acids, we strongly recommend that these tissue culture samples be processed in an area completely separate from other activities in the lab where environmental samples are concentrated, stool samples processed, and especially where CSF is processed. Thus, whenever L20B cultures are positive, the presumption is that poliovirus caused the CPE and that these tubes contain very high titers of poliovirus. These tubes should only be opened in a separate BSL-2 laminar flow hood designated for this purpose and situated in a separate room. We used the KingFisher RNA extraction procedure for extracting RNA from these test tube cultures for cost considerations (the extractor is less expensive than the easyMAG extractor), convenience (a dedicated machine is in the same BSL-2 laminar flow hood as used to open the CPE-positive tube cultures) and for cross-contamination control (we extract clinical samples on the easyMAG extractor).
-
29.
Recommendation: add the sample to the sidewall of the plate rather than directly on the cell monolayer to avoid disturbing the monolayer.
-
30.
Unopened plates can be counted when held in the upright position or after inverting them. Don’t keep them at an angle or inverted for too long as the monolayer may tear and slide under the agar overlay and ruin the plaque assay or prevent picking plaques for amplification and characterization. For convenience when counting or designating plaques to pick, mark the positions of the plaques on the underside of each dish with a dot from a marker.
-
31.
Alternatives: use disposable glass Pasteur pipettes or 0.5 mL reusable glass pipettes that are cleaned with chlorine solutions and autoclaved for repeated use.
-
32.
There is a much lower chance of splashing if larger tubes (3–15 mL) are used to collect the agar plug; however, using larger tubes presents a disadvantage for short-term and long-term storage.
-
33.
Highest titers can be recovered after one pr two cycles of freeze thawing of the tube cultures. For ease in storage and conservation of storage space, supernatants may be transferred to small 1.5 or 2.0 mL tubes. RNA may be extracted immediately, after up to 4 days of storage at 4 °C, or at any time after storage at −20 or −70 °C.
-
34.
Preparation of secondary stocks (repassage) is necessary to ensure high enough titers for successful sequencing. Incubate the tube cultures at 37 °C until culture reaches full CPE, usually 1–3 days but may take as long as 5 days for some plaques.
-
35.
Pooling before extraction will reduce by up to fourfold the number of RNA extractions and the number of diagnostic rRT-PCR reactions that need to be performed without significantly reducing Ct for positive samples. Specifically if four samples are pooled the Ct of a semiquantitative qRT-PCR reaction will be increased by two cycles. If the workload is not excessive and sufficient storage space is available, store separate unpooled 1 mL samples from each tube that had CPE, otherwise store the pool (also see Note 27 ).
-
36.
Storing small single-use aliquots of the extraction positive control at −70 °C provides the best reproducibility between tests so that test performance can be evaluated using Westgard Rules [24, 25] and Levy Jennings plots [26]
-
37.
We recommend trying the primers designed by the CDC (Table 1) using your enzymes and buffers and compare results with the buffers in the kits supplied by the CDC. In some cases this may lower limits of detection by 3–4 orders of magnitude due to incompatibilities between the CDC buffer and your enzymes. In the future to avoid any such problems, CDC-designed primers and probes may be sent without buffer.
-
38.
Infecting cells at an MOI of >5 may produce secondary stocks with poorer ratios of infectious to noninfectious particles, while infection at a MOI < 0 may take more than 48 h for the virus in the culture to reach a high enough concentrations of virus for downstream molecular analyses. The ratio of infectious to noninfectious particles is important for the correlation of PFU per mL with qRT-PCR results since both infective and noninfective particles will produce positive signals in qRT-PCR. Two rounds of plaque purification are recommended since the original plaques occasionally contain more than one virus due to micro-clumping or overlapping of adjacent plaques. Prepare primary and secondary stocks from the original sample or if possible from different contemporary samples. Design of primers and probes using related samples makes it less likely that there will be sequence variation within the area chosen for the primers or probe. A miss-match in the 3′ base of primers will render the primer useless, and as variability increases in the target sequence complementary to the probe specific binding of the probe will decrease until detection is no longer reliable.
-
39.
We used Primer Express Software v3.0 (Life Technologies, Life Technologies, Thermo Fisher Scientific, Waltham, MA, USA) for designing compatible primers and probes for our ABI 7500 instrument. For our assay the specific probes for the poliovirus strain of interest were labeled with 6-carboxyfluorescein (FAM) and black hole quencher 2 (BHQ-2; Metabion, Planegg/Steinkirchen, Germany) so that we could use them in multiplex with our MS-2 primers and VIC labeled probe (Table 1) based on sequences previously published by Dreier et al. [18] that we purchased from Life Technologies (Life Technologies, NY, USA).
-
40.
If possible, design one primer pair and probe from the first third of VP1 and the other in the last third of the VP1 sequence. Validate both. Remember the primer sequence is nondegenerate. The virus of interest may mutate and render one of the primers or the probe ineffective. Two validated sets will also allow for longer overlapping sequences when using the forward primer form the first third with reverse generic primer Q8 and the reverse primer from the last third with generic forward primer Y7 (see Table 1 and the discussion on sequencing a virus of interest from RNA extracted form a homotypic mix in Subheading 3.5).
-
41.
We optimize results for this standard assay condition for all reactions with nondegenerate primers and probes so that we can run RT-PCR and even PCR reactions for many different viruses in the same qRT-PCR run. The only time we may need to vary run conditions is when degenerate primers are used such as for the Pan poliovirus qRT-PCR described here.
-
42.
Take all precautions to prevent contamination. Handle the 0.2 mL PCR strips for the qPCR machines by the lids, being careful not to contaminate the tubes. To decrease the chance for cross contamination, place a fresh sheet of absorbent lab mat on the work area and distribute reagents in a dead-air hood. Expel used aerosol barrier tips into a disposable waste cup or bag.
-
43.
If the cap is firmly closed there is an increased chance for splashing and contamination when the cap is briefly removed in Molecular Area 2 for addition of the RNA template.
-
44.
The final reaction volume is 50 μL. Add water to compensate for any decrease in volume of primers. The choice of primers depends on the identity of polioviruses in the tissue culture or concentrated sewage sample. The Samples may contain (A) just the virus of interest or mixtures of sequence variants of this virus, (B) a mixture of the virus of interest and viruses of other serotypes (heterotypic mix), or (C) mixtures of vaccine, vaccine-derived, and/or wild, polioviruses of the same serotype as the virus of interest (homotypic mixtures). qRT-PCR results from molecular ITD tests described in the previous section indicate which of these which of the three possibilities is the case and this information determine the choice of primers from among those listed in Table 1 for both amplification and sequencing. RNA. Amplification conditions may differ depending on whether the primers are nondegenerate or degenerate. For example the final concentration of primers in the reaction mix depends on whether or not they are degenerate and on how many degenerate bases they contain. For convenience this is taken into account (Table 1) when preparing the concentration of primer stock by adjusting the concentration so that the necessary amount of primer is present in 1 μL of stock.
-
45.
To create two specific amplicons with a long overlap from the two the hemi-nested RT-PCRs, choose as the forward specific primer, the forward specific qRT-PCR primer from the pair designed for the 5′ third of VP1 and the reverse specific qRT-PCR primer designed for the 3′ third of VP1. Specific primers from either one of the specific qRT-PCR primer pairs can also be used in the hemi-nested reactions. This will produce two amplicons with a much a shorter overlap and difficulty in reading sequences immediately adjacent to the sequencing primer may result in gaps where there is no overlap.
One or both hemi-nested reactions may not work. Try RT-PCRs with generic primers paired with type specific internal VP1 primers or just pairs of type specific primers from Table 1 (for example P1W2s with Q8 (900 bp) and Y7 with P1W1a (250 bp) for wild type one).
If it is not possible to obtain sequences from heterotypic mixes and it is important to characterize the virus of interest, the virus of interest may be enriched or be isolated by one or two rounds of tissue culture of dilutions of the mixture grown in the presence of neutralizing antibodies to the other virus serotypes in the mixture.
-
46.
If two sets of specific primers were prepared for the qRT-PCR than use the forward primer from the first third of VP1 and the reverse primer from the last third of VP1 to label amplicons with the longest overlap. Specific primers appropriate for the wild serotype (Table 1) can be substituted for the specific qRT-PCR.
An additional alternative is to use specific degenerate primers designed to be the CDC to identify SOAS type 1 or 3 poliovirus from currently endemic to south Asia or WEAF type 1 or 3 polioviruses endemic to western Africa.
-
47.
Sabin 2 is the OPV serotype that causes most live vaccine poliomyelitis outbreaks. If all else fails and it is important to characterize the type two sequence, isolate or enrich for type two virus by growing limiting dilutions of the mixture in the presence of neutralizing antibodies to the other virus serotypes in the mixture. Specific kits for rule-in type 1 and type 2 VDPVs by positive identification rather than rule-out are in the last stages of testing by the CDC. If it is not possible to obtain sequences from heterotypic mixes and it is important to characterize the virus of interest, the virus of interest may be enriched or be isolated by one or two rounds of tissue culture of dilutions of the mixture grown in the presence of neutralizing antibodies to the other virus serotypes in the mixture.
-
48.
Use the same tip a few times without resetting the volume. The maximum amount of RNA that can be loaded on a column is 400 mg.
-
49.
When forming the agarose gel rather than running the sample in two adjacent lanes, use cellophane tape to close the space between adjacent teeth of the slot (lane) to make a single slot long enough to accommodate all of the remaining volume of the reaction mix.
-
50.
When purifying a number of bands at the same time it is easier to bring all of the gel slices to the same weight by adding a volume of 1× E buffer equal to the difference in weight between each sample and the heaviest sample. If you do this you do not need to reset pipette volumes for each sample when dispensing reagents form the kit.
-
51.
Modification of manufacturer’s instructions to reduce chance of contamination with amplified PCR template.
-
52.
Cleaning between samples and especially after a sample with a high concentration of nucleic acids reduces the chance of carryover to a sample containing low levels. If necessary the sample arm can be decontaminated from potential biologically active materials with a freshly prepared solution of 0.5 % solution of sodium hypochlorite
-
53.
The ratio of sample absorbance at 260 and 280 nm assess the purity of the DNA and RNA a ratio of ~1.8 is generally accepted as “pure” for DNA and ~2.0 is considered “pure” for RNA. Lower rations may indicate the presence of protein or other contaminants some of which may affect downstream reactions.
-
54.
Suggested naming conventions: Name each sequencing reaction according to the following convention: Date (YY-MM-DD), organism (polio), the ID of the cDNA RT-PCR (example P1002-2), the primer (example Q8), and the position on the microtiter plate (example C10) combined into a single name without spaces by using the underline character. In this case the name is 14_07-08-POLIO-P1002-2-Q8C10.
-
55.
RT-PCR produces a double strand amplicon containing both primers sequences. Sequencing of one strand will read through the primer at the other end if the amplicon is short enough. These primer sequences must be trimmed off from the end of the sequence since the sequence of nucleotides in the primer may not be 100 % identical with actual genome sequence. Having them aligned on the sequence makes trimming easier.
-
56.
We use the following file naming convention before the serotype of the virus has been identified: pvn_isolate ID number that may include a hyphen and letter to indicate the sample type_three letter country code and two digits for the year of isolation (for example: pvn_8066-e_ISR13 identifies a poliovirus isolated from environmental sample 8066 collected in Israel in 2013). The n in pvn is changed to 1, 2, or 3 when the serotype becomes known. The word “polio” in the name of the copy of the generic “proj polio” file should be changed to reflect the ID of the sequence, e.g., “proj pv_8066-e”.
-
57.
Each raw sequence file should have the ID of the virus added at the beginning of the full name. This is helpful because the original raw data file name indicates the date of sequencing reaction, the amplicon sequenced, and the primer used for sequencing is preserved. Putting the ID name first enables the computer to use alphanumeric criteria to easily sort together raw sequence files from the same isolate using different sequencing primers especially when sequenced on different days.
-
58.
Correct ambiguous calls by the computer based on a clear call for a complementary strand or sequence from the same strand produced using different primers since in almost all cases the sequences should be identical, e.g., usually there is a unique base at each position of the sequence. Normally when sequences do not match 100 %, this is an indication of a mistake in identification, requiring reviewing all steps from sample to sequence. However multiple peaks can be observed at one or more position when there are mixtures of closely related viruses and a minority species accounts for >10 % of the population. In addition sequences may differ when overlapping amplicons are sequenced for closely related mixtures of viruses using nondegenerate primers and there is a mismatch in some of the sequences at the 3′ base of one of the primers. When this occurs the two amplicons are no longer equivalent as a sub-member of the mix may be overrepresented or underrepresented. See discussion on homotypic mixtures in environmental samples i.e. when environmentl samples contain mixtures of the same serotype.
The position of mismatches is indicated by a dot under the bottom line containing the consensus and in the consensus itself by a degenerate base or semicolon (for gaps after alignment). Place the cursor on the first base of the consensus sequence and press command N (Mac) or control N (PC) and the position will shift to the next mismatch to the right. After doing this twice the space bar becomes a shortcut. If the shift is depressed at the same time the shift is used, the program will scan for mismatches 5′ ( to the left). Pressing Command 4 or Control 4 with automatically reverse complement all of the sequences in the contig without affecting alignment. This is convenient since in Sequencher, restoring and deleting sequences in an alignment is easier when done at the 3′ end of a sequence.
At the 5′ end there may be problems with uniform distances between peaks (an electrophoresis artifact resulting from the influence of neighboring bases on secondary structures of short sequences) that results in two partially overlapping peaks with one computer base call. If so correct this by placing the cursor on the base to the right of the missing base above the line and pressing the tab key to introduce a semicolon, “:”. Move the cursor to the semicolon and type in the missing base. At the 3′ end, the periodicity of peaks and especially the delineation between peaks and troughs for repetitive bases may be disrupted resulting in stretched out peaks for which the program will insert an extra base.
Usually there is a unique base at each position of the sequence. However multiple peaks can be observed at one or more position when there are mixtures of closely related viruses and a minority species accounts for > 10 % of the population.
-
59.
Alternative: select all of one sequence and paste into the new sequence, return to the contig and highlight and copy additional nonoverlapping sequences from other strands starting with the first nonoverlapping base.
-
60.
Note when you use the default Assembly Parameters for overlap (20 nt) and minimum match Percentage the Sabin contigs for each serotype remain separate and the unknown will match with one of them if it is vaccine or vaccine-derived with <15 % sequence difference. The sequence will not form a contig with one of the Sabin serotypes if it is from a highly diverged (>15 %0) VDPV or form a wild poliovirus.
-
61.
When correcting, look especially for gaps, paired gaps between the sequence of interest and the Sabin sequence, and at all bases that are different from the Sabin sequences. The graphic file of the Sabin strain can help to resolve problems at the beginning and end of sequences where the periodicity of peaks may be nonuniform and lead to incorrect automatic base calls. During alignment, the penalty for gaps for a short segment may be less than the penalty for the number of mismatched bases and the program will introduce a pair of gaps one in the prototype and another in the unknown. When this occurs activate the segment of aligned sequences between and including the matched gaps, gather the gaps to the left or right (Sequence/gather gaps/right) and delete the aligned gap by putting the cursor on the gap on the consensus line and pressing the backspace key.
Abbreviations
- AFP:
-
Acute flaccid paralysis
- AR:
-
Analytic reagent grade chemicals
- bp:
-
Base pairs
- BSL-2:
-
Biological safety level two
- CDC:
-
Centers for Disease Control and Prevention, Atlanta, GA, USA
- CPE:
-
Cytopathic effect—morphological changes in cells resulting from infection
- Ct:
-
Cycle threshold, the cycle at which specific signal from the probe is first detected above the threshold of detection in qRT-PCR
- FBS:
-
Fetal bovine serum
- GPEI:
-
Global Poliovirus Eradication Initiative
- IPV:
-
Inactivated polio vaccine (trivalent contains all three Salk serotypes)
- ITD:
-
Intratypic differentiation, e.g., determining whether the isolate is vaccine-like, VDPV, or wild
- MOI:
-
Multiplicity of infection
- OPV:
-
Live oral poliovirus (trivalent contains all three Sabin serotypes bivalent contains Sabin serotypes 1 and 3, monovalent contains either serotype 1, 2, or 3, exclusively)
- PCR:
-
Polymerase chain reaction
- PFU:
-
Plaque forming units
- qRT-PCR:
-
Quantitative reverse transcription polymerase chain reaction
- RT-PCR:
-
Reverse transcription polymerase chain reaction
- SIA:
-
Supplementary Immunization Activities
- TD:
-
Typic differentiation determining the serotype of the virus of interest
- VDPV:
-
Vaccine-derived polio virus
- VP1:
-
Viral capsid protein 1
- VP2:
-
Viral capsid protein 2
- VP3:
-
Viral capsid protein 3
- VP4:
-
Viral capsid protein 4
- WHO:
-
World Health Organization
- 5′ UTR:
-
5′ untranslated region of polio and non-polio enteroviruses
References
Racaniello VR (2001) Picornaviridae: the viruses and their replication. In: Fields BN, Knipe N, Howley P (eds) Virology, 4th edn. Lippincott Williams & Wilkins, Philadelphia, PA, pp 685–722
Shulman LM (2012) Polio and its epidemiology. In: Meyers RA (ed) SpringerLink (Online service). Encyclopedia of sustainability science and technology. Springer, New York, pp 8123–8173
Dowdle W, van der Avoort H, de Gourville E, Delpeyroux F, Desphande J, Hovi T, Martin J, Pallansch M, Kew O, Wolff C (2006) Containment of polioviruses after eradication and OPV cessation: characterizing risks to improve management. Risk Anal 26(6):1449–1469
Kew OM, Sutter RW, de Gourville EM, Dowdle WR, Pallansch MA (2005) Vaccine-derived polioviruses and the endgame strategy for global polio eradication. Annu Rev Microbiol 59:587–635
Hovi T, Shulman LM, van der Avoort H, Deshpande J, Roivainen M, De Gourville EM (2012) Role of environmental poliovirus surveillance in global polio eradication and beyond. Epidemiol Infect 140(1):1–13
Ranta J, Hovi T, Arjas E (2001) Poliovirus surveillance by examining sewage water specimens: studies on detection probability using simulation models. Risk Anal 21(6):1087–1096
WHO (2003) Guidelines for environmental surveillance of poliovirus circulation: WHO, Dept of Vaccines and Biologicals. http://apps.who.int/iris/handle/10665/67854. Last accessed 23 July 2014
Hovi T, Stenvik M, Partanen H, Kangas A (2001) Poliovirus surveillance by examining sewage specimens. Quantitative recovery of virus after introduction into sewerage at remote upstream location. Epidemiol Infect 127(1):101–106
Hindiyeh MY, Moran-Gilad J, Manor Y, Ram D, Shulman LM, Sofer D, Mendelson E (2014) Development and validation of a quantitative RT-PCR assay for investigation of wild type poliovirus 1 (SOAS) reintroduced into Israel. Euro Surveill 19(7):pii: 207103800
Lodder WJ, Buisman AM, Rutjes SA, Heijne JC, Teunis PF, de Roda Husman AM (2012) Feasibility of quantitative environmental surveillance in poliovirus eradication strategies. Appl Environ Microbiol 78(11):3800–3805
Shulman LM, Manor Y, Sofer D, Mendelson E (2012) Bioterrorism and surveillance for infectious diseases—lessons from poliovirus and enteric virus surveillance. J Bioterr Biodef S4:004
WHO (2004) Polio laboratory manual, 4th ed. WHO/IVB/04.10, WHO
Anis E, Kopel E, Singer SR, Kaliner E, Moerman L, Moran-Gilad J, Sofer D, Manor Y, Shulman LM, Mendelson E, Gdalevich M, Lev B, Gamzu R, Grotto I (2013) Insidious reintroduction of wild poliovirus into Israel, 2013. Euro Surveill 18(38):pii: 20586
Shulman LM, Hindiyeh M, Muhsen K, Cohen D, Mendelson E, Sofer D (2012) Evaluation of four different systems for extraction of RNA from stool suspensions using MS-2 coliphage as an exogenous control for RT-PCR inhibition. PLoS One 7(7):e39455
Manor Y, Shulman LM, Hindiyeh M, Ram D, Sofer D, Moran-Gilad J, Lev B, Grotto I, Gamzu R, Mendelson E (2014) Intensified environmental surveillance supporting the response to wild-type poliovirus type 1 silent circulation in Israel, 2013. Euro Surveill 19(7):20708
Shulman LM, Gavrilin E, Jorba J, Martin J, Burns CC, Manor Y, Moran-Gilad J, Sofer D, Hindiyeh MY, Gamzu R, Mendelson E, Grotto I, Genotype-Phenotype_Identification_(GPI)_Group (2014) Molecular epidemiology of silent introduction and sustained transmission of wild poliovirus type 1, Israel, 2013. Euro Surveill 19(7):pii=20709, Available online: http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20709
Saunders N, Zambon M, Sharp I, Siddiqui R, Bermingham A, Ellis J, Vipond B, Sails A, Moran-Gilad J, Marsh P, Guiver M, Division HPAMS (2013) Guidance on the development and validation of diagnostic tests that depend on nucleic acid amplification and detection. J Clin Virol 56(3):260–270
Dreier J, Stormer M, Kleesiek K (2005) Use of bacteriophage MS2 as an internal control in viral reverse transcription-PCR assays. J Clin Microbiol 43(9):4551–4557
Kilpatrick DR, Yang CF, Ching K, Vincent A, Iber J, Campagnoli R, Mandelbaum M, De L, Yang SJ, Nix A, Kew OM (2009) Rapid group-, serotype-, and vaccine strain-specific identification of poliovirus isolates by real-time reverse transcription-PCR using degenerate primers and probes containing deoxyinosine residues. J Clin Microbiol 47(6):1939–1941
Sanger F, Coulson AR (1975) A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol 94(3):441–448
Kilpatrick DR, Ching K, Iber J, Campagnoli R, Freeman CJ, Mishrik N, Liu HM, Pallansch MA, Kew OM (2004) Multiplex PCR method for identifying recombinant vaccine-related polioviruses. J Clin Microbiol 42(9):4313–4315
Kilpatrick DR, Nottay B, Yang CF, Yang SJ, Da Silva E, Penaranda S, Pallansch M, Kew O (1998) Serotype-specific identification of polioviruses by PCR using primers containing mixed-base or deoxyinosine residues at positions of codon degeneracy. J Clin Microbiol 36(2):352–357
Kilpatrick DR, Nottay B, Yang CF, Yang SJ, Mulders MN, Holloway BP, Pallansch MA, Kew OM (1996) Group-specific identification of polioviruses by PCR using primers containing mixed-base or deoxyinosine residue at positions of codon degeneracy. J Clin Microbiol 34(12):2990–2996
Westgard JO, Groth T, Aronsson T, Falk H, de Verdier CH (1977) Performance characteristics of rules for internal quality control: probabilities for false rejection and error detection. Clin Chem 23(10):1857–1867
Westgard JO, Barry PL, Hunt MR, Groth T (1981) A multi-rule Shewhart chart for quality control in clinical chemistry. Clin Chem 27(3):493–501
Levey S, Jennings ER (1950) The use of control charts in the clinical laboratory. Am J Clin Pathol 20(11):1059–1066
Toyoda H, Kohara M, Kataoka Y, Suganuma T, Omata T, Imura N, Nomoto A (1984) Complete nucleotide sequences of all three poliovirus serotype genomes. Implication for genetic relationship, gene function and antigenic determinants. J Mol Biol 174:561–585
Kilpatrick DR, Iber JC, Chen Q, Ching K, Yang SJ, De L, Mandelbaum MD, Emery B, Campagnoli R, Burns CC, Kew O (2011) Poliovirus serotype-specific VP1 sequencing primers. J Virol Methods 174:128–130
Centers for Disease Control and Prevention, Atlanta, GA., Kit Instructions In: Poliovirus rRT-PCR ITD 4.0 Kit. A kit for serotyping of L20B positive cell cultures and intratypic differentiation of polioviruses in support of the Global Polio Eradication Initiative. Distributed by The WHO Collaborating Centre for Enteroviruses and Polioviruses, Centers for Disease Control and Prevention, 1600 Clifton Road NE, Mailstop G-10, Atlanta, Georgia 30333 USA, +1-404-639-1341, Fax: +1-404-639-4011. Email: MMandelbaum@cdc.gov, HSun@cdc.gov, SOberste@cdc.gov. November 20, 2014
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer Science+Business Media New York
About this protocol
Cite this protocol
Shulman, L.M., Manor, Y., Hindiyeh, M., Sofer, D., Mendelson, E. (2016). Molecular Characterization of Polio from Environmental Samples: ISSP, The Israeli Sewage Surveillance Protocol. In: Martín, J. (eds) Poliovirus. Methods in Molecular Biology, vol 1387. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-3292-4_5
Download citation
DOI: https://doi.org/10.1007/978-1-4939-3292-4_5
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-3291-7
Online ISBN: 978-1-4939-3292-4
eBook Packages: Springer Protocols