Abstract
MicroRNAs (miRNAs) play an important role in the pathogenesis of neoplasm. Single nucleotide polymorphisms (SNPs) within miRNAs can change their phenotype and function. We attempted to analyze the relationship between two SNP loci in miRNAs and colorectal cancer (CRC) in Chinese Han population. We genotyped the polymorphism of two common miRNA SNPs, miR-146a (rs2910164 G > C) and miR-499 (rs3746444 T > C), in a case–control study of 276 CRC cases and 373 healthy controls using polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP). The genotypes and allele frequencies of the two SNP loci were first compared between patients and controls and then further analyzed among subgroups of patients with different clinicopathological profiles. The rs2910164 CG genotype was significantly associated with a decreased risk of CRC [CG versus GG, odds ratio (OR) = 0.567; 95 % confidence intervals (CIs) = 0.338–0.952; p = 0.031]. No significant differences of miR-499 genotype and allele distribution were detected between patients and controls. Comparison between groups divided by clinicopathologic features showed that the polymorphism of miR-146a was associated with the degree of tumor differentiation (p = 0.014), and the G allele of rs2910164 trended to a mature differentiation (OR = 0.553; 95 % CI = 0.315–0.971; p = 0.038). MiR-146a (rs2910164 G > C) polymorphism is associated with CRC susceptibility and histological differentiation in Chinese Han population.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) is a common gastrointestinal cancer. Its incidence ranks after lung cancer in malignant tumors in the western developed countries and after gastric and esophageal neoplasm in China. The American Cancer Society estimated that, in 2012 only, approximately 143,460 new cases of CRC were diagnosed, and 51,690 people died from CRC in the USA [1]. While liver metastasis is the main cause of death for CRC, the prognosis of all advanced CRC patients with or without metastasis is very poor. Epidemiological evidences suggest that risk factors for colorectal neoplasm include a positive family history, excessive meat consumption, smoking status, alcohol consumption, and certain environmental exposures. Moreover, host susceptibilities in molecular pathways have an implication for CRC screening, treatment, surveillance, and prevention [2].
MiRNAs regulate a variety of biological and pathological processes related to carcinogenesis and tumor progression by translational repression or degradation of target mRNAs [3, 4]. Since Michael et al. first described the roles of miRNA in CRC [6], more emerging evidences have shown that the dysregulation of miRNAs is present in CRC, including overexpression, silencing, and/or switching off [6–10]. As the most common genetic marker, single nucleotide polymorphisms (SNPs) located in pre-miRNAs or miRNAs may affect the expression and function of miRNAs and eventually contribute to the susceptibility of cancer development [11, 12]. A large number of studies had highlighted several associations between SNPs in miRNAs and the risk of CRC [13–18]. However, controversial experimental data were obtained. For example, Chae et al. reported that miR-146a rs2910164 polymorphism of genotype CC may contribute to a higher risk of CRC [15], whereas Hezova et al. [17] failed to find any significant association between rs2910164 and CRC. Therefore, more valuable replication studies about the possible association between SNPs and CRC risk are still urgently needed. In order to investigate the effect of genetic polymorphism in miRNAs on CRC risk, we genotyped two miRNA SNPs, miR-146a (rs2910164 G > C) and miR-499 (rs3746444 T > C), and assessed their associations with risk of CRC in the present case–control study (276 CRC patients and 373 controls) in Chinese Han ethnic population.
Material and methods
Study population
Two hundred seventy-six patients with CRC and 373 normal control subjects were recruited into the current study. The diagnosis of CRC was made in accordance with the clinical criteria and histopathological confirmation. The samples of the healthy controls without a history of any medical illness were collected from a physical examination population. Both of cases and controls were collected in the same period from April to August 2010. The participants with blood relationship were excluded. Written Informed consents were obtained from all included individuals, and this study was approved by the Ethical Committee of West China Hospital, Sichuan University.
Genotyping of SNP loci
Genomic DNA was extracted from the peripheral blood of all participants using the Chelex-100 procedure. The SNP genotypes of miR-146a and miR-499 gene were determined using polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP) and DNA sequencing analysis. The primers described previously [11] were used in this study. The thermal condition for miR-146a rs2910164 PCR reaction consisted of 94 °C for 5 min, followed by 40 cycles of 94 °C for 30 s, 60 °C for 20 s, and 72 °C for 20 min, with a final extension at 72 °C for 5 min. Both miR-146a and miR-499 SNP loci were amplified in a PCR reaction system with a total of 25 μl PCR reaction mix, and miR-146a SNP PCR reaction system includes 2.0 μl genomic DNA, 1.5 μl MgCl2 (25 mmol/l), 2.5 μl 10× buffer, 2.0 μl deoxynucleotide triphosphate (dNTP) (2.5 mmol/l), 0.14 μl Taq polymerase (0.7 U), 1.0 μl DMSO, and 2.5 μl primers (2 pmol/l). For miR-499 SNP, the reaction system consisted of 2.0 μl genomic DNA, 1.5 μl MgCl2 (25 mmol/l), 2.5 μl 10× buffer, 2.0 μl dNTP (2.5 mmol/l), 0.14 μl Taq polymerase (0.7 U), and 5.0 μl primers (2 pmol/l). The thermal condition for miR-449 SNP (rs3746444 T > C) PCR reaction consisted of 94 °C for 5 min, followed by 40 cycles of 94 °C for 30 s, 62 °C for 20 s, and 72 °C for 20 min, with a final extension at 72 °C for 5 min. After the PCR amplification, 5-μl PCR products were added onto a 2 % agarose gel, stained with ethidium bromide, and subjected to electrophoresis at 150 V for 30 min. In the meantime, 2.5-μl PCR products of miR-146a locus were digested with restriction enzyme SacI (Fermentas, USA) at 37 °C overnight. PCR products of miR-499 locus were digested with restriction enzyme BcII (Fermentas, USA) at 56 °C overnight. Five microliters of the resultant products was also subjected to a 3 % agarose gel electrophoresis at 150 V for 1 h. The gels were photographed under ultraviolet light with Gel Doc (Bio-Rad, USA). Some of the PCR products were randomly selected for DNA sequencing through the ABI 3130 sequencing system (ABI, USA) to validate the results.
Tumor marker tests
Electrochemiluminescence (ECL) was used for the quantitative determination of human serum carcinoembryonic antigen (CEA) and CA19-9. The test was conducted automatically by ROCHE-E170 with commercial kits (F. Hoffmann-La Roche Ltd., Switzerland), according to the manufacturers' instructions. The quality control (Bio-Rad, USA) and result audit were done by an experienced medical technologist. All assays were performed in the College of American Pathologists (CAP) certified lab.
Statistical analysis
Statistical analyses were performed with SPSS software (version 13.0; SPSS, Inc., USA). The allelic and genotype frequencies of the SNP loci were calculated by direct count. Chi-square (χ 2) test was used to compare allele/genotype frequencies between groups and Hardy–Weinberg equilibrium of genotype distribution. Odds ratio (OR) and 95 % confidence intervals (CIs) were calculated using logistic regression. F test and nonparametric test were used for the comparison of the serum level of tumor markers. Statistical significance was established at an alpha level of 0.05.
Results
Demographic characteristics of CRC patients
We analyzed the polymorphism of two SNP loci, miR-146a (rs2910164 G > C) and miR-499 (rs3746444 T > C), in 276 CRC patients (65.2 % male and 34.8 % female) and 373 healthy controls (67.8 % male and 32.2 % female) from Chinese Han ethnic population. Table 1 provided the demographic characteristics of CRC patients and controls. There were no significant differences in age, sex, smoking status, and drinking status between the patients and the controls.
Definition of miR-146a and miR-499 SNP genotypes
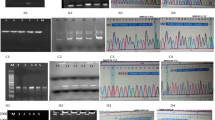
With the specific primers, PCR amplification generated a 147-bp product for miR-146a SNP locus. Following enzyme SacI digestion, the product gave rise to two smaller fragments with a size of 125 and 22 bp, respectively. According to the specificity of the enzyme-targeting sequence, the length polymorphism was defined as follows: one band (147 bp, no digestion) = genotype GG, two bands (125 + 22 bp, complete digestion) = genotype CC, and three bands (147 + 125 + 22 bp, partial digestion) = genotype CG.
The length of PCR product of miR-499 SNP locus before enzyme digestion was 146 bp. After digestion with BcII, two smaller fragments were observed with a size of 120 and 26 bp, respectively. The length polymorphism was defined as follows: one band (146 bp, no digestion) = genotype CC, two bands (120 + 26 bp, complete digestion) = genotype TT, and three bands (146 + 120 + 26 bp, partial digestion) = genotype CT. For confirmatory purpose, PCR products before enzyme digestion were randomly selected for DNA sequencing.
MiR-146a and miR-499 gene polymorphisms in patients and normal controls
Based on the above definition of the genotype for miR-146a and miR-499 SNPs, the genotype distributions and allele frequencies of the two SNPs were counted, calculated, and summarized in Table 2. The genotype frequencies of miR-146a and miR-499 SNPs in patients and normal control group were all in accordance with the Hardy–Weinberg equilibrium (p > 0.05).
The overall distributions of the genotype and allele of miR-146a SNP were not significantly different between patients and controls (p > 0.05). However, the heterozygote CG showed about 1.76-fold decrease risk of developing CRC (OR = 0.567; 95 % CI = 0.338–0.952; p = 0.031) when compared to the homozygote genotype GG in the miR-146a rs2910164. That is to say, genotype GG was significantly associated with a high risk of CRC. The genotype and allele frequencies of miR-499 SNP distributed similarly in patients and controls (p > 0.05).
MiRNA polymorphism and tumor markers
Tumor markers, CEA and CA19-9, have been widely used for CRC diagnosis. It has been found that the serum level of these two markers also have a great value in assessing the tumor stage and differentiation [19, 20]. Thus, we further determined the relationship between the two SNP loci and serum levels of CEA and CA19-9 in CRC patients. As shown in Table 3, neither CEA nor CA19-9 showed significant difference among patients with genotype of two studied SNP loci.
MiR-146a polymorphism and clinicopathological characteristics
Pathologic stage is the most accurate predictor of clinical outcomes of CRC patients. To determine whether there was any association between miR-146a SNP and certain clinicopathological characteristics, we further divided CRC patients into subgroups according to the differentiation degree, morphology type, invasiveness, and tumor pathologic stage. Comparison of the miR-146a polymorphism among the subgroups of CRC patients was summarized in Table 4. It was found that both genotype and allele distributions were significantly different in the two differentiation types (p = 0.014 and 0.038, respectively), and the G allele trended to a mature differentiation (OR = 0.553, 95 % CI = 0.315–0.971). We did not find any significant association between miR-146a SNP and other characteristics of CRC, such as morphology type, invasiveness, and tumor stage (p = 0.722, 0.516, and 0.655, respectively).
Discussion
The identification of genetic factors responsible for susceptibility to CRC is necessary to understand carcinogenesis and discover effective prevention and therapeutic approaches. In the present study, we examined the possible association between polymorphism of two SNP loci in microRNA genes (miR-146a rs2910164 G > C and miR-499 rs3746444 T > C) and risk of CRC in Chinese Han population. The main finding of the study was that the genetic polymorphism of SNP rs2910164 G > C in miR-146a gene was associated with the susceptibility and differentiation of CRC, as well.
MiR-146a rs2910164 can affect the production of mature miRNA through a change from G:U pair to C:U mismatch in the stem region of miR-146a [15–18]. Previous studies have shown that the abnormal expression of miR-146a is present in a variety of human malignancies, such as breast cancer [21] and pancreatic cancer [21]. Jazdzewski et al. found that the rs2910164 SNP could reduce mature miR-146a expression and affect target mRNA binding, and the SNP status affected target genes expression [22]. Shen et al. demonstrated that this variation in the pre-miR-146a resulted in an increased expression of mature miRNAs in breast cancer [23] and decreased expression in hepatocellular carcinoma [24] and PTC [22]. Tang et al. reported that miR-146a could target interferon (IFN) regulatory factor 5 and may directly repressed the activation downstream of type I IFN [25]. Further study by Paik et al. demonstrated that miR-146a acted as a tumor suppressor to downregulate NF-κB activity via targeting TRAF6 [26]. Therefore, we postulate that the polymorphism of rs2910164G > C predisposes to CRC via target selection and/or alteration of the expression of miRNAs. It may affect tumor immune response and, ultimately, the progression of CRC.
Our findings showed that rs2910164 GC genotype conferred a significantly reduced risk of CRC. Recently, several studies from worldwide yielded conflicting results as to the association between miR-146a rs2910164 and the susceptibility of CRC. Chae et al. [15] reported that the rs2910164 CC genotype in Korean showed significant susceptibility toward CRC when compared with the health control. A study from Europe did not found any significant association of miR-146a rs2910164 with CRC [17]. Besides, rs2910164 G allele has different frequencies among different populations (i.e., 0.763 in European and 0.535 in Chinese). These results suggest that the disaccord among current studies may be ascribed to ethnicity differences, and genetic variant may have different roles in various ethnic backgrounds. Ma et al. [16] demonstrated that the GC/CC genotype of miR-146a rs2910164 was associated with a decreased risk of CRC and reduced susceptibility in intermediately differentiated CRC in Chinese population. These findings were basically consisted with our results. On the contrary, another study of Chinese population from Sichuan Province displayed opposite results [18]. Lv et al. [18] reported that the rs2910164 GC genotype was associated with an increased risk of CRC. Possible explanations for the disaccords may be different case choices and/or limited sample size. The participants in our study were mainly advanced male CRC patients with ulcerative morphology, while patients in Lv's research were in a relatively mild condition. They may be in different etiologies and disease statuses. Further, both of the two Chinese studies supported that individuals with rs2910164 C allele had a protective effect in Chinese population, but this was not obvious in our study population. Above all, the miR-146a variant rs2910164 related to the susceptibility to CRC, nevertheless, has not yet been fully understood, large-scale and well-designed prospective studies are necessary to illuminate these problems.
Subsequently, in the analysis of histological differentiation, miR-146a SNP had a significant effect on CRC histological differentiation. Genotype GG and allele G were associated with a better differentiation. Mechanistic considerations of the SNP contributing to phenotypic changes were limited to protein-coding genes, while polymorphisms in miRNAs genes, namely, via nonprotein-coding RNA intermediaries interfering with the biological processing of miRNAs and/or target selection of miRNAs, have effects on phenotypes of human diseases [27]. Because of a relatively small sample size after stratifying by differentiated degree, further studies are still needed to validate these findings.
MiR-499 may play important roles in regulating different senescence induction mechanisms [28]. The miR-499 rs3746444 polymorphism manifests a change from A:U pair to G:U mismatch in the stem structure of pre-miR-499. Two meta-analyses supported the view that the miR-499 rs3746444 polymorphism was associated with breast cancer, and the C allele could increase cancer susceptibility in Asian [29, 30]. Liu et al. pointed out that overexpression of miR-499-5p promoted cell migration and invasion in CRC by targeting FOXO4 and PDCD4 [31]. Vinci et al. from Italy [14] demonstrated that the GG frequency of miR-499 rs3746444 was connected to a reduced miR-499 expression and a significant higher risk of CRC. Our data revealed a lack of association between the miR-499 rs3746444 polymorphism and the colorectal cancer risk in Chinese population. It was coincided with the previous study from China [18]. Although our results did not indicate any significant relationship between miR-499 rs3746444 and risk of CRC, we believe that our work will improve the understanding of the miRNAs involved in CRC onset and progression.
Further analysis showed that neither CEA nor CA19-9 was significantly different among patients with genotype of two studied SNP loci, suggesting that miR-146a and miR-499 SNPs may have no impact on CEA and CA19-9 serum level after surgery.
In conclusion, we confirmed that the genetic variants rs2910164 G > C of miR-146a was associated with CRC susceptibility and tumor differentiation. However, miR-499 SNP rs3746444 was neither associated with CRC nor with tumor maker. Our findings suggest the miR-146a rs2910164 variant as a biomarker in CRC onset and progression. Further larger population-based prospective and functional studies are warranted to validate our findings.
References
Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29.
Potter JD. Colorectal cancer: molecules and populations. J Natl Cancer Inst. 1999;91:916–32.
Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69.
Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5.
Michael MZ, O' Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res 2003;1:882–891.
Yang L, Belaguli N, Berger DH. MicroRNA and colorectal cancer. World J Surg. 2009;33:638–46.
Slaby O, Svoboda M, Fabian P, Smerdova T, Knoflickova D, Bednarikova M, et al. Altered expression of miR21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007;72:397–402.
Aaron JS, Suet YL, Jane JS, Krista AZ, Elise DB, Nozomu Y, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–36.
Motoyama K, Inoue H, Takatsuno Y, Tanaka F, Mimori K, Uetake H, et al. Over- and under-expressed microRNAs in human colorectal cancer. Int J Oncol. 2009;34:1069–75.
Tang JT, Fang JY. MicroRNA regulatory network in human colorectal cancer. Mini Rev Med Chem. 2009;9:921–6.
Hu Z, Chen J, Tian T, Zhou X, Gu H, Xu L, et al. Genetic variants of miRNA sequences and non-small cell lung cancer survival. Clin Invest. 2008;118:2600–8.
Hu ZB, Liang J, Wang ZW, Tian T, Zhou XY, Chen JP, et al. Common genetic variants in pre-MicroRNAs were associated with increased risk of breast cancer in Chinese women. Hum Mutat. 2009;30:79–84.
Naccarati A, Pardini B, Stefano L, Landi D, Slyskova J, Novotny J, et al. Polymorphisms in miRNA-binding sites of nucleotide excision repair genes and colorectal cancer risk. Carcinogenesis. 2012;33:1346–51.
Vinci S, Gelmini S, Mancini I, Malentacchi F, Pazzagli M, Beltrami C, et al. Genetic and epigenetic factors in regulation of microRNA in colorectal cancers. Methods. 2013;59:138–46.
Chae YS, Kim JG, Lee SJ, Kang BW, Lee YJ, Park JY, et al. A miR-146a polymorphism (rs2910164) predicts risk of and survival from colorectal cancer. Anticancer Res. 2013;33:3233–9.
Ma L, Zhu L, Gu D, Chu H, Tong N, Chen J, et al. A genetic variant in miR-146a modifies colorectal cancer susceptibility in a Chinese population. Arch Toxicol. 2013;87:825–33.
Hezova R, Kovarikova A, Bienertova-Vasku J, Sachlova M, Redova M, Vasku A, et al. Evaluation of SNPs in miR-196-a2, miR-27a and miR-146a as risk factors of colorectal cancer. World J Gastroenterol. 2012;18:2827–31.
Lv M, Dong W, Li L, Zhang L, Su X, Wang L, et al. Association between genetic variants in pre-miRNA and colorectal cancer risk in a Chinese population. J Cancer Res Clin Oncol. 2013;139:1405–10.
Chen CC, Yang SH, Lin JK, Lin TC, Chen WS, Jiang JK, et al. Is it reasonable to add preoperative serum level of CEA and CA19-9 to staging for colorectal cancer? J Surg Res. 2005;124:169–74.
Nozoe T, Rikimaru T, Mori E, Okuyama T, Takahashi I. Increase in both CEA and CA19-9 in sera is an independent prognostic indicator in colorectal carcinoma. J Surg Oncol. 2006;94:132–7.
Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–61.
Jazdzewski K, Murray EL, Franssila K, Jarzab B, Schoenberg DR, de la Chapelle A. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2008;105:7269–74.
Shen J, Ambrosone CB, DiCioccio RA, Odunsi K, Lele SB, Zhao H. A functional polymorphism in the miR-146a gene and age of familial breast/ovarian cancer diagnosis. Carcinogenesis. 2008;29:1963–6.
Xu T, Zhu Y, Wei QK, Yuan YF, Zhou F, Ge YY, et al. A functional polymorphism in the miR-146a gene is associated with the risk for hepatocellular carcinoma. Carcinogenesis. 2008;29:2126–31.
Tang YJ, Luo XB, Cui HJ, Ni XM, Yuan M, Guo YZ, et al. MicroRNA-146a contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum. 2009;60:1065–75.
Paik JH, Jang JY, Jeon YK, Kim WY, Kim TM, Heo DS. MicroRNA-146a downregulates NFκB activity via targeting TRAF6 and functions as a tumor suppressor having strong prognostic implications in NK/T cell lymphoma. Clin Cancer Res. 2011;17:4761–71.
Saunders MA, Liang H, Li WH. Human polymorphism at microRNAs and microRNA target sits. Proc Natl Acad Sci U S A. 2007;104:3300–5.
Lafferty-Whyte K, Cairney CJ, Jamieson NB, Oien KA, Nicol KW. Pathway analysis of senescence-associated miRNA targets reveals common processes to different senescence induction mechanisms. Biochim Biophys Acta. 2009;1792:341–52.
Wang F, Sun G, Zou Y, Li Y, Hao L, Pan F. Association of microRNA-499 rs3746444 polymorphism with cancer risk: evidence from 7188 cases and 8548 controls. PLoS One. 2012;7:e45042.
Fan C, Chen C, Wu D. The association between common genetic variant of microRNA-499 and cancer susceptibility: a meta-analysis. Mol Biol Rep. 2013;40:3389–94.
Liu X, Zhang Z, Sun L, Chai N, Tang S, Jin J, et al. MicroRNA-499-5p promotes cellular invasion and tumor metastasis in colorectal cancer by targeting FOXO4 and PDCD4. Carcinogenesis. 2011;32:1798–805.
Acknowledgments
We thank Dr. Junping Xin (Loyola University Medical Center) for critical review and editorial assistance during manuscript preparation.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Xuejiao Hu and Lixin Li contributed equally to this article.
Rights and permissions
About this article
Cite this article
Hu, X., Li, L., Shang, M. et al. Association between microRNA genetic variants and susceptibility to colorectal cancer in Chinese population. Tumor Biol. 35, 2151–2156 (2014). https://doi.org/10.1007/s13277-013-1285-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-013-1285-y