Abstract
Background
MicroRNAs (miRNAs) are a family of endogenous, small and non-coding RNAs that regulate gene expression negatively at the post-transcriptional level by suppressing translation or degrading target mRNAs, and are involved in diverse biological and pathological processes. Single nucleotide polymorphisms (SNPs) which are located in the miRNA-coding genes may participate in the process of development and diseases by altering the expression of mature miRNA. Recent studies investigating the association between hsa-mir-499 polymorphism (rs3746444) and cancer risk have yielded conflicting results.
Methods
In this meta-analysis, we conducted a search of case–control studies on the associations of SNP rs3746444 with susceptibility to cancer in electronic databases. A total of 31 studies involving 12799 cases and 14507 controls were retrieved and the strength of the association was estimated by pooled odds ratios (ORs) and 95% confidence intervals (CIs). Hardy-Weinberg equilibrium (HWE) was assessed by the goodness-of-fit chi-square test in controls. Subgroup analyses were done by racial descent and cancer type. Publication bias of literatures was evaluated by visual inspection of funnel plots and the linear regression asymmetry test by Egger et al. Sensitivity analysis was conducted by excluding one study at a time to examine the influence of individual data set on the pooled ORs.
Results
Overall, significant association between rs3746444 polymorphism and susceptibility to cancer was identified in TC versus TT and TC/CC versus TT (dominant) models. In the stratified analyses, increased risks were found in Asians, but not in Caucasians in all comparison models tested. Moreover, significant association with an increased risk was found in Chinese population. Also, much higher significant association with increased cancer risks were found in Iranian population. In different cancer types, a decreased risk was found in esophageal cancer.
Conclusion
Our meta-analysis suggested that hsa-mir-499 rs3746444 T > C polymorphism is associated with the risk of cancer in Asians, mainly in Iranian and Chinese population. However, rs3746444 T > C polymorphism is negatively associated with the risk of esophageal cancer.
Similar content being viewed by others
Background
In early 1990s, microRNAs (miRNAs) were first discovered through analysis of developmental timing mutants in C. elegans by several groups, simultaneously [1],[2]. In the last decade, the study of miRNA biology has attracted remarkable attention, resulting in rapid advances, and revealing miRNAs as key gene regulators in diverse biological pathways [3]. miRNAs participate in regulation of stem cell functions [4], development [5], drug resistance [6], metabolism and metabolic disorders [7], cardiovascular [8] and malignant diseases [9],[10].
miRNAs are hairpin-derived RNAs ~20-24 nucleotides long, which post-transcriptionally repress the expression of target genes usually by binding to the 3′-untranslated region (3′ UTR) of messenger RNA (mRNA) in a broad range of organisms in both normal physiological and disease contexts [11],[12]. The majority of human miRNA loci are located within intronic regions and are transcribed by RNA polymerase II as part of their hosting transcription units [13]. Typically, following RNA polymerase II-mediated transcription, the long primary precursors (pri-miRNAs) are cleaved by the nuclear RNase III (Drosha) to release ~70 nt pre-miRNAs [14]. The resulting transcripts adopt a stem-loop structure and are then exported to the cytoplasm where they are subsequently processed by another RNase III (Dicer) to generate mature double-stranded ~22 nt miRNAs [15]. Subsequently, one strand of this duplex is incorporated into an Argonaute-containing RNA-induced silencing complex (RISC), resulting in the translational repression and/or degradation of their target mRNAs [16].
Recent evidences support that single nucleotide polymorphisms (SNPs), occurring in DNA sequences of miRNA-coding genes or in miRNA-binding site in mRNAs, contribute to the biogenesis and functions of miRNAs [17]. Gain/loss-of-function of miRNA polymorphisms may result in enhancing of the combination of the miRNA to the targets or losing control of the mRNAs, which may be associated with diseases [18].
Reports on the associations between SNPs in pre-miRNA or SNPs in binding sites in 3′ UTR of miRNA targeting mRNAs and human diseases have provided new insights into the molecular mechanisms of pathophysiological processes in humans. rs4846049 (G > T) of MTHFR gene is associated with increased risk for coronary heart diseases, and the potentially pathogenetic mechanism may be SNP-modified posttranscriptional gene regulation by miRNA-149 to MTHFR [19]. MiR-196a binding-site SNP regulates RAP1A expression contributing to esophageal squamous cell carcinoma risk and metastasis [20]. A miRNA-binding SNP (1010A/G) located within 3′-UTR of HOXB5 is associated with gene expression and may be a promising prognostic factor for bladder cancer [21]. The A to G base change of rs999885 may provide a protective effect against chronic HBV infection but an increased risk for HCC in HBV persistent carriers by altering the expression of the miR-106b-25 cluster [22]. miR-200b/200c/429-binding site polymorphism in the 3′-UTR of AP-2α gene is associated with cisplatin resistance, suggesting that SNP (rs1045385) A > C variation may be a potential prognostic marker for cisplatin treatment [23].
To date, many studies explored the association between rs3746444 T > C SNP in hsa-mir-499 and susceptibility to diseases, such as breast cancer [24]–[26], lung cancer [27], congenital heart disease [28], dilated cardiomyopathy [29], gallbladder cancer [30], squamous cell carcinoma of the head and neck [31], chronic obstructive pulmonary disease [32], liver cancer [33], rheumatoid arthritis [34], coronary artery disease [35], and colorectal cancer [36]. Especially, one study suggested that miR-499 rs3746444*T alleles might be protective for breast cancer [37], but another found that the miR-499 rs3746444 C allele increased cancer risk in the allelic contrast model and in the dominant model, especially in breast cancer [38], while another study showed that no significant associations were observed between rs3746444 in miR-499 and breast cancer susceptibility [39]. Recently, a meta-analysis suggested that polymorphism of hsa-mir-499 rs3746444 T > C was not associated with increased susceptibility to cancers [40],[41], while other systematic analysis supported that hsa-mir-499 rs3746444 polymorphism contributed to the susceptibility to cancers [42]–[45].
The results of these observations remain controversial and inconclusive. In the present study, we conducted a meta-analysis in order to derive more precise and more comprehensive estimation of the associations between the SNP hsa-mir-499 rs3746444 T > C and susceptibility to cancers to quantify the potential between-study heterogeneity.
Methods
Study selection
We performed a publication search in PubMed, EMBASE, ISI Web of Science, The Cochrane Library, ScienceDirect, EBSCO, Ovid, Wiley Online Library, and HighWire databases with the following search terms: (miR-499 OR rs3746444) AND (cancer), by two independent investigators (Chen Chen and Shenglan Yang, last search update: June 29, 2014). Hand searches were also performed to identify additional articles in the reference lists of included articles not retrieved by initial electronic search. Publication date and publication language were not restricted in our search. All studies matching the inclusion criteria were retrieved for further examination and data extraction. All of the investigators have received training in literature search, statistics and evidence-based medicine.
Inclusion and exclusion criteria
Studies included in current meta-analysis had to meet all the following criteria: (1) evaluated the associations between the hsa-mir-499 rs3746444 polymorphism and cancer risk; (2) studied on human beings; (3) diseases were confirmed by histology, imaging or pathology; (4) a case–control design; (5) detailed genotype data were provided for the calculation of odds ratio (OR) and 95% confidence interval (CIs); (6) if serial studies of the same population from the same group were reported, the latest study was included. Studies were excluded when they represented duplicates of previous publications, or were meta -analyses, meeting abstracts, letters, reviews, or editorial articles.
Data extraction
Two investigators (Chen Chen and Shenglan Yang) independently extracted data from the included studies using a standard protocol and data-collection form according to the inclusion criteria listed above, and reached consensus on all items. Data extracted from eligible studies included the first author’s name, year of publication, country of origin, ethnicity, cancer type, genotyping method, total numbers of cases and controls, and genotype frequencies of cases and controls. For study including subjects of different countries of origin group from same ethnicity, we combined them together. The ethnic descents were categorized as Caucasian or Asian. If different results were generated, the two authors would check the data and have had a discussion to come to an agreement. Two senior investigators (Yan Wang and Dao Wen Wang) were invited to the discussion if disagreement still existed.
Statistical analysis
For each study, the departure of frequencies of hsa-mir-499 polymorphism from expectation under Hardy-Weinberg equilibrium (HWE) was assessed by the goodness-of-fit chi-square test in controls. P < 0.05 was considered representative of a departure from HWE.
OR corresponding to 95% CI was used to assess the strength of association between hsa-miR-499 rs3746444 T > C polymorphism and susceptibility to cancer risk. The significance of the pooled OR was determined by the Z-test, and P < 0.05 was considered as statistically significant. Pooled ORs were calculated for allele frequency comparison (C versus T, TC versus TT, CC versus TT, TC/CC versus TT (dominant) and CC versus TC/TT (recessive)), respectively. Subgroup analyses were done by racial descent and cancer type. Statistical heterogeneity among the studies was estimated using chi-square-based Q-test, a P value greater than 0.1 indicates no significant heterogeneity and the pooled OR was estimated by the fixed-effects model (the Mantel-Haenszel method); otherwise, the random-effects model (the DerSimonian and Laird method) was employed [46]. Publication bias of literatures was evaluated by funnel plots and the linear regression asymmetry test by Egger et al. [47]. An asymmetric plot suggests a possible publication bias and the P value of Egger’s test less than 0.05 was considered representative of statistically significant publication bias.Sensitivity analysis was conducted by deleting one study at a time to examine the influence of individual data set on the pooled ORs.
All of the statistical tests were performed with Review Manage (v.4.2; Oxford, England) and STATA software version 11.0 (STATA Corporation, College Station, TX, USA). All the P values are two-sided.
Results
Study characteristics
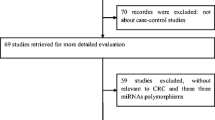
After first search in PubMed, EMBASE, ISI Web of Science, The Cochrane Library, ScienceDirect, EBSCO, Ovid, Wiley Online Library, and HighWire databases, 70, 76, 116, 0, 2080, 86, 23, 104, and 202 articles, respectively, were retrieved. Of these, 55 articles were relevant to the search words (Figure 1). After further manual search of reference lists, one more study was included. Then, 25 studies were excluded (7 meeting abstracts, 13 meta-analyses, 1 without detailed allele frequency data, and 4 non-case–control studies) and finally 31 studies involving a total of 12799 disease cases and 14507 controls met the inclusion criteria and were subjected to further examination. Characteristics of included studies are summarized in Table 1, including ethnicity, genotype detection method and cancer type. All studies were case–control studies, including 8 liver cancer studies [33],[48]–[54], 4 breast cancer studies [24]–[26],[55], 4 gastric cancer studies [56]–[59], 3 colorectal cancer studies [36],[60],[61], 2 lung cancer studies [27],[62], 2 esophageal cancer studies [63],[64], and others (1 cervical cancer study [65], 1 bladder cancer study [66], 1 prostate cancer study [67], 1 head and neck cancer study [31], 1 gallbladder cancer study [30], 1 childhood acute lymphoblastic leukemia [68], 1 renal cell carcinoma [69] and 1 oral squamous cell carcinoma [70]). Cancers were histologically or pathologically diagnosed in all studies. There were 28 studies of Asian descents (16 Chinese, 4 Indian, 3 Korean, 2 Iranians and 3 others) and 3 studies of Caucasian descents. Several genotyping methods were employed in the studies including Taqman, polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP), DNA sequencing, high resolution melting analysis (HRMA), T-ARMS-PCR, MALDI-TOF MS and Sequenom MassARRAY. PRISMA checklist was generated to provide detailed description of this meta-analysis (Additional file 1: Table S1).
Meta-analysis results
The association between hsa-mir-499 rs3746444 polymorphism and susceptibility to cancer was analyzed in 31 independent studies. The results are shown in Table 2 and Additional file 2: Figures S1, S2 and S3. Significant association between rs3746444 polymorphism and susceptibility to cancer was identified in TC versus TT and TC/CC versus TT (dominant) models, when all the eligible studies were pooled (C versus T: OR = 1.08, 95% CI 0.99-1.17, P = 0.08; TC versus TT: OR = 1.11, 95% CI 1.00-1.23, P = 0.04; CC versus TT: OR = 1.02, 95% CI 0.85-1.22, P = 0.85; TC/CC versus TT (dominant): OR = 1.11, 95% CI 1.01-1.23, P = 0.03; CC versus TC/TT (recessive): OR = 0.98, 95% CI 0.81-1.17, P = 0.79). Next, subgroup analyses were performed. Twenty-eight out of the thirty-one included studies were conducted in Asian population. In ethnicity subgroup analysis, significantly increased risks were found in Asians (TC versus TT: OR = 1.14; 95% CI = 1.02-1.27, P = 0.02; TC/CC versus TT (dominant): OR = 1.14; 95% CI = 1.02-1.27, P = 0.02), consistently. This increased risk of cancer was especially significant in Iranian (C versus T: OR = 1.57; 95% CI = 1.23-2.01, P = 0.0003; CC versus TT: OR = 2.23; 95% CI = 1.42-3.51, P = 0.0005; CC versus TC/TT (recessive): OR = 2.23; 95% CI = 1.44-3.45, P = 0.0003) and Chinese population (TC versus TT: OR = 1.18; 95% CI = 1.02-1.35, P = 0.02). However, no significant association between hsa-mir-499 rs3746444 T > C polymorphism and cancer risk was found in Caucasians in any of the genetic models tested. Further subgroup analysis based on cancer type revealed a decreased risk for esophageal cancer (C versus T: OR = 0.80; 95% CI = 0.66-0.98, P = 0.03).
Publication bias
Begg’s funnel plot and Egger’s test were performed to assess the publication bias of included studies. Symmetrical funnel plots were obtained in all comparison models (Figure 2). Further, Egger’s test confirmed the absence of publication bias in all genetic models (P > 0.05).
Sensitivity analysis
Exclusion of one study at a time was performed to examine the influence of the individual data set to the pooled ORs. Analyses were performed using STATA by means of the metainf program. Results showed that the pooled OR estimates were similar with those of the excluded studies (Figure 3).
Sensitivity analysis of the influence of a single study on the overall meta-analysis estimate. (A) C versus T; (B) TC versus TT; (C) CC versus TT; (D) TC/CC versus TT (dominant) and (E) CC versus TC/TT (recessive). The solid lines correspond to the pooled OR and 95% CI. Circles and dashed lines correspond to the specific OR and 95% CI without omitted study.
Discussion
In the present study, hsa-mir-499 rs3746444 polymorphism and susceptibility to cancer was evaluated in different genetic models in 12799 cancer cases and 14507 controls. Significant association between rs3746444 polymorphism and susceptibility to cancer was identified in TC versus TT and TC/CC versus TT (dominant) models. Subgroup analysis absed on ethnicity revealed significantly increased risk in Asians (TC versus TT and TC/CC versus TT (dominant), but there was no significant association in Caucasians in all genetic models. This association was especially marked in Iranians (C versus T; CC versus TT and CC versus TC/TT (recessive) and a significant association was also identified in Chinese (TC versus TT) However, subgroup analysis based on cancer type revealed a decreased risk for esophageal cancer (C versus T).
Many studies investigated the role of SNPs presented in precursor and mature miRNAs, and their influences on susceptibility and progression of various diseases. miRNA-associated SNPs can have direct or indirect effects: 1) direct effects are produced by the SNPs in the pri-miRNA, pre-miRNA or mature miRNA that impair or enhance miRNA processing or function; 2) indirect effects are derived from the SNPs in miRNA promoters that affect transcription and SNPs in an mRNA that create or destroy a target site [71].
Genome-wide patterns of human polymorphisms in miRNAs and miRNA target sites in 3′ UTRs of mRNAs revealed that only ~10% of human pre-miRNAs have documented SNPs, and <1% of mature miRNAs have SNPs in the functional seed region [72]. hsa-mir-499 rs3746444 polymorphism is located in the seed region of the mature miR-499 sequence. This T > C polymorphism-resulted mismatch may affect target mRNA expression. Several case–control studies have investigated the association between rs3746444 polymorphism and risks of various cancers. However, the results were controversial.
Different from past studies, we analyzed the specific association among different ethnic groups throughout Asia. In this meta-analysis, we first found that rs3746444 polymorphism was associated with increased cancer risk mainly in Asian population, especially in Chinese and Iranians. It is important to notice that, although rs3746444 polymorphism contributes to cancer risk in Chinese and Iranians, the genetic models are different. The genetic variations between Chinese Han population and Arabian population may contribute to the differences. Interestingly, we also found that rs3746444 polymorphism showed a decreased risk in esophageal cancer. This suggests that potentially different mechanisms may underlie tumorgenesis in different pathological backgrounds and the environment variations.
It has been suggested that miRNAs may influence gene expression of ~30% of protein-coding genes by binding to the mRNA incompletely, and a miRNA may affect multiple target mRNAs involved in same pathophysiological progress [3],[73]. This means that rs3746444 polymorphism may contribute to single gene function in certain disease model at molecular level. When taken together in epidemiological studies of populations, various effects of rs3746444 polymorphism on different genes may result in different associations with diseases at phenotype level.
Our meta-analysis pooled the largest numbers of cases and controls from included studies, which significantly increases the statistical power but some limitations should be considered. Firstly, lack of the consideration of combined genetic factors together with environmental exposures, while a more precise analysis needs to be conducted if more detailed data are available. Secondly, although sensitivity analysis showed that each study can not affect the overall results, some heterogeneity was evident in some of the comparisons. This may be due to different ethnicities and different pathology. And expanding the sample size may yield a better result. Thirdly, some unpublished data (such as negative report, not written in English and lack of studies in African population) may potentially influence the results of our meta-analysis. At last, different genotyping strategies may contribute to the bias in the analysis.
Recently several meta-analyses systematically reviewed the potential association of rs3746444 polymorphism with susceptibility to diseases [40]–[45],[74]–[77]. However, the results were controversial across these meta-analyses. Such discrepancies in the results of these systematic reviews may due to different search periods, databases searched, inclusion and exclusion criteria. Our efforts to search the maximum range of databases yielded the most comprehensive studies in the similar time period especially the most recent publications [53],[59]. Further, diagnosis of cancer is largely relied on confirmation by histology, imaging or pathology. Some of the other studies omitted this fundamental condition. Most importantly, deviations from Hardy-Weinberg equilibrium can inflate the chance of a false-positive association, and sensitivity analysis should be performed, pooling with and without studies not in HWE, to test the robustness of the results [78],[79]. Only few of other studies performed sensitivity analysis, which may limit the reliability of their conclusions. Although some studies claimed that their studies were in HWE, a careful examination of the studies revealed that the data did not reach HWE. In conclusion, in our study, relevant literatures selected from broad databases with stringent standards would be expected to provide a more reliable conclusion.
Conclusions
In summary, though with limitations, our meta-analysis suggested that hsa-mir-499 rs3746444 T > C polymorphism is associated with the risk of cancer, especially in Asians, mainly in Chinese and Iranians. However, rs3746444 T > C polymorphism is negatively associated with the risk of esophageal cancer. Larger studies from different ethnic groups and studying different types of cancers with detailed information are needed to further clarify the association between rs3746444 polymorphism and cancer risk.
Additional files
References
Lee RC, Feinbaum RL, Ambros V: The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993, 75 (5): 843-854.
Wightman B, Ha I, Ruvkun G: Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993, 75 (5): 855-862.
Mendell JT, Olson EN: MicroRNAs in stress signaling and human disease. Cell. 2012, 148 (6): 1172-1187.
Heinrich EM, Dimmeler S: MicroRNAs and stem cells: control of pluripotency, reprogramming, and lineage commitment. Circ Res. 2012, 110 (7): 1014-1022.
Boettger T, Braun T: A new level of complexity: the role of microRNAs in cardiovascular development. Circ Res. 2012, 110 (7): 1000-1013.
Cheng W, Liu T, Wan X, Gao Y, Wang H: MicroRNA-199a targets CD44 to suppress the tumorigenicity and multidrug resistance of ovarian cancer-initiating cells. FEBS J. 2012, 279 (11): 2047-2059.
Rottiers V, Naar AM: MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol. 2012, 13 (4): 239-250.
Grueter CE, van Rooij E, Johnson BA, DeLeon SM, Sutherland LB, Qi X, Gautron L, Elmquist JK, Bassel-Duby R, Olson EN: A cardiac microRNA governs systemic energy homeostasis by regulation of MED13. Cell. 2012, 149 (3): 671-683.
Akbari Moqadam F, Pieters R, den Boer ML: The hunting of targets: challenge in miRNA research. Leukemia. 2013, 27 (1): 16-23.
Vira D, Basak SK, Veena MS, Wang MB, Batra RK, Srivatsan ES: Cancer stem cells, microRNAs, and therapeutic strategies including natural products. Cancer Metastasis Rev. 2012, 31 (3–4): 733-751.
Ebert MS, Sharp PA: Roles for microRNAs in conferring robustness to biological processes. Cell. 2012, 149 (3): 515-524.
Pritchard CC, Cheng HH, Tewari M: MicroRNA profiling: approaches and considerations. Nat Rev Genet. 2012, 13 (5): 358-369.
Kim YK, Kim VN: Processing of intronic microRNAs. EMBO J. 2007, 26 (3): 775-783.
Meister G, Tuschl T: Mechanisms of gene silencing by double-stranded RNA. Nature. 2004, 431 (7006): 343-349.
Yang JS, Lai EC: Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol Cell. 2011, 43 (6): 892-903.
Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ: A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010, 465 (7298): 584-589.
Cai Y, Yu X, Hu S, Yu J: A brief review on the mechanisms of miRNA regulation. Genomics Proteomics Bioinformatics. 2009, 7 (4): 147-154.
Mishra PJ, Banerjee D, Bertino JR: MiRSNPs or MiR-polymorphisms, new players in microRNA mediated regulation of the cell: Introducing microRNA pharmacogenomics. Cell Cycle. 2008, 7 (7): 853-858.
Wu C, Gong Y, Sun A, Zhang Y, Zhang C, Zhang W, Zhao G, Zou Y, Ge J: The human MTHFR rs4846049 polymorphism increases coronary heart disease risk through modifying miRNA binding. Nutr Metab Cardiovasc Dis. 2013, 23 (7): 693-698.
Wang K, Li J, Guo H, Xu X, Xiong G, Guan X, Liu B, Chen X, Yang K, Bai Y: MiR-196a binding-site SNP regulates RAP1A expression contributing to esophageal squamous cell carcinoma risk and metastasis. Carcinogenesis. 2012, 33 (11): 2147-2154.
Luo J, Cai Q, Wang W, Huang H, Zeng H, He W, Deng W, Yu H, Chan E, Ng CF, Huang J, Lin T: A microRNA-7 binding site polymorphism in HOXB5 leads to differential gene expression in bladder cancer. PLoS One. 2012, 7 (6): e40127-
Liu Y, Zhang Y, Wen J, Liu L, Zhai X, Liu J, Pan S, Chen J, Shen H, Hu Z: A genetic variant in the promoter region of miR-106b-25 cluster and risk of HBV infection and hepatocellular carcinoma. PLoS One. 2012, 7 (2): e32230-
Wu Y, Xiao Y, Ding X, Zhuo Y, Ren P, Zhou C, Zhou J: A miR-200b/200c/429-binding site polymorphism in the 3′ untranslated region of the AP-2alpha gene is associated with cisplatin resistance. PLoS One. 2011, 6 (12): e29043-
Hu Z, Liang J, Wang Z, Tian T, Zhou X, Chen J, Miao R, Wang Y, Wang X, Shen H: Common genetic variants in pre-microRNAs were associated with increased risk of breast cancer in Chinese women. Hum Mutat. 2009, 30 (1): 79-84.
Catucci I, Yang R, Verderio P, Pizzamiglio S, Heesen L, Hemminki K, Sutter C, Wappenschmidt B, Dick M, Arnold N, Bugert P, Niederacher D, Meindl A, Schmutzler RK, Bartram CC, Ficarazzi F, Tizzoni L, Zaffaroni D, Manoukian S, Barile M, Pierotti MA, Radice P, Burwinkel B, Peterlongo P: Evaluation of SNPs in miR-146a, miR196a2 and miR-499 as low-penetrance alleles in German and Italian familial breast cancer cases. Hum Mutat. 2010, 31 (1): E1052-E1057.
Omrani M, Hashemi M, Eskandari-Nasab E, Hasani SS, Mashhadi MA, Arbabi F, Taheri M: hsa-mir-499 rs3746444 gene polymorphism is associated with susceptibility to breast cancer in an Iranian population. Biomark Med. 2014, 8 (2): 259-267.
Tian T, Shu Y, Chen J, Hu Z, Xu L, Jin G, Liang J, Liu P, Zhou X, Miao R, Ma H, Chen Y, Shen H: A functional genetic variant in microRNA-196a2 is associated with increased susceptibility of lung cancer in Chinese. Cancer Epidemiol Biomarkers Prev. 2009, 18 (4): 1183-1187.
Xu J, Hu Z, Xu Z, Gu H, Yi L, Cao H, Chen J, Tian T, Liang J, Lin Y, Qiu W, Ma H, Shen H, Chen Y: Functional variant in microRNA-196a2 contributes to the susceptibility of congenital heart disease in a Chinese population. Hum Mutat. 2009, 30 (8): 1231-1236.
Zhou B, Rao L, Peng Y, Wang Y, Chen Y, Song Y, Zhang L: Common genetic polymorphisms in pre-microRNAs were associated with increased risk of dilated cardiomyopathy. Clin Chim Acta. 2010, 411 (17–18): 1287-1290.
Srivastava K, Srivastava A, Mittal B: Common genetic variants in pre-microRNAs and risk of gallbladder cancer in North Indian population. J Hum Genet. 2010, 55 (8): 495-499.
Liu Z, Li G, Wei S, Niu J, El-Naggar AK, Sturgis EM, Wei Q: Genetic variants in selected pre-microRNA genes and the risk of squamous cell carcinoma of the head and neck. Cancer. 2010, 116 (20): 4753-4760.
Li LJ, Gao LB, Lv ML, Dong W, Su XW, Liang WB, Zhang L: Association between SNPs in pre-miRNA and risk of chronic obstructive pulmonary disease. Clin Biochem. 2011, 44 (10–11): 813-816.
Zhou J, Lv R, Song X, Li D, Hu X, Ying B, Wei Y, Wang L: Association between two genetic variants in miRNA and primary liver cancer risk in the Chinese population. DNA Cell Biol. 2012, 31 (4): 524-530.
Yang B, Chen J, Li Y, Zhang J, Li D, Huang Z, Cai B, Li L, Shi Y, Ying B, Wang L: Association of polymorphisms in pre-miRNA with inflammatory biomarkers in rheumatoid arthritis in the Chinese Han population. Hum Immunol. 2012, 73 (1): 101-106.
Zhi H, Wang L, Ma G, Ye X, Yu X, Zhu Y, Zhang Y, Zhang J, Wang B: Polymorphisms of miRNAs genes are associated with the risk and prognosis of coronary artery disease. Clin Res Cardiol. 2012, 101 (4): 289-296.
Min KT, Kim JW, Jeon YJ, Jang MJ, Chong SY, Oh D, Kim NK: Association of the miR-146aC > G, 149C > T, 196a2C > T, and 499A > G polymorphisms with colorectal cancer in the Korean population. Mol Carcinog. 2012, 51 (Suppl 1): E65-E73.
Chen QH, Wang QB, Zhang B: Ethnicity modifies the association between functional microRNA polymorphisms and breast cancer risk: a HuGE meta-analysis. Tumour Biol. 2014, 35 (1): 529-543.
Ma XP, Zhang T, Peng B, Yu L, Jiang de K: Association between microRNA polymorphisms and cancer risk based on the findings of 66 case–control studies. PLoS One. 2013, 8 (11): e79584-
Wang PY, Gao ZH, Jiang ZH, Li XX, Jiang BF, Xie SY: The associations of single nucleotide polymorphisms in miR-146a, miR-196a and miR-499 with breast cancer susceptibility. PLoS One. 2013, 8 (9): e70656-
Wang L, Qian S, Zhi H, Zhang Y, Wang B, Lu Z: The association between hsa-miR-499 T > C polymorphism and cancer risk: A meta-analysis. Gene. 2012, 508 (1): 9-14.
Du W, Ma XL, Zhao C, Liu T, Du YL, Kong WQ, Wei BL, Yu JY, Li YY, Huang JW, Li ZK, Liu L: Associations of single nucleotide polymorphisms in miR-146a, miR-196a, miR-149 and miR-499 with colorectal cancer susceptibility. Asian Pac J Cancer Prev. 2014, 15 (2): 1047-1055.
Wang Y, Yang B, Ren X: Hsa-miR-499 polymorphism (rs3746444) and cancer risk: a meta-analysis of 17 case–control studies. Gene. 2012, 509 (2): 267-272.
Wang F, Sun G, Zou Y, Li Y, Hao L, Pan F: Association of microRNA-499 rs3746444 polymorphism with cancer risk: evidence from 7188 cases and 8548 controls. PLoS One. 2012, 7 (9): e45042-
Chen P, Zhang J, Zhou F: miR-499 rs3746444 polymorphism is associated with cancer development among Asians and related to breast cancer susceptibility. Mol Biol Rep. 2012, 39 (12): 10433-10438.
He B, Pan Y, Cho WC, Xu Y, Gu L, Nie Z, Chen L, Song G, Gao T, Li R, Wang S: The association between four genetic variants in MicroRNAs (rs11614913, rs2910164, rs3746444, rs2292832) and cancer risk: evidence from published studies. PLoS One. 2012, 7 (11): e49032-
Wang Z, Cao Y, Jiang C, Yang G, Wu J, Ding Y: Lack of association of two common polymorphisms rs2910164 and rs11614913 with susceptibility to hepatocellular carcinoma: a meta-analysis. PLoS One. 2012, 7 (6): e40039-
Egger M, Davey Smith G, Schneider M, Minder C: Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997, 315 (7109): 629-634.
Xiang Y, Fan S, Cao J, Huang S, Zhang LP: Association of the microRNA-499 variants with susceptibility to hepatocellular carcinoma in a Chinese population. Mol Biol Rep. 2012, 39 (6): 7019-7023.
Akkiz H, Bayram S, Bekar A, Akgollu E, Uskudar O: Genetic variation in the microRNA-499 gene and hepatocellular carcinoma risk in a Turkish population: lack of any association in a case–control study. Asian Pac J Cancer Prev. 2011, 12 (11): 3107-3112.
Kim WH, Min KT, Jeon YJ, Kwon CI, Ko KH, Park PW, Hong SP, Rim KS, Kwon SW, Hwang SG, Kim NK: Association study of microRNA polymorphisms with hepatocellular carcinoma in Korean population. Gene. 2012, 504 (1): 92-97.
Zou HZ, Zhao YQ: Positive association between miR-499A > G and hepatocellular carcinoma risk in a Chinese population. Asian Pac J Cancer Prev. 2013, 14 (3): 1769-1772.
Shan YF, Huang YH, Chen ZK, Huang KT, Zhou MT, Shi HQ, Song QT, Yu ZP, Deng AM, Zhang QY: miR-499A > G rs3746444 and miR-146aG > C expression and hepatocellular carcinoma risk in the Chinese population. Genet Mol Res. 2013, 12 (4): 5365-5371.
Ma Y, Wang R, Zhang J, Li W, Gao C, Liu J, Wang J: Identification of miR-423 and miR-499 polymorphisms on affecting the risk of hepatocellular carcinoma in a large-scale population. Genet Test Mol Biomarkers. 2014, 18 (7): 516-524.
Chu YH, Hsieh MJ, Chiou HL, Liou YS, Yang CC, Yang SF, Kuo WH: MicroRNA gene polymorphisms and environmental factors increase patient susceptibility to hepatocellular carcinoma. PLoS One. 2014, 9 (2): e89930-
Alshatwi AA, Shafi G, Hasan TN, Syed NA, Al-Hazzani AA, Alsaif MA, Alsaif AA: Differential expression profile and genetic variants of microRNAs sequences in breast cancer patients. PLoS One. 2012, 7 (2): e30049-
Ahn DH, Rah H, Choi YK, Jeon YJ, Min KT, Kwack K, Hong SP, Hwang SG, Kim NK: Association of the miR-146aC > G, miR-149 T > C, miR-196a2T > C, and miR-499A > G polymorphisms with gastric cancer risk and survival in the Korean population. Mol Carcinog. 2013, 52 (Suppl 1): E39-E51.
Okubo M, Tahara T, Shibata T, Yamashita H, Nakamura M, Yoshioka D, Yonemura J, Ishizuka T, Arisawa T, Hirata I: Association between common genetic variants in pre-microRNAs and gastric cancer risk in Japanese population. Helicobacter. 2010, 15 (6): 524-531.
Wu XJ, Mi YY, Yang H, Hu AK, Li C, Li XD, Zhang QG: Association of the hsa-mir-499 (rs3746444) polymorphisms with gastric cancer risk in the Chinese population. Onkologie. 2013, 36 (10): 573-576.
Pu JY, Dong W, Zhang L, Liang WB, Yang Y, Lv ML: No association between single nucleotide polymorphisms in pre-mirnas and the risk of gastric cancer in Chinese population. Iranian J Basic Med Sci. 2014, 17 (2): 128-133.
Lv M, Dong W, Li L, Zhang L, Su X, Wang L, Gao L, Zhang L: Association between genetic variants in pre-miRNA and colorectal cancer risk in a Chinese population. J Cancer Res Clin Oncol. 2013, 139 (8): 1405-1410.
Hu X, Li L, Shang M, Zhou J, Song X, Lu X, Wang J, Ying B, Wang L: Association between microRNA genetic variants and susceptibility to colorectal cancer in Chinese population. Tumour Biol. 2014, 35 (3): 2151-2156.
Vinci S, Gelmini S, Pratesi N, Conti S, Malentacchi F, Simi L, Pazzagli M, Orlando C: Genetic variants in miR-146a, miR-149, miR-196a2, miR-499 and their influence on relative expression in lung cancers. Clin Chem Lab Med. 2011, 49 (12): 2073-2080.
Umar M, Upadhyay R, Prakash G, Kumar S, Ghoshal UC, Mittal B: Evaluation of common genetic variants in pre-microRNA in susceptibility and prognosis of esophageal cancer. Mol Carcinog. 2013, 52 (Suppl 1): E10-E18.
Wei J, Zheng L, Liu S, Yin J, Wang L, Wang X, Shi Y, Shao A, Tang W, Ding G, Liu C, Chen S, Gu H: MiR-196a2 rs11614913 T > C polymorphism and risk of esophageal cancer in a Chinese population. Hum Immunol. 2013, 74 (9): 1199-1205.
Zhou B, Wang K, Wang Y, Xi M, Zhang Z, Song Y, Zhang L: Common genetic polymorphisms in pre-microRNAs and risk of cervical squamous cell carcinoma. Mol Carcinog. 2011, 50 (7): 499-505.
Mittal RD, Gangwar R, George GP, Mittal T, Kapoor R: Investigative role of pre-microRNAs in bladder cancer patients: a case–control study in North India. DNA Cell Biol. 2011, 30 (6): 401-406.
George GP, Gangwar R, Mandal RK, Sankhwar SN, Mittal RD: Genetic variation in microRNA genes and prostate cancer risk in North Indian population. Mol Biol Rep. 2011, 38 (3): 1609-1615.
Hasani SS, Hashemi M, Eskandari-Nasab E, Naderi M, Omrani M, Sheybani-Nasab M: A functional polymorphism in the miR-146a gene is associated with the risk of childhood acute lymphoblastic leukemia: a preliminary report. Tumour Biol. 2014, 35 (1): 219-225.
Du M, Lu D, Wang Q, Chu H, Tong N, Pan X, Qin C, Yin C, Wang M, Zhang Z: Genetic variations in microRNAs and the risk and survival of renal cell cancer. Carcinogenesis. 2014, 35 (7): 1629-1635.
Hou YY, Lee JH, Chen HC, Yang CM, Huang SJ, Liou HH, Chi CC, Tsai KW, Ger LP: The association between miR-499a polymorphism and oral squamous cell carcinoma progression. Oral Dis. 2014 Apr 2. doi:10.1111/odi.12241. [Epub ahead of print]
Salzman DW, Weidhaas JB: SNPing cancer in the bud: MicroRNA and microRNA-target site polymorphisms as diagnostic and prognostic biomarkers in cancer. Pharmacol Ther. 2013, 137 (1): 55-63.
Saunders MA, Liang H, Li WH: Human polymorphism at microRNAs and microRNA target sites. Proc Natl Acad Sci U S A. 2007, 104 (9): 3300-3305.
Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM: Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005, 433 (7027): 769-773.
Fan C, Chen C, Wu D: The association between common genetic variant of microRNA-499 and cancer susceptibility: a meta-analysis. Mol Biol Rep. 2013, 40 (4): 3389-3394.
Yin Z, Yan L, Cui Z, Li X, Ren Y, Zhou B: Effects of common polymorphisms rs2910164 in miR-146a and rs3746444 in miR-499 on cancer susceptibility: a meta-analysis. Mol Biol Rep. 2013, 40 (4): 3003-3013.
Hu M, Zhao L, Hu S, Yang J: The association between two common polymorphisms in MicroRNAs and hepatocellular carcinoma risk in Asian population. PLoS One. 2013, 8 (2): e57012-
Qiu MT, Hu JW, Ding XX, Yang X, Zhang Z, Yin R, Xu L: Hsa-miR-499 rs3746444 polymorphism contributes to cancer risk: a meta-analysis of 12 studies. PLoS One. 2012, 7 (12): e50887-
Schaid DJ, Jacobsen SJ: Biased tests of association: comparisons of allele frequencies when departing from Hardy-Weinberg proportions. Am J Epidemiol. 1999, 149 (8): 706-711.
Attia J, Thakkinstian A, D’Este C: Meta-analyses of molecular association studies: methodologic lessons for genetic epidemiology. J Clin Epidemiol. 2003, 56 (4): 297-303.
Acknowledgements
This work was supported by grant from the National Natural Science Foundation of China (No. 81070236 and 31200594) and Key Project of Ministry of Health of the People's Republic of China. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
CC and SY carried out the studies and drafted the manuscript. SC and YW helped to draft the manuscript. CC and DWW participated in the design of the study and performed the statistical analysis and final editing of the manuscript. All authors read and approved the final manuscript.
Electronic supplementary material
12881_2014_126_MOESM2_ESM.doc
Additional file 2: Forest plot of ORs for the association of hsa-miR-499 rs3746444 T>C polymorphism with cancer risk in different situations. Figure S1. Forest plot of ORs for the association of hsa-miR-499 rs3746444 T>C polymorphism with cancer risk is illustrated by ethnicity. (A) C versus T; (B) TC versus TT; (C) CC versus TT; (D) TC/CC versus TT (dominant) and (E) CC versus TC/TT (recessive). Figure S2. Forest plot of ORs for the association of hsa-miR-499 rs3746444 T>C polymorphism with cancer risk is illustrated by cancer type. (A) C versus T; (B) TC versus TT; (C) CC versus TT; (D) TC/CC versus TT (dominant) and (E) CC versus TC/TT (recessive). Figure S3. Forest plot of ORs for the association of hsa-miR-499 rs3746444 T>C polymorphism with cancer risk is illustrated by country in Asia. (A) C versus T; (B) TC versus TT; (C) CC versus TT; (D) TC/CC versus TT (dominant) and (E) CC versus TC/TT (recessive). (DOC 862 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Chen, C., Yang, S., Chaugai, S. et al. Meta-analysis of Hsa-mir-499 polymorphism (rs3746444) for cancer risk: evidence from 31 case-control studies. BMC Med Genet 15, 126 (2014). https://doi.org/10.1186/s12881-014-0126-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12881-014-0126-1