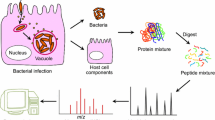
Abstract
Host–pathogen protein–protein interactions are highly complex and dynamic and mediate key steps in pathogen adhesion to host, host invasion, and colonization as well as immune evasion. In bacteria, these interactions most often involve specialized virulence factors or effector proteins that specifically target central host proteins. Here, I present a mass spectrometry-based proteomics approach starting with the identification of host–pathogen interactions by affinity-purification followed by mapping the specific host–pathogen protein–protein interaction interfaces by crosslinking mass spectrometry and structural modeling of the complexes.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Happonen L, Hauri S, Birkedal GS et al (2019) A quantitative Streptococcus pyogenes–human protein–protein interaction map reveals localization of opsonizing antibodies. Nat Commun 10:2727. https://doi.org/10.1038/s41467-019-10583-5
Liu X, Huuskonen S, Laitinen T et al (2021) SARS-CoV-2–host proteome interactions for antiviral drug discovery. Mol Syst Biol 17:e10396. https://doi.org/10.15252/msb.202110396
Penn BH, Netter Z, Johnson JR et al (2018) An Mtb-human protein-protein interaction map identifies a switch between host antiviral and antibacterial responses. Mol Cell 71:637–648.e5. https://doi.org/10.1016/j.molcel.2018.07.010
Jäger S, Cimermancic P, Gulbahce N et al (2012) Global landscape of HIV–human protein complexes. Nature 481:365–370. https://doi.org/10.1038/nature10719
D’Costa VM, Coyaud E, Boddy KC et al (2019) BioID screen of Salmonella type 3 secreted effectors reveals host factors involved in vacuole positioning and stability during infection. Nat Microbiol 4:2511–2522. https://doi.org/10.1038/s41564-019-0580-9
Olson MG, Widner RE, Jorgenson LM et al (2019) Proximity labeling to map host-pathogen interactions at the membrane of a bacterium-containing vacuole in chlamydia trachomatis-infected human cells. Infect Immun 87:e00537–e00519. https://doi.org/10.1128/iai.00537-19
Dickinson MS, Anderson LN, Webb-Robertson B-JM et al (2019) Proximity-dependent proteomics of the chlamydia trachomatis inclusion membrane reveals functional interactions with endoplasmic reticulum exit sites. PLoS Pathog 15:e1007698. https://doi.org/10.1371/journal.ppat.1007698
V’kovski P, Gerber M, Kelly J et al (2019) Determination of host proteins composing the microenvironment of coronavirus replicase complexes by proximity-labeling. elife 8:e42037. https://doi.org/10.7554/elife.42037
Breton ML, Meyniel-Schicklin L, Deloire A et al (2011) Flavivirus NS3 and NS5 proteins interaction network: a high-throughput yeast two-hybrid screen. BMC Microbiol 11:234. https://doi.org/10.1186/1471-2180-11-234
Qin L, Wang X, Gao Y et al (2020) Roles of EvpP in Edwardsiella piscicida-macrophage interactions. Front Cell Infect Mi 10:53. https://doi.org/10.3389/fcimb.2020.00053
Margarit I, Bonacci S, Pietrocola G et al (2009) Capturing host-pathogen interactions by protein microarrays: identification of novel streptococcal proteins binding to human fibronectin, fibrinogen, and C4BP. FASEB J 23:3100–3112. https://doi.org/10.1096/fj.09-131458
Hauri S, Khakzad H, Happonen L et al (2019) Rapid determination of quaternary protein structures in complex biological samples. Nat Commun 10:192. https://doi.org/10.1038/s41467-018-07986-1
Chowdhury S, Khakzad H, Bergdahl GE et al (2021) Streptococcus pyogenes forms serotype- and local environment-dependent interspecies protein complexes. Msystems 6:e0027121. https://doi.org/10.1128/msystems.00271-21
Schweppe DK, Harding C, Chavez JD et al (2015) Host-microbe protein interactions during bacterial infection. Chem Biol 22:1521–1530. https://doi.org/10.1016/j.chembiol.2015.09.015
Tyanova S, Temu T, Cox J (2016) The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc 11:2301–2319. https://doi.org/10.1038/nprot.2016.136
Tyanova S, Temu T, Sinitcyn P et al (2016) The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13:731–740. https://doi.org/10.1038/nmeth.3901
Chen Z-L, Meng J-M, Cao Y et al (2019) A high-speed search engine pLink 2 with systematic evaluation for proteome-scale identification of cross-linked peptides. Nat Commun 10:3404. https://doi.org/10.1038/s41467-019-11337-z
Grimm M, Zimniak T, Kahraman A, Herzog F (2015) xVis: a web server for the schematic visualization and interpretation of crosslink-derived spatial restraints. Nucleic Acids Res 43:W362–W369. https://doi.org/10.1093/nar/gkv463
Zundert GCP v, Rodrigues JPGLM, Trellet M et al (2016) The HADDOCK2.2 web server: user-friendly integrative modeling of biomolecular complexes. J Mol Biol 428:720–725. https://doi.org/10.1016/j.jmb.2015.09.014
Yang B, Wu Y-J, Zhu M et al (2012) Identification of cross-linked peptides from complex samples. Nat Methods 9:904–906. https://doi.org/10.1038/nmeth.2099
Jumper J, Evans R, Pritzel A et al (2021) Highly accurate protein structure prediction with alpha fold. Nature 596:583–589. https://doi.org/10.1038/s41586-021-03819-2
Chambers MC, Maclean B, Burke R et al (2012) A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotechnol 30:918–920. https://doi.org/10.1038/nbt.2377
Schiffrin B, Radford SE, Brockwell DJ, Calabrese AN (2020) PyXlinkViewer: a flexible tool for visualization of protein chemical crosslinking data within the PyMOL molecular graphics system. Protein Sci 29:1851–1857. https://doi.org/10.1002/pro.3902
Acknowledgments
Support from the Swedish National Infrastructure for Biological Mass Spectrometry (BioMS) and the SciLifeLab, Integrated Structural Biology platform (ISB), is gratefully acknowledged. The cloning, expression, and purification of ISP and GFP were performed at the Lund Protein Production Platform, Lund University, Sweden (http://www.lu.se/lp3). This work was supported by the Crafoord Foundation, the Royal Physiographic Society of Lund, Stiftelsen Clas Groschinskys Minnesfond, Åke Wibergs Stiftelse, and Alfred Österlunds Stiftelse. Emil Tykesson is acknowledged for generating the AlphaFold model of ISP, and Simon Ekström and Esko Oksanen for critically reviewing this chapter.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Happonen, L.J. (2023). Affinity-Purification Combined with Crosslinking Mass Spectrometry for Identification and Structural Modeling of Host–Pathogen Protein–Protein Complexes. In: Nordenfelt, P., Collin, M. (eds) Bacterial Pathogenesis. Methods in Molecular Biology, vol 2674. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-3243-7_12
Download citation
DOI: https://doi.org/10.1007/978-1-0716-3243-7_12
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-3242-0
Online ISBN: 978-1-0716-3243-7
eBook Packages: Springer Protocols