Abstract
Background
Pantoea spp. are frequently isolated from a wide range of ecological niches and have various biological roles, as plant epi- or endophytes, biocontrol agents, plant-growth promoters or as pathogens of both plant and animal hosts. This suggests that members of this genus have undergone extensive genotypic diversification. One means by which this occurs among bacteria is through the acquisition and maintenance of plasmids. Here, we have analyzed and compared the sequences of a large plasmid common to all sequenced Pantoea spp.
Results and discussion
The L arge P antoea P lasmids (LPP-1) of twenty strains encompassing seven different Pantoea species, including pathogens and endo-/epiphytes of a wide range of plant hosts as well as insect-associated strains, were compared. The LPP-1 plasmid sequences range in size from ~281 to 794 kb and carry between 238 and 750 protein coding sequences (CDS). A core set of 46 proteins, encompassing 2.2% of the total pan-plasmid (2,095 CDS), conserved among all LPP-1 plasmid sequences, includes those required for thiamine and pigment biosynthesis. Phylogenetic analysis reveals that these plasmids have arisen from an ancestral plasmid, which has undergone extensive diversification. Analysis of the proteins encoded on LPP-1 also showed that these plasmids contribute to a wide range of Pantoea phenotypes, including the transport and catabolism of various substrates, inorganic ion assimilation, resistance to antibiotics and heavy metals, colonization and persistence in the host and environment, pathogenesis and antibiosis.
Conclusions
LPP-1 is universal to all Pantoea spp. whose genomes have been sequenced to date and is derived from an ancestral plasmid. LPP-1 encodes a large array of proteins that have played a major role in the adaptation of the different Pantoea spp. to their various ecological niches and their specialization as pathogens, biocontrol agents or benign saprophytes found in many diverse environments.
Similar content being viewed by others
Background
The enterobacterial genus Pantoea currently comprises nineteen species of Gram-negative, yellow or beige pigmented, motile rods [1]. Members of this genus have been isolated from a wide range of environments including soil, water, dust, dairy products, meat, fish, insects, humans and animals [2]. Most frequently they are found associated with a broad range of plant hosts, as non-pathogenic endophytes or epiphytes, colonizing the leaves, stems and roots [2, 3]. In this context, some Pantoea strains can be beneficial to the plant host by contributing to growth promotion through processes such as the production of the plant-growth hormone indole-acetic acid (IAA), phosphate solubilization or nitrogen fixation [4, 5]. Some Pantoea strains also provide effective protection to plants against various bacterioses as well as fungal diseases and postharvest fruit rots [6–8]. However, strains of several Pantoea species represent major plant pathogens themselves. Pantoea stewartii subsp. stewartii causes the devastating disease Stewart’s wilt of maize, and pathovars of Pantoea agglomerans which induce galls on gypsophila, beet, Douglas firs and wisteria [9, 10]. Pantoea spp. are also frequently found associated with insects [11, 12]. For example, the corn flea beetle Chaetocnema pulicaria, serves as the vector of the Stewart’s wilt pathogen P. stewartii subsp. stewartii[9, 13]. Strains of some Pantoea species have been implicated as opportunistic human pathogens in cases of septicemia following penetrating trauma with plant material, nosocomial infections due to exposure to contaminated hospital materials and secondary pathogens complicating pre-existing illnesses [3, 14, 15].
The various ecological niches occupied by Pantoea species including both plant and animal hosts, and their distinctive lifestyles as epi- and endophytes, plant-growth promoters, biological control agents, or as pathogens of plant and animal hosts are indicative of extensive diversification within the genus Pantoea and even among individual strains belonging to the various Pantoea species. One means by which this diversification may be accomplished in bacteria is through the acquisition of plasmids, extra-chromosomal genetic elements capable of transfer between strains, species and genera and subsequent stable vertical transmission within the bacterial cell line [16]. These plasmids carry genes that can confer various phenotypes on the bacterium, including toxin, hormone production and virulence factors contributing to pathogenesis and host specificity, resistance to antibiotics and heavy metals and survival in adverse conditions, catabolism of amino and organic acids, carbohydrates and inorganic ions, colonization and dissemination [16, 17]. Plasmid acquisition can thus contribute to both the survival of a bacterium in an existing environment and colonization of novel niches [17].
Plasmids are common features among Pantoea species. Strains of P. stewartii subsp. stewartii carry up to 13 plasmids which may contribute 25% of the total genome content [18]. Pantoea sp. At-9b, which is associated with leaf-cutter ants, carries five large plasmids which encompass approximately 31% of the total genome content [12]. The importance of these plasmids to the biology of Pantoea species is evident in P. agglomerans pvs. betae and gypsophilae, which carry a 150 kb pPATH plasmid which is absent from other, non-pathogenic, P. agglomerans strains. This plasmid encodes a Type III secretion system as well as enzymes for the synthesis of indole acetic acid which confer the ability on these pathovars to infect and induce tumorigenesis on various plant hosts [10]. The biological control agent Pantoea vagans C9-1, registered in the United States and Canada for control of the fire blight pathogen Erwinia amylovora, carries a plasmid, pPag2, which encodes for the biosynthesis of the antimicrobial peptide herbicolin I [8, 19].
During early experiments with strains of several Pantoea species it was observed that extensive culturing and growth at supra-optimal temperatures result in the development of non-pigmented variants [20–22]. Biochemical analyses showed that these variants also lost the ability to synthesize the essential co-factor thiamine and could no longer utilize carbohydrate sources such as citrate and maltose [20, 22, 23]. The lack of reversion to a wild-type phenotype along with further basic molecular characterization indicated that these characteristics are encoded on a single large plasmid which has been lost in the non-pigmented variants [20, 23]. With the availability of the complete genome sequence, Smits et al. [23] were able to characterize the 530-kb plasmid carrying these factors in P. vagans C9-1. This plasmid also carries genes for the biosynthesis of the siderophore desferrioxamine E, acyl-homoserine lactone biosynthesis and quorum sensing, ampicillin resistance and carbohydrate utilization pathways which may contribute to the ecological fitness and efficacy of this biocontrol strain [8, 23].
The complete or draft genomes of 20 Pantoea strains belonging to seven distinct species have been sequenced to date. Here we elucidate and compare the sequences of the pigment biosynthesis and thiamine autotrophy-associated plasmid which we have given the collective name Large Pantoea Plasmid-1 (LPP-1). Our analyses reveal that LPP-1, which is common to these sequenced Pantoea strains, has been derived from an ancestral plasmid that has undergone extensive diversification. The variation in the LPP-1 plasmid sequences may be linked to various phenotypes that suggest this plasmid is an important driver of the biological, ecological and lifestyle diversification observed among the Pantoea species.
Results and discussion
Characteristics of the Large Pantoea Plasmid
The LPP-1 plasmid sequences of 20 strains encompassing seven species within the genus Pantoea were analyzed (Table 1). These plasmids represent the largest extra-chromosomal elements in each of the sequenced organisms. The LPP-1 genomes range in size from 281 to 794 kb in size and thus contribute between 5.6% (P. stewartii subsp. stewartii DC283) and 12.6% (Pantoea sp. At-9b) of the total genome content. This indicates that LPP-1 contributes significantly to the Pantoea genotype. On the basis of a phylogeny using the amino acid sequences of four chromosomal house-keeping genes, the Pantoea strains used in this study could be separated into three distinct groups, Group I-III (Figure 1). Similarly, a pattern in terms of the LPP-1 size and G+C content (%) could be observed within each distinct group (Table 1). Group I encompasses members of the species P. agglomerans, Pantoea eucalypti, P. vagans and Pantoea anthophila. The Group I LPP-1 plasmid sequences have an average size 524-kb and G+C content of 53.90%. One notable exception is the P. anthophila Sc1 LPP-1 which is only 411 kb in size, with a G+C content of 56.27%. The LPP-1 of group II organisms Pantoea ananatis and P. stewartii has an average size of 310 kb with a G+C content of 51.75%. Group III incorporates the leaf-cutter ant-associated Pantoea sp. At-9b and the orchid pathogen Pantoea cypripedii LMG2657T and their LPP-1 plasmid sequences have an average size of 720 kb and G+C content of 54.18%.
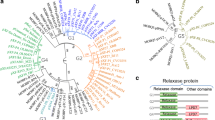
Phylogeny of the Pantoea spp. based on chromosomal house-keeping genes and LPP-1 plasmid genes. A neighbour-joining phylogeny constructed on the basis of the concatenated amino acid sequences of four chromosomal house-keeping genes, atpD, infB, gyrB and rpoB is shown on the left hand side. The type strains of those species for which no type strain genome sequences are available, were included. The neighbour-joining tree on the right hand represents the phylogeny constructed using the concatenated amino acid sequences for 20 proteins conserved on all Group I-III Pantoea spp. Bootstrap values are shown. The branches in both trees are coloured according to the groups, with Group I branches in blue, Group II in red and Group III in green
The large Pantoea plasmids are derived from a common ancestor
The LPP-1 CDS sets for each organism within each separate group were compared by BlastP analyses. By this means, the LPP-1 pan-genome for each group, as well as the conserved core and flexible portions were determined (Figure 2). The core CDS sets for each group were then further compared in order to determine those CDS common to all LPP-1 plasmids. A total of 46 CDS are conserved among the LPP-1 plasmids of all 20 strains (Additional file 1: Table S1, Figure 2). Included among these are five genes, crtEYIBZ, whose translation products are involved in the production of the yellow carotenoid pigment zeaxanthin [24]. This secondary metabolite has been shown to protect the producing organism against UV irradiation and phototoxic damage [25, 26]. An additional gene, crtX, whose product adds a diglucoside group to the pigment is absent from the LPP-1 plasmids of Group III. Also common on all LPP-1 plasmids are the thiOSGF genes required for biosynthesis of the essential co-factor thiamine [27]. Pantoea strains lacking this plasmid have been demonstrated to require thiamine supplementation for growth [23]. Other conserved genes include those encoding AscBFG involved in the transport and catabolism of arbutin, salicin and cellobiose, the Dat and Ddc enzymes intermediate in the the biosynthesis of 1,3-diaminopropane and the enzyme malate:quinone oxidoreductase (Mqo) involved in the tricarboxylic acid cycle.
Venn diagram of the Pantoea spp. LPP-1 pan-genome. The outer circles represent the LPP-1 pan-genomes for Group I (blue), Group II (red) and Group III (green) and show the conserved (core) and non-conserved (flexible) CDS for each group. The core CDS were further compared as shown in the inner Venn diagram, with underlined value in the centre representing the conserved core CDS for all LPP-1 genomes
A phylogeny based on the concatenated amino acid sequences of 20 LPP-1 core CDS was constructed (Figure 1). This phylogeny shows a good correlation with that of the concatenated amino acid sequences of the four chromosomal house-keeping markers atpD, gyrB, infB and rpoB in terms of the clear taxon delineation into Groups I-III observed in both phylogenies (Figure 1). Furthermore, the taxon separation within each group in the LPP-1 tree largely matches those based on house-keeping genes present on the chromosome. This indicates that the LPP-1 plasmid may have been derived from an ancestral plasmid which shares a similar evolutionary history as the chromosomes of the associated Pantoea spp. As it has been observed that LPP-1 carries genes necessary for the production of the essential co-factor thiamine, and the selective advantage which would be provided by the carotenoid pigment in organisms which are frequently associated with UV-exposed surfaces of plants, strengthens the argument for vertical inheritance.
LPP-1 has undergone extensive diversification among the Pantoea spp
While there are 46 CDS conserved among the LPP-1 of all 20 Pantoea strains, this only represents a small fraction (2.2%) of the total pan-plasmid (2,095 CDS) (Figure 2). This indicates that although they are derived from a common ancestral plasmid, the LPP-1 plasmids of the different Pantoea strains have undergone massive genetic diversification. Even within the different groups, the extensive plasticity of the LPP-1 plasmid sequence can be observed. Among Pantoea Group I strains, the flexible portion of the pan-plasmid (709 CDS) (73.8%) makes up 73.8% of the pan-plasmid (Figures 2 and 3), while in Group II, this portion contributes 66.7% (318 CDS) of the pan-plasmid CDS (Figures 2 and 4). In Group III, where only two closely related strains were compared, the flexible CDS encompasses 44.8% (382 CDS) of the total (Figures 2 and 5). The extensive mosaicism of the Group I-III pan-plasmids can also be observed in Figures 35, with multiple non-conserved genomic regions inserted into the LPP-1 backbone. Many of these non-conserved regions show evidence that they represent mobile genetic elements, such as transposable and insertion elements. Furthermore, phage elements are integrated into the Group I, II and III LPP-1 plasmids of P. agglomerans IG1, P. ananatis BD442 and Pantoea sp. At-9b (Figures 35). There is also evidence of the exchange of genetic elements between the chromosomes and the LPP-1 plasmids of the Pantoea strains of the different groups. The maltose locus, malGFEKLMQPT, located on the LPP-1 of all Group I and III strains is maintained on the chromosome of Group II strains. Type VI secretion systems showing high sequence identity to those encoded on the LPP-1 plasmid of three Group II strains and P. vagans MP7 (Figures 3 and 4) have also been observed on the chromosomes of P. eucalypti αB, P. agglomerans E325 and Pantoea sp. At-9b [28]. There is also evidence for chromosome/LPP-1 exchange within the groups. For example, the pagRI genes, required for the production of the acyl-homoserine lactone autoinducer PagI and the transcriptional regulator PagR involved in the regulation of ecological fitness and virulence genes in various Pantoea spp. and related taxa, are chromosomally localized in most of the Group I strains, but are located on the plasmid of P. vagans strains C9-1 and MP7 [23].
Genome diagram of the Group I LPP-1 pan-genome. Diagram showing the major phenotypes encoded on the Group I LPP-1 pan-genome. The circles represent the LPP-1 genomes of the Pantoea strains (from inside to outside): 1. P. agglomerans E325, 2. P. agglomerans MP2, 3. P. agglomerans IG1, 4. P. agglomerans SL1_M5, 5. P. vagans MP7, 6. P. vagans C9-1, 7. P. eucalypti αB and 8. P. anthophila Sc1. CDS in the flexible portion of Group I pan-genome are colored dark blue, while those in conserved among all Group I LPP-1 genomes are colored light blue. Black arrows indicate those CDS conserved among all 20 LPP-1 genomes
Genome diagram of the Group II LPP-1 pan-genome. Diagram showing the major phenotypes encoded on the Group II LPP-1 pan-genome. The circles represent the LPP-1 genomes of the Pantoea strains (from inside to outside): 1. P. stewartii subsp. stewartii DC283, 2. P. stewartii subsp. indologenes LMG2632T, 3. P. ananatis PA4, 4. P. ananatis LMG5342, 5. P. ananatis B1-9, 6. P. ananatis BD442, 7. P. ananatis AJ13355, 8. P. ananatis PA13, 9. P. ananatis LMG20103 and 10. P. ananatis LMG2665T. CDS in the flexible portion of Group II pan-genome are colored maroon, while those in conserved among all Group II LPP-1 genomes are colored red. Black arrows indicate those CDS conserved among all 20 LPP-1 genomes
Genome diagram of the Group III LPP-1 pan-genome. This diagram shows the major phenotypes encoded on the Group III LPP-1 pan-genome. The inner circular track represents Pantoea sp. At-9b, while the outer track represents the LPP-1 genome of P. cypripedii LMG2657T. CDS in the flexible portion of Group III pan-genome are colored dark green, while those in conserved among all Group III LPP-1 genomes are colored light green. Black arrows indicate those CDS conserved among all 20 LPP-1 genomes
LPP-1 plays a major role in the diversification of Pantoea spp
The functions of the core and flexible CDS encoded on the various LPP-1 plasmids were determined on the basis of sequence similarity to characterized proteins. This indicated that the LPP-1 plasmids carry genes which may influence a variety of phenotypes. These phenotypes, which are discussed in further detail below, include the ability to take up and utilize a wide range of carbohydrate, amino acid and organic acid substrates as well as inorganic ions, resistance to antibiotics and heavy metals, as well as encoding potential pathogenicity and antibiosis factors. The LPP-1 plasmid may thus contribute extensively to the ability of Pantoea spp. to colonize the various ecological niches and hosts from which they have been isolated, as well as their different lifestyles as plant endo- or epiphytes, biocontrol agents and pathogens (Additional file 1: Table S1).
LPP-1 contributes to Pantoea metabolic versatility
White, plasmid-cured variants of P. vagans C9-1 have been shown to be unable to efficiently utilize a number of carbohydrate, amino and organic acid substrates, thus indicating a role for the plasmid in the metabolic versatility of this organism [23]. Analysis of the LPP-1 plasmid sequences of the 20 strains showed that this may represent a general trend for the plasmid, as a large number of loci for the transport and utilization of these metabolic substrates are encoded on LPP-1 (Additional file 1: Table S1). Common to all LPP-1 plasmids is a locus, ascBFG, which plays a role in the transport and catabolism of the sugars arbutin, salicin and cellobiose [29]. Other transport and metabolism-associated loci are more restricted in their distribution among the LPP-1 plasmids. An additional locus for the transport of these sugars, bglBFG, is found in most Group I and III strains, but not in strains of P. ananatis, P. eucalypti αB or P. vagans MP7. A maltose/maltodextrin transport and catabolism locus is found in all Group I and III organisms [23], but not those of Group II, where a locus with high sequence identity is located on the chromosome. Other carbohydrate transport and metabolism-associated loci encoded on the LPP-1 plasmid sequence, include those for galactan (ganKEFGABCLR), a major constituent of the pectic cell wall polymers found in plant cell walls [30], found in all Group I strains except P. anthophila Sc1, malonate (mdcABCDEFGH) encoded on Group III LPP-1 plasmids and a locus (frlABDR) for the catabolism of the glucose/amine agglomerates fructoselysine and psicoselysine [31] encoded on the P. agglomerans MP2 LPP-1. The LPP-1 of all Pantoea Group II strains also encodes a glucoamylase (Gca), which can be used to release glucose from starch, a major energy storage compound in plants [32].
A number of loci for the transport and catabolism of amino acid and organic acid substrates are also encoded on the Group I-III LPP-1 plasmids, including those for the transport of branched amino acids (azlCD) found in Group I-III, tyrosine (tutB-tpl) in P. ananatis strains and a tryptophan-specific transporter (mtr) encoded on the LPP-1 plasmids of P. vagans MP7 and P. eucalypti αB. A D-methionine transporter is also encoded on the LPP-1 of most Pantoea Group I strains. While these represent loci which show extensive homology to characterized transport and catabolism systems for known substrates, the uptake and utilization of a large number of other uncharacterized substrates may be linked to the LPP-1 plasmids. For example, a further 41 putative transport systems for unknown substrates were identified in the LPP-1 plasmid of Pantoea sp. At-9b. This further supports the important role of LPP-1 in the metabolic versatility of Pantoea spp.
LPP-1 plays a role in iron and nitrogen assimilation
The LPP-1 plasmid sequences also encode several proteins involved in the assimilation of the essential elements iron and nitrogen, which are commonly found in limited supply in the environment. In order to scavenge iron from the environment or from their host, bacteria use high-affinity iron-binding molecules known as siderophores [8, 33]. A desferrioxamine E siderophore biosynthetic locus, dfoJACS is encoded on all Group I LPP-1 plasmids, while this locus is found on the chromosome of Pantoea Group II strains [33]. Iron-siderophore complexes are subsequently captured at the bacterial cell surface by high-affinity TonB-dependent receptor proteins, and translocated into the periplasm by the Ton system before being transported into the cytoplasm via inner membrane-associated ABC transporters [34]. Nine outer membrane receptor proteins without cognate siderophore biosynthetic systems are encoded on the LPP-1 plasmids (Additional file 1: Table S1). These can be utilized to scavenge iron-loaded siderophores produced by other microorganisms occupying the same ecological niches. This xenosiderophore scavenging will thus allow the acquisition of siderophore-bound iron without the energy cost of siderophore production [35]. An inner membrane iron and manganese transporter, SitABCD is also encoded on the LPP-1 of all Group I and II strains, with the exception of P. stewartii subsp. stewartii DC283.
The Rut pathway has been shown to release nitrogen through the catabolism of pyrimidine nucleotides [36]. The rutRABCDEFG locus encoded on the LPP-1 of all Group II strains may thus aid in the assimilation of nitrogen. A locus, narIJHGK, in the Group III strain P. cypripedii LMG2657T can reduce nitrate to nitrite which can act as terminal respiratory electron acceptor [37]. An ortholog of NorV, a flavorubredoxin involved in the detoxification of the nitric oxide, a cytotoxic byproduct of nitrate reduction [37] is also encoded on the LPP-1 of this organism.
LPP-1 contributes to antibiotic and heavy metal resistance
A β-lactamase bla and its cognate transcriptional regulator ampR have been identified on the LPP-1 plasmid of P. vagans C9-1, and their translation products were shown to be required for ampicillin resistance [8, 23]. Genes showing extensive sequence identity to bla-ampR are present on the LPP-1 plasmids of the Group I strains P. agglomerans IG1, P. anthophila Sc1 and P. vagans MP7. The LPP-1 of P. vagans MP7 also carries a locus terZABCDEF, which is localized on the pPag1 plasmid of P. vagans C9-1 [19]. This locus is required for resistance to tellurite, a rare oxide mineral which has a long history as antimicrobial agent [38]. Six distinct RND multidrug efflux pumps are also encoded on the Group I-III LPP-1 plasmids (Additional file 1: Table S1). Furthermore, all Group II and III LPP-1 plasmids carry a locus (arnBCATEF) and a further protein PagO encoded on the LPP-1 of Group III Pantoea spp., which are involved in the addition of 4-amino-4-deoxy-L-arabinose and palmitate side chains to the lipopolysaccharide lipid A moiety, respectively. These modifications confer resistance to polymyxin and cationic antimicrobial peptides [39, 40]. Thus LPP-1 encodes various proteins which may play a role in protecting the different Pantoea species against various antimicrobials. LPP-1 also carries loci which can confer resistance to heavy metals. Loci showing extensive sequence conservation to those involved in resistance to arsenate (arsCBR) are located on the LPP-1 plasmids of four Group I Pantoea strains, while a chromate resistance locus (chrAB) is carried on the P. vagans MP7 LPP-1.
LPP-1 may play a role in host colonization and persistence
A total of seven distinct fimbriae are encoded among the twenty LPP-1 plasmids which were compared, with fimbrial loci present on the LPP-1 plasmids of representatives of all three groups (Additional file 1: Table S1). P. stewartii subsp. indologenes LMG2632T LPP-1 encodes a cblABCD locus required for the synthesis of cable pili, which have been shown to play an important role in the attachment of the cystic fibrosis pathogen Burkholderia cepacia to epithelial cells and persistence in the airways [41]. The LPP-1 plasmid of P. agglomerans IG1 also carries a gene encoding a putative AidA-like autotransporter adhesin. The presence of genes for these adhesive structures on LPP-1, suggests this plasmid plays a role in the attachment and persistence of Pantoea in the different environmental niches they occupy, including potentially the various plant, insect and vertebrate hosts from which they have been isolated.
The Group II LPP-1 also carries two fliC genes encoding flagellin, the major subunit of flagella. These fliC genes are not located adjacent to the remainder of the fli locus required for flagellum biosynthesis, which is present on the chromosome next to a further fliC copy. Multiple fliC copies have been found encoded on the plasmids of a number of bacteria, including Salmonella enterica and Escherichia coli[42, 43]. As the flagellin protein represents one of the major surface antigens in Gram-negative bacteria which when recognized by the host triggers defense responses, the alternate expression of different copies of fliC, termed phase variation, can enable the bacterium to evade host defenses [42, 43]. The LPP-1 plasmid sequences of the Group II P. ananatis strains as well as P. vagans C9-1 encode a protein NdvB required for the synthesis the extracellular cyclic β-glucan polymer [44]. In the phytopathogen Xanthomonas campestris pv. campestris, this glucan has been shown to be spread throughout the host plant and suppress both systemic and localized host defenses [44]. By contrast, with the exception of P. stewartii subsp. stewartii DC283, all Group II Pantoea strains carry a locus, budABCR, involved in the biosynthesis of the bacterial volatile 2,3-butanediol, which has been demonstrated to trigger systemic resistance in plants [45, 46]. This may correlate with the role as plant-growth promoter which has been identified for some Pantoea spp. [4, 5].
The LPP-1 plasmid sequence of Pantoea sp. At-9b also carries an extensive locus for the synthesis and maturation of a formate hydrogenase (Additional file 1: Table S1, Figure 5). This enzyme enables a bacterium to exploit hydrogen as an energy source under anaerobic conditions and has been found to play a major role in gut colonization and virulence in enteric pathogens [47]. As Pantoea sp. At-9b has been found in close association with leaf-cutter ants [12], this hydrogenase may play a role in its colonization of an insect host.
LPP-1 encodes putative pathogenicity and antibiosis factors
The Hrp Type III secretion system represents one of the main pathogenicity determinants of the corn pathogen P. stewartii subsp. stewartii, playing a role in both systemic infection and the development of the water-soaked lesions typical of Stewart’s wilt of corn [9, 13]. The LPP-1 plasmid sequences of the Group II strains P. stewartii subsp. stewartii DC283 and P. stewartii subsp. indologenes LMG2632T incorporate a Hrp locus. The latter organism is thought to cause leaf spot of pearl and foxtail millet and rot of pineapple [48]. The presence of the Hrp T3SS on the P. stewartii LPP-1 suggests this plasmid is thus required for pathogenesis on their different plant hosts.
Encoded on the LPP-1 of three P. ananatis strains (Additional file 1: Table S1 and Figure 4) as well as termite symbiont P. vagans MP7 is a Type VI secretion system (T6SS). Early studies indicated that this secretion system plays a major role in virulence in both animal and plant pathogens [28, 49]. However, the presence in many potentially non-pathogenic bacteria and inconsistencies in experimental data have led to a number of different functions being postulated for this secretion system. One function recently identified for the T6SS of a number of different bacterial taxa is the secretion of antimicrobial effectors, which suggests it may play a role in antibiosis and inter-bacterial competition [49]. The function of the T6SS in the various Pantoea spp. still needs to be elucidated.
Located on the LPP-1 of P. agglomerans IG1 and Pantoea sp. At-9b are genes encoding a predicted bacteriocin and its respective immunity protein showing highest sequence identity to Alveicin A (aat-aai) produced by Hafnia alvei and Klebicin B (kba-kbi) produced by Klebsiella pneumoniae, respectively [50, 51]. These bacteriocins are small toxic proteins, frequently encoded on plasmids, which can kill closely related bacteria [51]. The LPP-1 plasmids of the Group II strains P. ananatis B1-9 and LMG2665T also carry a ~25.5-kb locus encoding proteins showing low sequence similarity to polyketide synthases (PKS)/non-ribosomal peptide synthases (NRPS). These proteins are involved in the synthesis of a wide range of structural variable secondary metabolites including antibiotics (e.g. vancomycin and penicillin), siderophores (e.g. yersiniabactin and pyoverdine) as well as toxins (e.g. the Pseudomonas syringae phytotoxin syringomycin) [52]. However, due to the low sequence similarity to characterized PKS/NRPS proteins, further characterization of the compounds produced by this locus in the P. ananatis strains will need to be performed before a definitive function can be assigned.
Conclusions
The acquisition and maintenance of plasmids by bacteria has been demonstrated to play a key role in their adaptation to novel ecological niches and their development as symbiont, plant-growth promoters, saprophytes, biocontrol agents or pathogens [16]. Our analysis of the Large Pantoea plasmid LPP-1 common to all members of the genus Pantoea sequenced to date, showed that similarly, this plasmid is likely to have played a major role in the ecological and biological adaptation of Pantoea spp. A core set of genes conserved on all LPP-1 plasmids include those required for thiamine biosynthesis, an essential cofactor for bacterial growth and survival. The maintenance of these essential genes on LPP-1, correlate with our phylogenetic analysis, suggesting that this plasmid must be stably inherited and maintained by Pantoea spp. and has thus evolved from a common ancestral plasmid. The backbone of LPP-1 has however been permeated, through the incorporation of a multitude of genetic elements to give rise to the extremely variable LPP-1 plasmids which can be observed for the various Pantoea spp. These variable elements include genes for the transport and catabolism of various metabolic substrates, the assimilation of inorganic ions, and resistance to antimicrobial peptides and heavy metals. These factors may in turn have provided the Pantoea spp. with the selective advantage to thrive in the vast array of ecological niches they occupy. Furthermore, the LPP-1 plasmid sequences encode proteins for the synthesis of several putative plant colonization, plant-growth promoting, antimicrobial and pathogenicity factors, which may have enabled their specialization as epi- and endophytes, effective biocontrol agents, as insect endosymbionts and as pathogens of plant and animal hosts. The LPP-1 plasmid thus represents a major evolutionary driver among the ecologically and biologically robust Pantoea spp.
Methods
Assembly and annotation of LPP-1
The genome sequences of 20 Pantoea spp. isolated from different environmental sources and hosts were used in this study (Table 1). These included nine published complete and high-quality draft genomes and eleven unpublished draft genomes sequenced, assembled and annotated in our laboratories or which are publically available [11, 12, 53–60]. The LPP-1 nucleotide sequences for the complete Pantoea genomes were used to assemble the LPP-1 plasmid for those organisms for which draft genomes are available. Draft contigs were aligned against the complete LPP-1 sequences of the nearest phylogenetic relatives using progressiveMauve [61]. Co-linear contigs were assembled to high quality draft LPP-1 sequences. Local BlastN analyses with the BioEdit v 7.0.5.3 software package [62] were performed using the thiamine biosynthesis locus thiOSFG and the carotenoid biosynthetic locus crtEXYBIZ to confirm that the plasmids belonged to the LPP-1 group. The CDS for complete LPP-1 sequences were downloaded from the National Center for Biotechnology Information (NCBI) database [63] and those encoded on the draft plasmids were predicted using FgenesB [64]. The CDS sets for each LPP-1 plasmid were standardized by performing reciprocal local BlastN analyses using BioEdit [62] to identify and add CDS which were not predicted in a particular LPP-1 plasmid sequence. BlastP analysis [65] against the NCBI protein database [63], as well as annotation of the LPP-1 CDS using the GenDB pipeline [66], were used to predict their function on the basis of sequence similarity to characterized proteins. Transport-associated proteins were further characterized by comparison against the Transporter Classification Database (TCDB) [67].
Comparative analyses of the LPP-1 plasmids
The LPP-1 plasmids were categorized into three groups consistent with the phylogenetic clusters identified for the producing Pantoea strains on the basis of four chromosomal loci as described below. The LPP-1 pan-genomes for each of these groups were determined. This was done by local BlastP analysis of the LPP-1 CDS sets using a reciprocal best hit (RBH) approach [68]. Homologs were considered when CDS shared more than 70% amino acid identity over 70% of the average length of the two proteins. Comparative diagrams for each group incorporating the pan-genomes and CDS sets for all LPP-1 plasmids were constructed using GenomeDiagram [69]. The core CDS set, those that are common to all LPP-1 plasmids in each group, as well as the flexible CDS set were determined. The core CDS sets for each group were further compared using the RBH parameters described above. By this means, the CDS set which is core to all LPP-1 plasmids was determined.
Phylogenetic analyses
A phylogeny based on the amino acid sequences for four chromosomal house-keeping genes, atpD, gyrB, infB and rpoB was constructed. Similarly, the amino acid sequences of 20 distinct plasmid-borne genes which are found in the overall LPP-1 core were used to determine the phylogenetic relationships of the large Pantoea plasmids. These included the thiamine biosynthetic locus thiOSGF, pigment biosynthesis locus crtEYIBZ, carbohydrate transport and utilization locus ascBFG, branched amino acid transporter azlCD, malate:quinone oxidoreductase mqo, 1,3-diaminopropane biosynthetic genes dat-ddc and the replication genes repA and parAB. A ClustalW alignment for the concatenated amino acid sequences was performed using the parameters as previously described [28]. Neighbour-joining phylogenetic trees (Poisson correction; complete gap deletion; bootstrap analysis – n = 1,000) were constructed using the Molecular Evolution Genetics Analysis (MEGA) v 5.0.3 software package [70].
References
Brady CL, Cleenwerck I, Van Der Westhuizen L, Venter SN, Coutinho TA, De Vos P: Pantoea rodasii sp. nov., Pantoea rwandensis sp. nov. and Pantoea wallisii sp. nov. isolated from Eucalyptus. Int J Syst Evol Microbiol. 2012, 62: 1457-1464. 10.1099/ijs.0.032615-0.
Grimont PAD, Grimont F: Genus XXIII Pantoea. Bergey’s Manual of Systematic Bacteriology. Volume 2B. Edited by: Brenner DJ, Krieg NR, Staley JT, Garrity GM. 2005, Springer, New York, 713-720. 2
Coutinho TA, Venter SN: Pantoea ananatis: an unconventional plant pathogen. Mol Plant Pathol. 2009, 10: 325-335. 10.1111/j.1364-3703.2009.00542.x.
Loiret FG, Ortega E, Kleiner D, Ortega-Rodés P, Rodes R, Dong Z: A putative new endophytic nitrogen-fixing bacterium Pantoea sp. from sugarcane. J Appl Microbiol. 2004, 97: 504-511. 10.1111/j.1365-2672.2004.02329.x.
Mishra A, Chauhan PS, Chaudry V, Tripathi M, Nautiyal CS: Rhizosphere competent Pantoea agglomerans enhances maize (Zea mays) and chickpea (Cicer arietinum L.) growth, without altering the rhizosphere functional diversity. Antonie Van Leeuwenhoek. 2011, 100: 405-413. 10.1007/s10482-011-9596-8.
Frances J, Bonaterra A, Moreno MC, Cabrefiga J, Badosa E, Montesinos E: Pathogen aggressiveness and postharvest biocontrol efficiency in Pantoea agglomerans. Postharvest Biol Technol. 2006, 39: 299-307. 10.1016/j.postharvbio.2005.11.002.
Pusey PL, Stockwell VO, Reardon CL, Smits THM, Duffy B: Antibiosis activity of Pantoea agglomerans biocontrol strain E325 against Erwinia amylovora on apple flower stigmas. Phytopathology. 2011, 101: 1234-1241. 10.1094/PHYTO-09-10-0253.
Smits THM, Rezzonico F, Kamber T, Blom J, Goesmann A, Ishimaru CA, Frey JE, Stockwell VO, Duffy B: Metabolic versatility and antibacterial metabolite biosynthesis are distinguishing genomic features of the fire blight antagonist Pantoea vagans C9-1. PLoS One. 2011, 6: e22247-10.1371/journal.pone.0022247.
Roper CM: Pantoea stewartii subsp. stewartii: lessons learned from a xylem-dwelling pathogen of sweet corn. Mol Plant Pathol. 2011, 12: 628-637. 10.1111/j.1364-3703.2010.00698.x.
Barash I, Manulis-Sasson S: Recent evolution of bacterial pathogens: the gall-forming Pantoea agglomerans case. Ann Rev Phytopathol. 2009, 47: 133-152. 10.1146/annurev-phyto-080508-081803.
Adams AS, Jordan MS, Adams SM, Suen G, Goodwin LA, Davenport KW, Currie CR, Raffa KF: Cellulose-degrading bacteria associated with the invasive woodwasp Sirex noctilio. ISME J. 2011, 5: 1323-1331. 10.1038/ismej.2011.14.
Suen G, Scott JJ, Aylward FO, Adams SM, Tringe SG, Pinto-Tomas AA, Foster CE, Pauly M, Weimer PJ, Barry KW, Goodwin LA, Bouffard P, Li L, Osterberger J, Harkins TT, Slater SC, Donohue TH, Currie CR: An insect herbivore microbiome with high plant biomass-degrading capacity. PLoS Genet. 2010, 6: e1001129-10.1371/journal.pgen.1001129.
Correa V, Majerczak DR, Ammar E-D, Merighi M, Pratt R, Hogenhout S, Coplin DL, Redinbaugh MG: The bacterium Pantoea stewartii uses two different type III secretion systems for colonizing its plant host and insect vector. Appl Environ Microbiol. 2012, 78 (17): 6327-6336. 10.1128/AEM.00892-12.
Cruz AT, Cazacu AC, Allen CH: Pantoea agglomerans, a plant pathogen causing human disease. J Clin Microbiol. 2007, 45: 1989-1992. 10.1128/JCM.00632-07.
Rezzonico F, Smits THM, Montesinos E, Frey JE, Duffy B: Genotypic comparison of Pantoea agglomerans plant and clinical strains. BMC Microbiol. 2009, 9: 204-10.1186/1471-2180-9-204.
Vivian A, Murillo J, Jackson RW: The roles of plasmids in phytopathogenic bacteria: mobile arsenals?. Microbiology. 2001, 147: 763-780.
Kado CI: Origin and evolution of plasmids. Antonie Van Leeuwenhoek. 1998, 73: 117-126. 10.1023/A:1000652513822.
Coplin DL, Rowan RG, Chisholm DA, Whitmoyer RE: Characterization of plasmids in Erwinia stewartii. Appl Environ Microbiol. 1981, 42: 599-604.
Kamber T, Lansdell TA, Stockwell VO, Ishimaru CA, Smits THM, Duffy B: Characterization of the biosynthetic operon for the antibacterial peptide herbicolin in Pantoea vagans biocontrol strain C9-1 and incidence in Pantoea species. Appl Environ Microbiol. 2012, 78: 4412-4419. 10.1128/AEM.07351-11.
Chatterjee AK, Gibbins LN: Induction of nonpigmented variants of Erwinia herbicola by incubation at supraoptimal temperatures. J Bacteriol. 1971, 105: 107-112.
Gantotti BV, Beer SV: Plasmid-borne determinants of pigmentation and thiamine prototrophy in Erwinia herbicola. J Bacteriol. 1982, 151: 1627-1629.
Thiry G: Plasmids of the epiphytic bacterium Erwinia ureduvora. J Gen Microbiol. 1984, 130: 1623-1631.
Smits THM, Rezzonico F, Pelludat C, Goesmann A, Frey JE, Duffy B: Genomic and phenotypic characterization of a nonpigmented variant of Pantoea vagans biocontrol strain C9-1 lacking the 530-kb megaplasmid pPag3. FEMS Microbiol Lett. 2010, 308: 48-54. 10.1111/j.1574-6968.2010.01994.x.
Misawa N, Nakagawa M, Kobayashi K, Yamano S, Izawa Y, Nakamura K, Harashima K: Elucidation of the Erwinia uredovora carotenoid biosynthetic pathway by functional analysis of gene products expressed in Escherichia coli. J Bacteriol. 1990, 172: 6704-6712.
Tuveson RW, Larson RA, Kagan J: Role of cloned carotenoid genes expressed in Escherichia coli in protecting against inactivation by near-UV light and specific phototoxic molecules. J Bacteriol. 1988, 170: 4675-4680.
Dussault D, Caillet S, Tien CL, Lacroix M: Carotenoids’ influence on radiotolerance of Pantoea agglomerans, a plant pathogen. Lett Appl Microbiol. 2008, 47: 208-213. 10.1111/j.1472-765X.2008.02410.x.
Park J-H, Dorrestein PC, Zhai H, Kinsland C, McLafferty FW, Begley TP: Biosynthesis of the thiazole moiety of thiamin pyrophosphate (vitamin B1). Biochemistry. 2003, 42: 12430-12438. 10.1021/bi034902z.
De Maayer P, Venter SN, Kamber T, Duffy B, Coutinho TA, Smits THM: Comparative genomics of the type VI secretion systems of Pantoea and Erwinia species reveals the presence of putative effector islands that may be translocated by the VgrG and Hcp proteins. BMC Genomics. 2011, 12: 576-10.1186/1471-2164-12-576.
An CL, Lim WJ, Hong SY, Shin EC, Kim MK, Lee JR, Park SR, Woo JG, Lim YP, Yun HD: Structural and biochemical analysis of the asc operon encoding 6-phospho-β-glucosidase in Pectobacterium carotovorum subsp. carotovorum LY34. Res Microbiol. 2005, 156: 145-153. 10.1016/j.resmic.2004.09.010.
Delangle A, Prouvost A-F, Cogez V, Bohin J-P, Lacroix J-M, Hugouvieux Cotte-Pattat N: Characterization of the Erwinia chrysanthemi gan locus, involved in galactan catabolism. J Bacteriol. 2007, 189: 7053-7061. 10.1128/JB.00845-07.
Wiame E, Van Schaftingen E: Fructoselysine 3-epimerase, an enzyme involved in the metabolism of the unusual Amadori compound psicoselysine in Escherichia coli. Biochem J. 2004, 378: 1047-1052. 10.1042/BJ20031527.
Marín-Navarro J, Polaina J: Glucoamylases: structural and biotechnological aspects. Appl Microbiol Biotechnol. 2011, 89: 1267-1273. 10.1007/s00253-010-3034-0.
Smits THM, Duffy B: Genomics of iron-acquisition in the plant pathogen Erwinia amylovora: insights in the biosynthetic pathway of the siderophore desferrioxamine E. Arch Microbiol. 2011, 193: 693-699. 10.1007/s00203-011-0739-0.
Expert D: Withholding and exchanging iron: interactions between Erwinia spp. and their plant hosts. Annu Rev Phytopathol. 1999, 37: 307-334. 10.1146/annurev.phyto.37.1.307.
Brickman TJ, Anderson MT, Armstrong SK: Bordetella iron transport and virulence. Biometals. 2007, 20: 303-322. 10.1007/s10534-006-9031-1.
Parales RE, Ingraham JL: The surprising Rut pathway: an unexpected way to derive nitrogen from pyrimidines. J Bacteriol. 2010, 192: 4086-4088. 10.1128/JB.00573-10.
Rowley G, Hensen D, Felgate H, Arkenberg A, Appia-Ayme C, Prior K, Harrington C, Fields SJ, Butt JN, Baggs E, Richardson DJ: Resolving the contributions of the membrane-bound and periplasmic nitrate reductase systems to nitric oxide and nitrous oxide production in Salmonella enterica serovar Typhimurium. J Biochem. 2012, 441: 755-762. 10.1042/BJ20110971.
Taylor DE: Bacterial tellurite resistance. Trends Microbiol. 1999, 7: 111-115. 10.1016/S0966-842X(99)01454-7.
Breazeale SD, Ribeiro AA, McClerren AL, Raetz CRH: A formyltransferase required for polymyxin resistance in Escherichia coli and the modification of lipid A with 4-amino-4-deoxy-L-arabinose. Biochem. 2005, 280: 14154-14167.
Hwang PM, Choy W-Y, Lo EI, Chen L, Forman-Kay JD, Raetz CRH, Privé GG, Kay LE: Solution structure and dynamics of the outer membrane enzyme PagP by NMR. Proc Nat Acad Sci USA. 2002, 99: 13560-13565. 10.1073/pnas.212344499.
Goldberg JB, Ganesan S, Comstock AT, Zhao Y, Sajjan US: Cable pili and the associated 22 kDa adhesin contribute to Burkholderia cenocepacia persistence in vivo. PLoS One. 2011, 6: e22435-10.1371/journal.pone.0022435.
Ikeda JS, Schmitt CK, Darnell SC, Watson PR, Bispham J, Wallis TS, Weinstein DL, Metcalf ES, Adams P, O’Connor DC, O’Brien AD: Flagellar phase variation of Salmonella enterica serovar Typhimurium contributes to virulence in the murine typhoid infection model but does not influence Salmonella-induced enteropathogenesis. Infect Immun. 2001, 69: 3021-3030. 10.1128/IAI.69.5.3021-3030.2001.
Liu B, Hu B, Zhou Z, Guo D, Guo X, Ding P, Feng L, Wang L: A novel non-homologous recombination-mediated mechanism for Escherichia coli unilateral flagellar phase variation. Nucleic Acids Res. 2012, 40: 4530-4538. 10.1093/nar/gks040.
Rigano LA, Payette C, Brouillard G, Marano M-R, Abramowicz L, Torres PS, Yun M, Castagnaro AP, El Oirdi M, Dufour V, Malamud F, Dow M, Bouarab K, Vojnov AA: Bacterial cyclic β-(1,2)-glucan acts in systemic suppression of plant immune responses. Plant Cell. 2007, 19: 2077-2089. 10.1105/tpc.106.047944.
Blomqvist K, Nikkola M, Lehtovaara P, Suihko M-L, Airaksinen U, Stråby KB, Knowles JKC, Penttilä ME: Characterization of the genes of the 2,3-butanediol operons from Klebsiella terrigena and Enterobacter aerogenes. J Bacteriol. 1993, 175: 1392-1404.
Ryu C-M, Farag MA, Hu C-H, Reddy MS, Kloepper JW, Paré PW: Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol. 2004, 134: 1017-1026. 10.1104/pp.103.026583.
Maier RJ, Olczak A, Maier S, Soni S, Gunn J: Respiratory hydrogen use by Salmonella enterica serovar Typhimurium is essential for virulence. Infect Immun. 2004, 72: 6294-6299. 10.1128/IAI.72.11.6294-6299.2004.
Ahmad M, Majerczak DR, Pike S, Hoyos ME, Novacky A, Coplin DL: Biological activity of harpin produced by Pantoea stewartii subsp. stewartii. Mol Plant Microbe Interact. 2001, 14: 1223-1234. 10.1094/MPMI.2001.14.10.1223.
Schwarz S, Hood RD, Mougous JD: What is type VI secretion doing in all those bugs?. Trends Microbiol. 2010, 18: 531-537. 10.1016/j.tim.2010.09.001.
Riley MA, Pinou T, Wertz JE, Tan Y, Valletta CM: Molecular characterization of the klebicin B plasmid of Klebsiella pneumoniae. Plasmid. 2001, 45: 209-221. 10.1006/plas.2001.1519.
Wertz JE, Riley MA: Chimeric nature of two plasmids of Hafnia alvei encoding the bacteriocins Alveicins A and B. J Bacteriol. 2004, 186: 1598-1605. 10.1128/JB.186.6.1598-1605.2004.
Kleinkauf H, Von Döhren H: A nonribosomal system of peptide biosynthesis. Eur J Biochem. 1996, 236: 335-351. 10.1111/j.1432-1033.1996.00335.x.
Choi O, Lim JY, Seo Y-S, Hwang I, Kim J: Complete genome sequence of the rice pathogen Pantoea ananatis PA13. J Bacteriol. 2012, 194: 531-10.1128/JB.06450-11.
De Maayer P, Chan WY, Venter SN, Toth IK, Birch PRJ, Joubert F, Coutinho TA: Genome sequence of Pantoea ananatis LMG20103, the causative agent of Eucalyptus blight and dieback. J Bacteriol. 2010, 192: 2936-2937. 10.1128/JB.00060-10.
De Maayer P, Chan WY, Rezzonico F, Bühlmann A, Venter SN, Blom J, Goesmann A, Frey JE, Smits THM, Duffy B, Coutinho TA: Complete genome sequence of clinical isolate Pantoea ananatis LMG5342. J Bacteriol. 2012, 194: 1615-1616. 10.1128/JB.06715-11.
Hara Y, Kadotani N, Izui H, Katashkina JI, Kuvaeva T, Andreeva I, Golubeva LI, Malko DB, Makeev VJ, Mashko SV, Kozlov YI: The complete genome sequence of Pantoea ananatis AJ13355, an organism with great biotechnological potential. Appl Microbiol Biotechnol. 2011, 93: 331-341.
Kim HJ, Lee JH, Kang BR, Rong X, McSpadden Gardener BB, Ji HJ, Park C-S, Kim YC: Draft genome sequence of Pantoea ananatis B1-9, a nonpathogenic plant growth-promoting bacterium. J Bacteriol. 2012, 194: 729-10.1128/JB.06484-11.
Matsuzawa T, Mori K, Kadowaki T, Shimada M, Tashiro K, Kuhara S, Inagawa H, Soma G-I, Takegawa K: Genome sequence of Pantoea agglomerans strain IG1. J Bacteriol. 2012, 194: 1258-1259. 10.1128/JB.06674-11.
Medrano EG, Bell AA: Genome sequence of Pantoea sp. strain Sc1, an opportunistic cotton pathogen. J Bacteriol. 2012, 194: 3019-10.1128/JB.00450-12.
Smits THM, Rezzonico F, Kamber T, Goesmann A, Ishimaru CA, Stockwell VO, Frey JE, Duffy B: Genome sequence of the biocontrol agent Pantoea vagans C9-1. J Bacteriol. 2010, 192: 6486-6487. 10.1128/JB.01122-10.
Darling AE, Mau B, Perna NT: progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010, 5: e11147-10.1371/journal.pone.0011147.
Hall TA: BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999, 41: 95-98.
National Centre for Biotechnology Information Protein Database.http://www.ncbi.nlm.nih.gov/protein,
Softberry FgenesB bacterial operon and gene prediction server.http://linux1.softberry.com,
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ: Basic local alignment search tool. J Mol Biol. 1990, 215: 403-410.
Meyer F, Goesmann A, McHardy AC, Bartels D, Bekel T, Clausen J, Kalinowski J, Linke B, Rupp O, Giegerich R, Pühler A: GenDB – an open source genome annotation system for prokaryote genomes. Nucleic Acids Res. 2003, 31: 2187-2195. 10.1093/nar/gkg312.
Saier MH, Tran CV, Barabote RD: TCDB: the transporter classification database for membrane transport analyses and information. Nucleic Acid Res. 2006, 34: 181-186. 10.1093/nar/gkj001.
Pritchard L, White JA, Birch PR, Toth IK: GenomeDiagram: a python package for the visualization of large-scale genomic data. Bioinformatics. 2006, 22: 616-617. 10.1093/bioinformatics/btk021.
Moreno-Hagelsieb G, Latimer K: Choosing BLAST options for better detection of orthologs as reciprocal best hits. Bioinformatics. 2008, 24: 319-324. 10.1093/bioinformatics/btm585.
Kumar S, Dudley J, Nei M, Tamura K: MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008, 9: 299-306. 10.1093/bib/bbn017.
Acknowledgements
The authors acknowledge Michael Poulsen (University of Copenhagen, Denmark) for the use of the P. agglomerans MP2 and P. vagans MP7 genomes, Jürg E. Frey, Fabio Rezzonico and Markus Oggenfuss (ACW Wädenswil, Switzerland) for genome sequencing, as well as Don Cowan (Center for Microbial Ecology and Genomics, University of Pretoria, South Africa) for reviewing the manuscript. This study was partially supported by the National Research Foundation (NRF), the Tree Protection Co-operative Programme (TPCP), the NRF/Dept. of Science and Technology Centre of Excellence in Tree Health Biotechnology (CTHB), and the THRIP support program of the Department of Trade and Industry, South Africa, the Swiss Federal Office for Agriculture (BLW Fire Blight Research – Achilles), and the Swiss Secretariat for Education and Research (SBF C07.0038). It was conducted within the Swiss ProfiCrops program.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contribution
PDM, SNV, BD, THMS and TAC conceived the study. PDM, WYC, JB and THMS performed experiments and analysis, PDM, SNV, BD, THMS and TAC wrote the original manuscript. All authors contributed to the final version.
Electronic supplementary material
12864_2012_4393_MOESM1_ESM.xlsx
Additional file 1: Table S1. Phenotypes encoded on the LPP-1 genomes of Group I-III Pantoea spp. Table showing the phenotypes, as well as the genetic loci for these phenotypes, encoded on the LPP-1 genomes of Group I, II and III Pantoea strains. The presence/absence of a particularly genetic locus and phenotype in each Group is indicated by a + or -, respectively, and the strain number designations follow those indicated in Table 1. The phenotypes are further subdivided into groups according to their function, including those factors which are conserved (core) among all twenty Pantoea spp., and those potentially involved in transport and catabolism of metabolic substrates, inorganic ion assimilation, resistance to antimicrobials and heavy metal, host colonization and persistence and putative pathogenesis and antibiosis factors. (XLSX 12 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
De Maayer, P., Chan, WY., Blom, J. et al. The large universal Pantoea plasmid LPP-1 plays a major role in biological and ecological diversification. BMC Genomics 13, 625 (2012). https://doi.org/10.1186/1471-2164-13-625
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2164-13-625