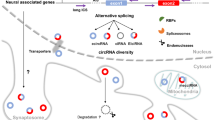
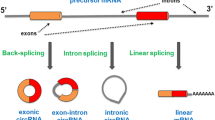
Abstract
In addition to expressing a large number of protein-coding transcripts, including alternatively spliced isoforms of the same mRNAs, neurons express a large number of noncoding RNAs. These include microRNAs (miRNAs), circular RNAs (circRNAs), and other regulatory RNAs. The isolation and quantitative analyses of diverse types of RNAs in neurons are critical to understand not only the posttranscriptional mechanisms regulating mRNA levels and their translation but also the potential of several RNAs expressed in the same neurons to regulate these processes by generating networks of competing endogenous RNAs (ceRNAs). This chapter will describe methods for the isolation and analyses of circRNA and miRNA levels from the same brain tissue sample.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162:156–159. https://doi.org/10.1016/0003-2697(87)90021-2
Glažar P, Papavasileiou P, Rajewsky N (2014) CircBase: a database for circular RNAs. RNA 20:1666–1670. https://doi.org/10.1261/rna.043687.113
Dudekula DB, Panda AC, Grammatikakis I et al (2016) CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol 13:34–42. https://doi.org/10.1080/15476286.2015.1128065
Untergasser A, Cutcutache I, Koressaar T et al (2012) Primer3-new capabilities and interfaces. Nucleic Acids Res 40. https://doi.org/10.1093/nar/gks596
Zimmerman AJAJ, Hafez AKAK, Amoah SKSK et al (2020) A psychiatric disease-related circular RNA controls synaptic gene expression and cognition. Mol Psychiatry 25. https://doi.org/10.1038/s41380-020-0653-4
Panda A, Gorospe M (2018) Detection and analysis of circular RNAs by RT-PCR. Bio-Protocol 8. https://doi.org/10.21769/bioprotoc.2775
Dell’Orco M, Oliver RJ, Perrone-Bizzozero N (2020) HuD binds to and regulates circular RNAs derived from neuronal development- and synaptic plasticity-associated genes. Front Genet 11. https://doi.org/10.3389/fgene.2020.00790
Livak LJ, Schmittgen TD (2001) Analysis of relative gene expression data using realtime quantitative PCR and the 22DDCT method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Bastle RM, Oliver RJ, Gardiner AS et al (2017) In silico identification and in vivo validation of miR-495 as a novel regulator of motivation for cocaine that targets multiple addiction-related networks in the nucleus accumbens. Mol Psychiatry 23:434–443. https://doi.org/10.1038/mp.2016.238
Acknowledgments
This work was supported by NIH, R01DA034097, and R21 DA048651 grants to N.P.B. as well as a mentored PI grant as part of an P20 grant from the NIGMS (1P20GM121176-01, N.M.), a NIAAA R01 grant (R01AA026583, N.M. and G.P.), a NARSAD Young Investigator Grant (FP00000839, N.M.) by the Brain & Behavior Research Foundation, and a high priority short-term R56 award from the NIMH (1R56MH119150-01, N.M.).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Dell’Orco, M., Papageorgiou, G., Mellios, N., Perrone-Bizzozero, N.I. (2023). Noncoding Regulatory RNAs: Isolation and Analysis of Neuronal Circular RNAs and MicroRNAs. In: Udvadia, A.J., Antczak, J.B. (eds) Axon Regeneration. Methods in Molecular Biology, vol 2636. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-3012-9_5
Download citation
DOI: https://doi.org/10.1007/978-1-0716-3012-9_5
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-3011-2
Online ISBN: 978-1-0716-3012-9
eBook Packages: Springer Protocols