Abstract
Genomics has clarified the biosynthetic pathway for desferrioxamine E critical for iron acquisition in the enterobacterial fire blight pathogen Erwinia amylovora. Evidence for each of the individual steps and the role of desferrioxamine E biosynthesis in pathogen virulence and cell protection from host defenses is presented. Using comparative genomics, it can be concluded that desferrioxamine biosynthesis is ancestral within the genera Erwinia and Pantoea.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Iron is an essential nutritional factor for every living organism as a cofactor for numerous proteins. Even though iron is one of the most abundant micronutrients in nature, iron bioavailability under physiological conditions is fairly low. Under aerobic conditions, at neutral and alkaline pH, the most prevalent form is ferric iron (FeIII), which is commonly precipitated to form iron minerals. In order to solubilize and acquire iron from their environment, living organisms utilize high-affinity uptake systems. Microorganisms excrete siderophores, usually low molecular weight molecules in the range of 400–1,000 Da, to increase the solubility of iron. Once an excreted siderophore has captured ferric iron, the resulting iron–siderophore complex is reabsorbed by the cell via highly selective membrane-associated ATP-dependent transport systems (Köster 1991).
Desferrioxamines (DFOs) consisting of alternating diamine and dicarboxylic acid building blocks linked by amide bonds are among the strongest hydroxamate siderophores known. Desferrioxamine E (DFO-E, also nocardamine) was first isolated from a Streptomyces sp. (Yang and Leong 1982) and subsequently found in many other bacteria, including Pantoea agglomerans (formerly Erwinia herbicola), Pseudomonas stutzeri and Hafnia alvei (Meyer and Abdallah 1980; Berner et al. 1988; Reissbrodt et al. 1990).
Biosynthesis of DFO by Erwinia amylovora
It has been reported that enterobacterial plant pathogenic E. amylovora strains produce DFOs (Feistner et al. 1993; Kachadourian et al. 1996). This pathogen is the major global threat to sustainable pome fruit production (i.e., apple, pear and quince), and it also causes economic, amenity and ecological damage to a wide range of ornamental, landscape and forest Rosaceae species (Bonn and van der Zwet 2000; Duffy et al. 2005; Rezzonico et al. 2011). The major product of DFO biosynthesis in E. amylovora is DFO-E, while minor amounts of other DFOs (D2, X1–7 and G1) are also produced (Feistner et al. 1993; Kachadourian et al. 1996). Using transposon mutagenesis, a DFO-negative mutant of E. amylovora CFBP 1430 was obtained (Kachadourian et al. 1996), in which the transposon was inserted in the dfoA gene (Dellagi et al. 1998). Additionally, a DFO uptake mutant with a transposon insertion in the TonB-dependent receptor gene foxR was also obtained. This mutant was able to synthesize DFO, but was deficient in its ability to use DFO as siderophore. Based on this, it was severely impaired in ability to grow under iron-limited conditions (Kachadourian et al. 1996; Dellagi et al. 1998). The foxR gene was subsequently cloned and sequenced (Kachadourian et al. 1996).
Using this foxR sequence, we searched the complete, annotated genome sequence of E. amylovora CFBP 1430 (Smits et al. 2010b) in order to localize the gene on the chromosome. As it is known that single cosmids complementing all dfo mutants in E. amylovora CFBP 1430 can be obtained, it was assumed that DFO biosynthetic genes are clustered on the E. amylovora genome (Dellagi et al. 1998). The flanking regions of the foxR gene were analyzed for the presence of potential DFO biosynthesis genes. Directly downstream of foxR and on the complementary strand (Fig. 1), three genes designated here as dfoJAC were identified and annotated as siderophore biosynthetic genes. They have significant sequence identity to the DFO biosynthesis genes from Streptomyces coelicolor A3(2) and to the alcaligin biosynthesis genes from Bordetella spp. The dfoA gene was identical to the previously unpublished dfoA sequence (Dellagi et al. 1998; Smits et al. 2010b).
The DFO biosynthetic gene clusters in closely related bacteria
The recent availability of genome sequences of related species and genera enables investigation into the dissemination of DFO biosynthesis genes within the enterobacteria. The DFO biosynthesis gene clusters in the genomes of all E. amylovora strains currently published (Sebaihia et al. 2010; Smits et al. 2010b; Powney et al. 2011; Smits et al. 2011a) have an identical arrangement (Fig. 1). Slightly lower sequence identities were observed between the Spiraeoideae isolates CFBP 1430 and Ea273 and the Rubus isolate ATCC BAA-2158. The same genetic arrangement was also found in the genomes of Erwinia pyrifoliae strains (Kube et al. 2010; Smits et al. 2010a), Erwinia sp. Ejp617 (Park et al. 2011) and Erwinia tasmaniensis (Kube et al. 2008; Fig. 1), species not previously reported to produce DFOs (Smits et al. 2010a).
However, the gene organization of the dfo gene cluster in the genome of Erwinia billingiae Eb661 (Kube et al. 2010; Fig. 1) is different in that a major facilitator superfamily (MFS) transporter, here designated DfoS, is present directly downstream of dfoC in the same transcriptional direction. An MFS transporter is also seen in the genomes of different Pantoea spp. (De Maayer et al. 2010; Smits et al. 2010c, 2011b) at the similar position (Fig. 1). Orthologues of this MFS transporter can also be identified in the genomes of E. amylovora CFBP 1430, E. pyrifoliae strains (Kube et al. 2010; Smits et al. 2010a), Erwinia sp. Ejp617 (Park et al. 2011) and E. tasmaniensis (Kube et al. 2008; Fig. 1), but located distantly from the dfoJAC cluster. It can be speculated that DfoS is responsible for cell export of DFO-E, analogous to the alcaligin transporter AlcS in Bordetella bronchiseptica (Brickman and Armstrong 2005; Fig. 1).
The closely related fire blight biocontrol agent Pantoea vagans C9-1 (Rezzonico et al. 2010; Smits et al. 2010c) produces DFOs (Feistner and Ishimaru 1996). In this antagonistic bacterium, the dfoJACS gene cluster is located on the megaplasmid pPag3, which can be lost under stressful, unfavorable growth conditions (Smits et al. 2010d). This gene cluster includes downstream of dfoC the dfoS gene, but not the gene encoding the TonB-dependent receptor for ferrioxamine. Examination of the genome for the foxA gene encoding the ferrioxamine TonB-dependent receptor of P. agglomerans K4 (Deiss et al. 1998) indicated that it is encoded on the chromosome (Smits et al. 2010c). The DFO biosynthesis clusters in the draft genomes of P. agglomerans E325 (Smits and Duffy, unpublished) and Pantoea eucalypti αB (Adams et al. 2009) have the same arrangement as in P. vagans C9-1. The chromosomally encoded DFO biosynthesis cluster of the Eucalyptus pathogen Pantoea ananatis LMG 20103 (De Maayer et al. 2010) also contains the foxA gene directly downstream of dfoS (Fig. 1). The genome of Pantoea sp. At-9b (Pinto-Tomás et al. 2009) lacks the DFO biosynthesis cluster. None of the former Erwinia spp. now included in the genera Pectobacterium, Brenneria and Dickeya or any other Enterobacteriaceae contain a DFO biosynthesis cluster although many of these produce alternative iron siderophores.
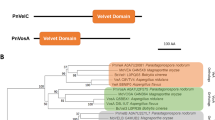
Phylogenetic analysis of concatenated DfoJAC (Fig. 2) revealed that the sequences from pathogenic Erwinia spp. are very closely related, whereas DfoJAC of E. billingiae Eb661 branched deeply within the tree. Additionally, the DfoJAC of three Pantoea spp., all previously incorrectly assigned to P. agglomerans (Ewing and Fife 1972; Gavini et al. 1989; Brady et al. 2009; Rezzonico et al. 2009), had a higher sequence identity to each other than compared with that of P. ananatis LMG 20103, which also deeply branched. However, pairwise distances are above 75% for all genes, indicating a common origin for the DFO biosynthesis cluster in Erwinia and Pantoea.
The putative pathway for DFO biosynthesis
Using cross-feeding experiments, Feistner (Feistner 1995) had proposed a pathway for DFO biosynthesis in E. amylovora (Fig. 3) that is essentially identical to the pathway as determined for S. coelicolor A3(2) (Challis 2005), for which some biochemical evidence is present (Schupp et al. 1987, 1988; Barona-Gómez et al. 2004). The individual steps and the enzymes involved are outlined below.
The putative first enzyme in the biosynthetic pathway for DFO is a pyridoxal-5’phosphate-dependent lysine/ornithine decarboxylase DfoJ (EC 4.1.1.-) (Expert et al. 2000). This enzyme has moderate sequence identity to DesA, the respective protein in the DFO biosynthesis pathway of S. coelicolor A3(2) (Schupp et al. 1988; Bentley et al. 2002), and therefore the putative function might be that of the lysine decarboxylase, yielding cadaverine (Fig. 3). Based on the incorporation of small amounts of putrescine into the DFO profile of E. amylovora (Feistner et al. 1993), it is very well possible that this enzyme can also decarboxylate ornithine to putrescine.
In Bergey’s manual of Systematic Microbiology, however, it is described that lysine/ornithine decarboxylase was not detected using Møller’s method in Erwinia spp. (Hauben and Swings 2005). As Møller’s assay is performed in a rich medium containing sufficient iron (Møller 1955), induction of the iron regulon will not have taken place, so that the result of this assay has to be taken with great care. The enzyme assays were never performed under low-iron conditions.
The second step of the proposed DFO biosynthetic pathway is an oxygenation on one of the terminal amino groups of cadaverine and, to minor extend, of putrescine to yield 1-amino-5-(N-hydroxy)-aminopentane and 1-amino-4-(N-hydroxy)-aminobutane, respectively (Fig. 3). The amine monooxygenase DfoA belongs to the IucD family of enzymes and is a flavin-dependent protein. Orthologues are present in many organisms that are able or hypothesized to produce hydroxamate-type siderophores (Challis 2005).
Theoretically, there is an alternative biosynthetic pathway over initial N-monooxygenation and subsequent decarboxylation (Feistner 1995), comparable to the pathway for aerobactin biosynthesis (Challis 2005). This alternative pathway (Fig. 3) corresponds to the current annotation of DfoA as a l-lysine N6-monooxygenase (EC 1.14.13.59). However, based on cross-feeding experiments (Feistner 1995), there is currently only evidence for an initial decarboxylation (Expert et al. 2000), indication that this enzyme has a hypothetical function as a cadaverine N-monooxygenase.
The protein encoded by dfoC is predicted to contain two domains: an acyl transferase domain of the AlcB family (PFAM 10331) within the first 150 amino acids and a siderophore synthase domain belonging to the IucA/IucC family (PFAM 04183). The latter domain belongs to the type C siderophore synthases (Challis 2005). The gene cluster encoding DFO biosynthesis in S. coelicolor A3(2) has two genes encoding the same functions as DfoC (Fig. 1): desC encodes the acyl transferase, while desD encodes the siderophore synthase (Barona-Gómez et al. 2004). DfoC is thus a natural fusion protein.
The first reaction performed by DfoC (Fig. 3) involves the coupling of succinyl-CoA to 1-amino-5-(N-hydroxy)-aminopentane to form the intermediate N-5-aminopentyl-N-(hydroxyl)-succinamic acid. This intermediate, described as being chemically instable (Barona-Gómez et al. 2004), is most probably not released into the medium, but is directly, in an ATP-dependent reaction, trimerized and cyclisized by DfoC to form DFO-E. The incorporation of one molecule of 1-amino-4-(N-hydroxy)-aminobutane will yield DFO-D2 that was detected as second major DFO in E. amylovora (Feistner et al. 1993). Based on the minor DFOs that were detected in cultures of E. amylovora (Feistner et al. 1993), we conclude that DfoC can accommodate a variety of precursors and ring sizes that are not known to be tolerated by Streptomyces spp.
The TonB-dependent ferrioxamine receptor FoxR
For the reabsorption of iron(III)-loaded siderophores, a TonB-dependent receptor is commonly employed, transporting the siderophores into the periplasmic space. For ferrioxamine, the respective receptors were named FoxR in E. amylovora (Kachadourian et al. 1996; Dellagi et al. 1998), FoxA in P. agglomerans (Deiss et al. 1998) and FauA in Bordetella. The gene encoding the FoxR receptor was cloned and characterized (Kachadourian et al. 1996). FoxR belongs to the TonB-dependent ferrihydroxamate receptors [TC 1.B.14.1.1] and may accept a broad spectrum of ferrioxamines, but not other siderophores (Deiss et al. 1998). In the E. amylovora CFBP 1430 genome (Smits et al. 2010b), the foxR gene is located on the complementary strand directly downstream of dfoC (Fig. 1).
In E. amylovora CFBP 1430, foxR mutants retain their ability to synthesize DFO but lose the ability to utilize this as an iron substrate, are impaired in growth under iron-limited conditions such as found on flower surfaces, have reduced virulence and are less resistant to plant defense responses (Kachadourian et al. 1996; Expert et al. 2000).
The genome of E. amylovora CFBP 1430 contains genes for three additional TonB-dependent receptors. One of these encodes a copper uptake receptor, while the other two encode for receptors with unknown specificity (Smits et al. 2010b). In the genome, remnants of an inactivated ferrichrome TonB-dependent receptor fhuA were identified, but it still contains its cognate ABC transporter fhuCDB. In contrast, the two closely related species E. pyrifoliae DSM 12163 and E. tasmaniensis Et1/99 contain a full fhuACDB system, indicating that they should be able to utilize this siderophore. Furthermore, the genome of E. tasmaniensis Et1/99 encodes a full iron(III) dicitrate uptake system, located downstream of the DFO gene cluster and an achromobactin-like siderophore uptake system (Kube et al. 2008; Smits et al. 2010b). The base for the limited utilization of siderophores by E. amylovora (Kachadourian et al. 1996) is thus determined by the lack of the corresponding systems.
Regulation
The DFO biosynthetic genes and the TonB-dependent receptor FoxR in E. amylovora CFBP 1430 are inducible under low-iron conditions (Kachadourian et al. 1996), indicating a global regulation by the ferric uptake receptor Fur (Hantke 1981; de Lorenzo et al. 1987), and the corresponding gene was identified in the genome (Smits et al. 2010b). A Fur box (de Lorenzo et al. 1987) was identified upstream of the E. amylovora CFBP 1430 foxR gene (Kachadourian et al. 1996), but could also be identified upstream of the dfoJ gene (Fig. 1), as in S. coelicolor A3(2) (Barona-Gómez et al. 2004). Based on the limited intergenic space between dfoJ and dfoA (10 bp) and between dfoA and dfoC (1 bp), it can be predicted that a single transcript spanning the dfoJAC region is generated.
Role of DFO in interaction with plants
The role of DFO biosynthesis in fire blight caused by E. amylovora has been well studied (Expert 1999; Expert et al. 2000). It has been shown that two outer membrane proteins with an apparent M w of 70 and 80 kDa are induced under low-iron conditions, one of which corresponds to FoxR (Dellagi et al. 1998). A foxR mutant of E. amylovora CFBP 1430 accumulated DFO molecules in its external medium, decreasing the iron availability (Dellagi et al. 1998) and inhibiting its own growth. A gene knockout of dfoA in E. amylovora CFBP 1430 completely abolished production of DFO-E in this strain (Dellagi et al. 1998). Additionally, it was noted that this enzyme not only had a critical function for iron acquisition, but also played a role in cell protection against the host oxidative burst elicited during infection (Dellagi et al. 1998). However, unlike non-pathogenic mutants inactivated in the hrp-type III secretion system or in the amylovoran biosynthetic cluster (Oh and Beer 2005), dfoA mutants still caused fire blight when inoculated at high cell titer (Dellagi et al. 1998) indicating a limited role in virulence. Both types of mutants were handicapped in ability to colonize floral tissues and cause necrotic symptoms on apple flowers, indicating that iron availability is limited in planta. Considering the limited iron uptake determinants available to E. amylovora (Smits et al. 2010b), DFO plays a primary role in iron scavenging and fitness of the organism in planta.
Conclusions
We have identified the biosynthetic genes for DFO in E. amylovora CFBP 1430 (Smits et al. 2010b) and provide genomic clarification for the individual gene products. High conservation of the DFO biosynthetic proteins observed in genomes of currently sequenced members of the related genera Erwinia and Pantoea indicates the ancestral nature of this feature. In contrast, the pectolytic ex. Erwinia group (e.g., Dickeya dadantii Ech703 and Pectobacterium atrosepticum SCRI1043) do not carry DFO biosynthetic genes.
The role of DFO in pathogenicity of E. amylovora is intimately related to the life cycle of the pathogen within its host. Evolution of DFOs fulfill a dual function for the pathogen as an efficient iron transporter to cope with iron-limited conditions within host tissues and also as a defense mechanism to protect against oxidative burst host reactions during early infection stages. Although not yet confirmed with mutational analysis, accumulating evidence anticipates a similar role for DFOs in closely related plant pathogenic and/or plant-associated Erwinia species.
References
Adams AS, Currie CR, Cardoza Y, Klepzig KD, Raffa KF (2009) Effects of symbiotic bacteria and tree chemistry on the growth and reproduction of bark beetle fungal symbionts. Can J For Res 39:1133–1147
Barona-Gómez F, Wong U, Giannakopulos AE, Derrick PJ, Challis GL (2004) Identification of a cluster of genes that directs desferrioxamine biosynthesis in Streptomyces coelicolor M145. J Am Chem Soc 126:16282–16283
Bentley SD, Chater KF, Cerdeño-Tárraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang C-H, Kieser T, Larke L, Murphy L, Oliver K, O’Neil S, Rabbinowitsch E, Rajandream M-A, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147
Berner I, Konetschny-Rapp S, Jung G, Winkelmann G (1988) Characterization of ferrioxamine E as the principle siderophore of Erwinia herbicola (Enterobacter agglomerans). Biol Met 1:51–56
Bonn WG, van der Zwet T (2000) Distribution and economic importance of fire blight. In: Vanneste JL (ed) Fire blight: the disease and its causative agent, Erwinia amylovora. CAB International, Wallingford, pp 37–53
Brady CL, Venter SN, Cleenwerck I, Engelbeen K, Vancanneyt M, Swings J, Coutinho TA (2009) Pantoea vagans sp. nov., Pantoea eucalypti sp. nov., Pantoea deleyi sp. nov. and Pantoea anthophila sp. nov. Int J Syst Evol Microbiol 59:2339–2345
Brickman TJ, Armstrong SK (2005) Bordetella AlcS transporter functions in alcaligin siderophore export and is central to inducer sensing in positive regulation of alcaligin system gene expression. J Bacteriol 187:3650–3661
Challis GL (2005) A widely distributed bacterial pathway for siderophore biosynthesis independent of nonribosomal peptide synthetases. ChemBioChem 6:601–611
de Lorenzo V, Wee S, Herrero M, Neilands JB (1987) Operator sequences of the aerobactin operon of plasmid ColV-K30: binding of the ferric uptake regulation (fur) repressor. J Bacteriol 169:2624–2630
De Maayer P, Chan WY, Venter SN, Toth IK, Birch PRJ, Joubert F, Coutinho TA (2010) The genome sequence of Pantoea ananatis LMG20103, the causative agent of Eucalyptus blight and dieback. J Bacteriol 192:2936–2937
Deiss K, Hantke K, Winkelmann G (1998) Molecular recognition of siderophores: a study with cloned ferrioxamine receptors (FoxA) from Erwinia herbicola and Yersinia enterocolitica. Biometals 11:131–137
Dellagi A, Brisset M-N, Paulin J-P, Expert D (1998) Dual role of desferrioxamine in Erwinia amylovora pathogenicity. Mol Plant-Microbe Interact 11:734–742
Duffy B, Schärer H-J, Bünter M, Klay A, Holliger E (2005) Regulatory measures against Erwinia amylovora in Switzerland. EPPO Bull 35:239–244
Ewing WH, Fife MA (1972) Enterobacter agglomerans (Beijerinck) comb. nov. (the Herbicola-Lathyri bacteria). Int J Syst Bacteriol 22:4–11
Expert D (1999) Withholding and exchanging iron: interactions between Erwinia spp. and their host plants. Annu Rev Phytopathol 37:307–334
Expert D, Dellagi A, Kachadourian R (2000) Iron and fire blight: role in pathogenicity of desferrioxamine E, the main siderophore of Erwinia amylovora. In: Vanneste JL (ed) Fire blight: the disease and its causative agent, Erwinia amylovora. CAB International, Wallingford, pp 179–195
Feistner GJ (1995) Proferrioxamine synthesis in Erwinia amylovora in response to precursor or hydroxylysine feeding: metabolic profiling with liquid chromatography-electrospray mass spectrometry. Biometals 8:318–327
Feistner GJ, Ishimaru C (1996) Proferrioxamine profiles of Erwinia herbicola and related bacteria. Biometals 9:337–344
Feistner GJ, Stahl DC, Gabrik AH (1993) Proferrioxamine siderophores of Erwinia amylovora. A capillary liquid chromatographic/electrospray tandem mass spectrometry study. Org Mass Spectrom 28:163–175
Gavini F, Mergaert J, Beji A, Mielcarek C, Izard D, Kersters K, De Ley J (1989) Transfer of Enterobacter agglomerans (Beijerinck 1888) Ewing and Fife 1972 to Pantoea gen. nov. as Pantoea agglomerans comb. nov. and description of Pantoea dispersa sp. nov. Int J Syst Bacteriol 39:337–345
Hantke K (1981) Regulation of ferric iron transport in Escherichia coli K12: isolation of a constitutive mutant. Mol Gen Genet 182:288–292
Hauben L, Swings J (2005) Genus XIII. Erwinia. In: Brenner DJ, Krieg NR, Staley JT, Garrity GM (eds) Bergey’s manual of systematic bacteriology. 2nd edn, Vol 2: The Proteobacteria, part B: the Gammaproteobacteria. Springer, New York, pp 670–679
Kachadourian R, Dellagi A, Laurent J, Bricard L, Kunesch G, Expert D (1996) Desferrioxamine-dependent iron transport in Erwinia amylovora CFBP 1430: cloning of the gene encoding the ferrioxamine receptor FoxR. Biometals 9:143–150
Köster W (1991) Iron(III) hydroxamate transport across the cytoplasmic membrane of Escherichia coli. Biol Met 4:23–32
Kube M, Migdoll AM, Müller I, Kuhl H, Beck A, Reinhardt R, Geider K (2008) The genome of Erwinia tasmaniensis strain Et1/99, a non-pathogenic bacterium in the genus Erwinia. Environ Microbiol 10:2211–2222
Kube M, Migdoll AM, Gehring I, Heitmann K, Mayer Y, Kuhl H, Knaust F, Geider K, Reinhardt R (2010) Genome comparison of the epiphytic bacteria Erwinia billingiae and E. tasmaniensis with the pear pathogen E. pyrifoliae. BMC Genomics 11:393
Meyer J-M, Abdallah M (1980) The siderochromes of non-fluorescent pseudomonads: production of nocardamine by Pseudomonas stutzeri. J Gen Microbiol 118:125–129
Møller V (1955) Simplified test for some amino acid decarboxylases and arginine dihydrolase system. Acta Pathol Microbiol Scand 36:158–172
Oh C-S, Beer SV (2005) Molecular genetics of Erwinia amylovora involved in the development of fire blight. FEMS Microbiol Lett 253:185–192
Park DH, Thapa SP, Choi B-S, Kim W-S, Hur JH, Cho JM, Lim J-S, Choi I-Y, Lim CK (2011) Complete genome sequence of Japanese Erwinia strain Ejp617, a bacterial shoot blight pathogen of pear. J Bacteriol 193:586–587
Pinto-Tomás AA, Anderson MA, Suen G, Stevenson DM, Chu FST, Cleland WW, Weimer PJ, Currie CR (2009) Symbiotic nitrogen fixation in the fungus gardens of leaf-cutter ants. Science 326:1120–1123
Powney R, Smits THM, Sawbridge T, Frey B, Blom J, Frey JE, Plummer KM, Beer SV, Luck J, Duffy B, Rodoni B (2011) Genome sequence of an Erwinia amylovora strain with restricted pathogenicity to Rubus plants. J Bacteriol 193:785–786
Reissbrodt R, Rabsch W, Chapeaurouge A, Jung G, Winkelmann G (1990) Isolation and identification of ferrioxamine G and E in Hafnia alvei. Biol Met 3:54–60
Rezzonico F, Smits THM, Montesinos E, Frey JE, Duffy B (2009) Genotypic comparison of Pantoea agglomerans plant and clinical strains. BMC Microbiol 9:204
Rezzonico F, Vogel G, Duffy B, Tonolla M (2010) Whole cell MALDI-TOF mass spectrometry application for rapid identification and clustering analysis of Pantoea species. Appl Environ Microbiol 76:4497–4509
Rezzonico F, Smits THM, Duffy B (2011) Diversity and functionality of CRISPR regions in fire blight pathogen Erwinia amylovora. Appl Environ Microbiol 77:3819–3829
Schupp T, Waldmeier U, Divers M (1987) Biosynthesis of desferrioxamine B in Streptomyces pilosus: evidence for the involvement of lysine decarboxylase. FEMS Microbiol Lett 42:135–139
Schupp T, Toupet C, Divers M (1988) Cloning and expression of two genes of Streptomyces pilosus involved in the biosynthesis of the siderophore desferrioxamine B. Gene 64:179–188
Sebaihia M, Bocsanczy AM, Biehl BS, Quail MA, Perna NT, Glasner JD, DeClerck GA, Cartinhour S, Schneider DJ, Bentley SD, Parkhill J, Beer SV (2010) Complete genome sequence of the plant pathogen Erwinia amylovora strain ATCC 49946. J Bacteriol 192:2020–2021
Smits THM, Jaenicke S, Rezzonico F, Kamber T, Goesmann A, Frey JE, Duffy B (2010a) Complete genome sequence of the fire blight pathogen Erwinia pyrifoliae DSM 12163T and comparative genomic insights into plant pathogenicity. BMC Genomics 11:2
Smits THM, Rezzonico F, Kamber T, Blom J, Goesmann A, Frey JE, Duffy B (2010b) Complete genome sequence of the fire blight pathogen Erwinia amylovora CFBP 1430 and comparison to other Erwinia spp. Mol Plant-Microbe Interact 23:384–393
Smits THM, Rezzonico F, Kamber T, Goesmann A, Ishimaru CA, Stockwell VO, Frey JE, Duffy B (2010c) The genome sequence of the biocontrol agent Pantoea vagans strain C9-1. J Bacteriol 192:6486–6487
Smits THM, Rezzonico F, Pelludat C, Goesmann A, Frey JE, Duffy B (2010d) Genomic and phenotypic characterization of a non-pigmented variant of Pantoea vagans biocontrol strain C9-1 lacking the 530 kb megaplasmid pPag3. FEMS Microbiol Lett 308:48–54
Smits THM, Rezzonico F, Duffy B (2011a) Evolutionary insights from Erwinia amylovora genomics. J Biotechnol. doi:10.1016/j.jbiotec.2010.1010.1075
Smits THM, Rezzonico F, Kamber T, Goesmann A, Ishimaru CA, Frey JE, Stockwell VO, Duffy B (2011b) Metabolic versatility and antibiotic biosynthesis are distinguishing genomic features of the commercial fire blight antagonist Pantoea vagans C9-1. PLoS One 6(7):e22247
Yang C, Leong J (1982) Production of deferriferrioxamines B and E from a ferroverdin-producing Streptomyces species. J Bacteriol 49:381–383
Acknowledgments
We thank Dr. Cosima Pelludat (ACW) for insightful discussion during preparation of this review. Funding was provided in part by the Swiss Secretariat for Education and Research (SBF C07.0038) and the Swiss Federal Office for Agriculture (BLW Fire Blight Research—Pathogen). This work was conducted within the Swiss ProfiCrops program and the European Science Foundation research network COST Action 864.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by John Helmann.
Rights and permissions
About this article
Cite this article
Smits, T.H.M., Duffy, B. Genomics of iron acquisition in the plant pathogen Erwinia amylovora: insights in the biosynthetic pathway of the siderophore desferrioxamine E. Arch Microbiol 193, 693–699 (2011). https://doi.org/10.1007/s00203-011-0739-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-011-0739-0