Abstract
Extensive utilization of the synthetic dyes in various industries is leading to water and soil contamination and ultimately impacting the humans. A research study was conducted for investigating the biodecolorization and biotransformation of Mordant Black 11 dye. For this purpose, potential of biofilm forming bacteria Klebsiella pneumoniae MB398 isolated from effluent outlets of Tops Food Industry, Hattar, Pakistan, was assessed to decolorize and transform Mordant Black 11 dye. Bacterial strain MB398 exhibited the capability of growing optimally at acidic pH (pH 6.0). Klebsiella pneumoniae MB398 efficiently decolorized Mordant Black 11 dye (64.55%) in aerobic environment at pH 6.0 and 37 °C over 24 h, which further increased to 75.35% over a period of 72 h of incubation. Strain MB398 also exhibited the capability of decolorizing Mordant Black 11 dye in the presence of cadmium (63.71%), chromium (61.78%), and copper (61.50%), respectively. UV-VIS spectrophotometric analysis, FTIR, and HPLC spectra were also indicative of biotransformation of dye molecules by Klebsiella pneumoniae MB398. GC-MS analysis of Mordant Black 11 dye revealed formation of 9 novel and unique metabolites including phenol,2,4-bis(1,1-dimethylethyl); 9-eicosene, (E); ethanol,2,2-(2-propenyloxy); acetic acid, benzene; 1-naphthol; methyl formate; valeraldehyde,2,4-dimethyl; and 7-hexadecene (Z). A possible metabolic pathway depicting the biotransformation of Mordant Black 11 dye by Klebsiella pneumoniae MB398 was projected. Findings of the current research study strongly suggest application of Klebsiella pneumoniae MB398 for developing large scale bioremediation strategies for the abatement of synthetic dyes to retain environmental sustainability in bioeconomic way.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Mordant Black 11 dye (3-hydroxy-4-(1-hydroxy-2-naphthylazo)-7-nitro-naphthalene sulfonic acid sodium salt) is one of the large azo dye. Mordant Black 11 is a mono-azo dye that is comprised of naphthol and sulfonate moieties. Mordant Black 11 dye is extensively applied in dyeing of wool, silk, multifibers, and nylon after processing with salts of chromium. Mordant Black 11 due to complex molecular structure and persistent nature is highly stable against chemicals and is resistant to fading via exposure to sunlight and water. Based on these characteristics the dye is applied in huge amount on commercial scales for dyeing in various industrial processes [1, 2]. The effluents generated form these dyeing processing units are enriched with residual and/or unbound Mordant Black 11 dye that may cause environmental damages to the living organisms and aquatic life [3]. On the other hand, its degradation products (e.g., Naphthoquinone) are more toxic and carcinogenic as both in solution are known to be absorbed in harmful amounts via intact skin and cause irritation. Its persistence in drinking water reservoirs might be lethal to humans and exerts severe environmental distresses; therefore, its segregation and removal from wastewater are essential prior to discharge into water bodies [4, 5, 7]. Most of the studies reported dye removal processes with reference to physical and chemical approaches [1, 6, 8, 9]. But there are installation cost and secondary pollutants generation constraints associated with these procedures. Therefore, application of biological approaches was found to be more effective, economical and environmentally sustainable alternatives for the detoxification and removal of dyes from industrial effluents.
Biological remediation is an approach where different types of microorganisms are applied for detoxification and transformation of chemically complex compounds into simpler harmless molecules [10, 11]. Literature survey has revealed that only few research studies have raised the removal of Mordant Black 11 [10, 12, 13] but none indicated its transformation/intermediate products. Present research study is focused on demonstrating the potential of acidophilic Klebsiella pneumoniae MB398 to decolorize/degrade Mordant Black 11 dye in the aqueous medium, so that the bacterial strain could possibly be utilized in the decolorization and removal of Mordant Black 11 dye from contaminated environmental compartments and industrial effluents. Moreover, in current research, HPLC and GC-MS tools have been applied for identifying the possible intermediates and/or degradation products of Mordant Black 11 dye formed after bacterial treatment.
Methodology
Bacterial strain
Biofilm forming bacterial strain identified as Klebsiella pneumoniae MB398 (KP886827 accession number, GenBank) is used in the current research study. The bacterial strain was isolated from effluent outlets of Tops Food Industry, Hattar, using spread plate method. The strain was purified via streak plate method and screened for decolorization potential for various dyes (data submitted for publication). The bacterial strain exhibited highest decolorization potential for Mordant Black 11 dye up to 1000 mg L−1. For identification, bacterial DNA extraction and PCR amplification protocol described by Kilany [14] was followed with some modifications, and submitted to Macrogen for 16S rRNA sequencing. The bacterial strain was identified by comparing 16S rRNA genes with those of other bacterial strains available in NCBI database via BLAST tool and MEGA6 software for phylogenetic analysis.
Bacterial inoculum preparation
Klebsiella pneumoniae MB398 was grown in M9 broth (composition provided in supplementary data) supplemented with Mordant Black 11 dye (100 mg L−1) for 24 h at 37 °C for achieving 106–108 CFUs mL−1. Two percent of this bacterial inoculum was utilized in all the decolorization and biotransformation experiments.
Quantification of Mordant Black 11 dye decolorization efficiency of Klebsiella pneumoniae MB398
For investigating the decolorization potential of Klebsiella pneumoniae MB398, experiments were carried out in Erlenmeyer flaks (100 mL) containing 50 mL of M9 broth (3.0 g of KH2PO4, 6.0 g of Na2HPO4.2H2O, 1.0 g of NH4Cl, 0.5 g of NaCl, 1 mL of 0.1M CaCl2, 10 mL of 20% C6H12O6 [glucose], 1 mL of 1M MgSO4.7H2O, 5.0 g L−1 of casein hydrolysate, pH 6.0±0.2) augmented with Mordant Black 11 dye (100 mg L−1). Experimental flasks after inoculation with bacterial inoculum (2%) were incubated for 3 consecutive days at 37 °C under shaking conditions (150 rev/min). Treated samples (10 mL) were aseptically withdrawn intermittently (i.e., after 24, 48 & 72 h) followed by quantitative analysis for decolorization efficiency of Klebsiella pneumoniae MB398 using UV-VIS spectrophotometer. Absorbance maximum (λmax) for dye was recorded at 610 nm. Supernatant (free of bacterial biomass) after centrifugation (3500 rev/min, 20 mins) was utilized for determining the decolorization proficiency of MB398. The decolorization of Mordant Black 11 dye (%) was estimated by observing decline in λmax against blank (medium deprived of dye and inoculum). The medium supplemented with dye but devoid of bacterial inoculum was used as control. Color removal efficiency (%) was estimated using formula presented in Eq. 1. Experiments were performed in triplicates for authentication of results, which are represented by mean ± standard error.
where MBi = absorbance value of Mordant Black 11 dye prior to bacterial decolorization and MBf = absorbance value of Mordant Black 11 dye after bacterial decolorization at regular intervals.
Analysis of influence of various environmental parameters affecting the decolorization of Mordant Black 11 dye by Klebsiella pneumoniae MB398
Influence of various environmental parameters on decolorization of Mordant Black 11 dye by acidophilic Klebsiella pneumoniae MB398 was also assessed using three different temperatures (30, 37 and 45 °C) and variable pHs (5, 6, 7, 8 and 9). M9 minimal broth supplemented with 100 mg L−1 dye was inoculated with bacterial culture and incubated for 24 h and analyzed spectrophotometrically. Effects of initial dye concentration (50, 75, 100 and 150 mg L−1) and increasing inoculum volumes (0.5, 1, 1.5, and 2%) on the dye decolorization efficacy of the bacterium were also determined. Dye decolorization potential of MB398 in the presence of different metals (chromium, cadmium and copper) at varying concentrations (50, 75, 100 and 150 mg L−1) containing fixed concentration of Mordant Black 11 dye (100 mg L−1) was also investigated and analyzed using UV-VIS spectrophotometer. All the experiments were conducted in triplicates and the results obtained from each set are representing mean ± standard error.
Analysis of metabolites formed after biodecolorization of Mordant Black 11 dye by Klebsiella pneumoniae MB398
Biotransformation of dye molecules analyzed through absorption spectroscopy
Samples treated with Klebsiella pneumoniae MB398 from the flasks were withdrawn after 24, 48, and 72 h following centrifugation (3500 rev/min, 20 min). Supernatants devoid of bacterial biomass were examined using UV-VIS absorption spectroscopy (ranging between 400 and 800 nm) against M9 medium as blank. The absorption spectra of the decolorized medium (after bacterial treatment) were compared with that of the control dye (without any bacterial treatment) for analyzing the apparent changes.
Fourier transform spectroscopy for analyzing the dye metabolites and bacterial biomass after decolorization
The degradation products of Mordant Black 11 dye were extracted using equal volume of ethyl acetate following centrifugation under abovementioned operating conditions. Dried extracts (via evaporation over anhydrous sodium sulfate in vacuum conditions) and control (Mordant Black 11 dye) were mixed and ground separately with KBr (spectroscopic grade, in ratio of 5:95). Finely powdered samples were pressed using hydraulic presser to get transparent pellets. These pellets were analyzed for obtaining the IR spectra using FTIR8400 spectrophotometer (Shimadzu) in region between 4000 and 400 cm−1 at scanning speed of 15. The obtained spectra of bacterial treated samples were compared with the spectrum of control (Mordant Black 11 dye) for tracing the changes occurred in peak patterns.
Klebsiella pneumoniae MB398 cultures grown in M9 broth augmented with Mordant Black 11 dye (100 mg L−1) after centrifugation settled (as biomass) at the bottom of falcon tubes were dried (in the desiccator). These dried samples (after bacterial treatment) and dye as control (without any bacterial treatment) were homogenized into a fine powder with KBr (5:95) for obtaining pellets [10]. Pellets were then analyzed using the instruments and conditions mentioned above and resulting spectra were compared with that of control.
High-performance liquid chromatography for analyzing the dye metabolites after decolorization
The decolorization of Mordant Black 11 dye by Klebsiella pneumoniae MB398 was analyzed through high-performance liquid chromatography (Shimadzu JapanLC-20AT) coupled with C18 column system and UV-VIS detector (SPD20A). Medium after incubation (24 h) was centrifuged for removing the bacterial biomass and the supernatant was treated with ethyl acetate (1:1) for extracting the degradation products. Microfiltered extracts were then evaporated in vacuum rotary evaporator. After evaporation, the obtained crystals were dissolved in HPLC grade methanol (1 mL) and analyzed using reverse phase HPLC (high-performance liquid chromatography) with operating conditions mentioned in Table 1. The retention time for Mordant Black 11 dye obtained via HPLC processed chromatograms was 5.045 min. Concentration of dye in control and residual dye in the bacterial treated samples through HPLC was calculated by comparing peak areas of chromatograms of the standard dye and the decolorized dye samples using formulae in Eqs. 2 and 3.
where CD0h = dye concentration (mg L−1) in control at 0 h, PAC = peak area of the control dye, PAS = peak area of the standard dye, CS = concentration of the standard dye.
where CD24h = residual dye concentration (mg L−1) in sample after 24 h, PAD = peak area of residual dye in bacterial treated sample after 24 h, PAS = peak area of the standard dye, CS = concentration of the standard dye.
The calculated dye concentrations (of control and bacterial treated samples) were applied for measuring the percentage biodegradation of Mordant Black 11 dye using Eq. 4.
where MB (%) = percentage biodegradation of Mordant Black 11, MBC = dye concentration in control, MBS = residual dye concentration in sample.
GC-MS for analyzing the dye metabolites/products after decolorization
GC-MSQP5050A gas chromatography integrated with the mass spectrometer was utilized for detection of degradation metabolites and/or products of Mordant Black 11 dye under the operating conditions mentioned in Table 2. Samples via auto-sampler were injected for recording the respective chromatograms. Spectral library software NIST1.10 (Shimadzu) was applied for matching the mass spectra and retention times of the degradation metabolites and/or products.
Results and discussion
Bacteria belonging to Klebsiella genus are widely distributed in nature with different strains that are suitable for conducting the bioremediation studies owing to the presence of thick capsule, faster growth rates and resistance to different contaminants [15]. Numerous research studies have utilized and reported Klebsiella pneumoniae strains for the remediation of aromatic hydrocarbons and heavy metal removal [16,17,18,19,20,21]. Since indigenous polluted sites and the microorganisms dwelling over there, are potential sources of pollution degraders, as they are widely diverse, well adapted to survive in the contaminated environments, thus serve as the best players of degradation and remediation, considering these aspects we have utilized Klebsiella pneumoniae MB398 for the bioremediation purpose. Klebsiella pneumoniae MB398 was isolated from the biofilms grown over effluent outlets of Tops Food Industry, Hattar and displayed resistance to wider range of metals, antibiotics and dyes as well (data submitted for publication).
Decolorization of Mordant Black 11 dye
Acidophilic Klebsiella pneumoniae MB398 displayed an increase in the dye decolorization potential from 64 to 75% over a period of 24 to 72 h (Fig. 1). Maximum decolorization of Mordant Black 11 dye (75.35%) by the strain MB398 was noticed after 72 h of incubation under acidic environment (pH 6.0). Ali and associates [12] reported 60% decolorization of Mordant Black 11 dye over 24 h, under aerobic conditions using Sphingomonas sp. Likewise, Klebsiella sp. Y3 isolated from textile dyeing industry displayed 73% removal of Mordant Black 11 dye within 48 h of incubation period as documented by Cui and colleagues [13].
Analysis of influence of various environmental parameters affecting the decolorization of Mordant Black 11 dye by Klebsiella pneumoniae MB398
Influence of various environmental parameters for optimizing the Mordant Black 11 dye decolorization conditions by Klebsiella pneumoniae MB398 was also scrutinized. The rate of decolorization of Mordant Black 11 dye was directly proportional to the increase in inoculum volume (as evident in Fig. 2a). Klebsiella pneumoniae MB398 displayed the highest rate of dye decolorization (68.69%) with an inoculum volume of 2%. Ajaz and his colleagues [22] assessed the capability of Alcaligenes aquatilis using 2, 4, 6, 8 and 10% inoculum for decolorizing Synazol Red 6HBN and documented that the bacterium optimally decolorized 60–65% of dye at 6% inoculum volume. Another study reported 87% removal of Acid Red 337 in 10% volume of Bacillus megaterium utilizing different concentrations of bacterial inoculum volumes ranging between 2, 4, 6, 8, and 10% [23]. In a study, Lysinibacillus fusiformis displayed 95% of Methyl Red dye removal at 20% inoculum volume [24].
A reduction in decolorization of Mordant Black 11 dye by Klebsiella pneumoniae MB398 was noticed upon increasing the dye concentration from 50 to 150 mg L−1 (Fig. 2b). Klebsiella pneumoniae MB398 efficiently decolorized 72.60% of Mordant Black 11 dye at 50 mg L−1 concentration, while it declined to 49.65% with the increase in concentration of Mordant Black 11 dye to 150 mg L−1. The decline in dye decolorization rates by the bacteria at increased dye concentrations might be attributed to the presence of sulfonic groups, which act as detergents and may lead to bacterial growth inhibition [25]. Saratale with associates [26] and Shah [27] documented that the reduction in rates of dye decolorization at higher concentrations might be attributed to the saturation and/or occupation of the adsorption sites on to the bacterial cell surfaces, and toxicity of the dye exerted on the bacterium via inhibition of cellular metabolism. Intwala and Disha [11] reported 98% color reduction of Mordant Black 11 (at 50 mg L−1) by Bacillus lentus over a duration of 98 h. In a study Lysinibacillus fusiformis W1B6 displayed increased rates of Methyl Red decolorization ranging between 10 and 100 mg L−1, and the decolorization percentage declined significantly with the increase in concentration of dye to 1000 mg L−1 [24]. Similarly, another research study reported reduction in the decolorization of Acid Red 337 dye from 85 to 22% upon increase in the dye concentration (500–1000 mg L−1) by Bacillus megaterium [23].
Klebsiella pneumoniae MB398 exhibited maximum decolorization of Mordant Black 11 dye (70.58%) at pH 6.0 (acidic pH) followed by a significant reduction in the decolorization potential of the strain MB398 under alkaline conditions (Fig. 2c). Moraxella osloensis exhibited increase in Mordant Black 17 dye decolorization to 78% at acidic pH (pH 6.0) which reduced at alkaline pH [28]. Similar results were reported in another research study where Lysinibacillus fusiformis decolorized 90% of Methyl Red dye at pH range of 5.5, which declined sharply to 34% at pH 9.0 and 30% at pH 10.0 [24]. The results indicate pH exerts a dynamically vital impact on the growth of the microorganisms and their metabolism via regulating the transportation of nutrients and different substrates across the cellular membranes as documented by Patel and associates [29] regarding the degradation of phenanthrene by Pseudoxanthomonas sp. DMVP2.
Mordant Black 11 dye decolorization at different temperature ranges showed that Klebsiella pneumoniae MB398 could optimally decolorize 61.03% of the dye at 37°C, which declined to 39.82% upon increase in the temperature, i.e., 45 °C (Fig. 2d). Likewise, Intwala and Disha [11] reported highest decolorization of the dye by Bacillus lentus at 30 °C, which declined upon increase in the temperature to 40°C. Chang and coworkers [30] stated that the temperature required for maximum color removal has a direct relation with optimal growth temperature of the bacterial culture, and the reduction in color removal proficiency of the bacterium upon increase in temperature range might be attributed to denaturation of the enzymes involved in the dye degradation and decolorization, and loss in the cell viability.
Usually dyes are applied in combination with certain metal salts for instance chromium, cadmium etc., therefore, the influence of selected metals on Mordant Black 11 dye decolorization potential of Klebsiella pneumoniae MB398 was also studied. Klebsiella pneumoniae MB398 showed a reduction in the dye decolorization potential upon increasing the chromium concentration ranging from 50 to 150 mg L−1. Average decolorization rate ranged between 49.42 and 61.78% as shown in Fig. 3. Percentage decolorization of Mordant Black 11 dye declined with the addition of copper in the medium (Fig. 3). Highest dye decolorization rates for MB398 (62.43%) were observed with 75 mg L−1 of added copper. Almost a similar trend in discoloration of Mordant Black 11 dye was detected upon adding cadmium to the dye containing medium as the one recorded for chromium. Dye decolorization rates of more than 60% were noticed in combination of Mordant Black 11 dye with 50 mg L−1 cadmium (Fig. 3).
The assessment of various environmental parameters revealed that any deflection and fluctuation above and below the optimum condition in any factor might have significant impact on the dye decolorization proficiency of Klebsiella pneumoniae MB398.
Analysis of metabolites formed after biodecolorization of Mordant Black 11 dye by Klebsiella pneumoniae MB398
Biotransformation of dye molecules analyzed through absorption spectroscopy
The absorption spectra of Mordant Black 11 dye by acidophilic Klebsiella pneumoniae MB398 represented assorted patterns of the changes induced by the bacterium within the dye molecules compared to untreated dye (control). λmax for Mordant Black 11 dye was obtained at 610 nm with 2.216 OD (before treatment with the bacterial strain). Spectral changes disclosed a shifting in peak by MB398 towards 582–584 nm (0.946 OD) after 24 h, 566–570 nm (0.758 OD) after 48 h and 554–568 nm (0.690 OD) after 72 h of treatment (Fig. 4). Disappearance of existing peaks in addition to the formation of newer ones and reduction in absorbance maxima of Mordant Black 11 dye after treatment with Klebsiella pneumoniae MB398 were indicative of molecular rearrangements in the structure of Mordant Black 11 dye and its transformation of into different metabolites. Similar findings have been documented by Shah [27] regarding molecular rearrangements in the structure of azo dyes and their subsequent degradation. Asad and associates [31] stated that the decline in λmax of peaks in proportion to one another occurs due to the adsorption of dye, whereas, disappearance of the entire peak preexisting at λmax and appearance of newer ones is attributed to the biodegradation of dye.
Fourier transform spectroscopy (FTIR) for analyzing the dye metabolites and bacterial biomass after decolorization
Mordant Black 11 dye prior to the bacterial treatment (control) showed peaks representing sulfonic acid (1338.64 cm−1), C-H out-of-plane deformations for di- and tri-substituted aromatic compounds (between 883.43 and 738.76 cm−1), C-O stretching vibrations of H-bonded alcohols and phenols (between 1209.41 and 1051.24cm−1) and O-H in-plane deformations for secondary and primary alcohols (1404.22 cm−1). The peak showing NO2 stretching vibrations for nitro compounds were recorded at 1502.60 cm−1, for N=N stretching of azo bonds at 1566.25 cm−1, and N–H bond stretching vibrations of hydrogen bonded amines (3452.70 cm−1). Peaks for C=C stretching vibrations associated with aromatic skeleton (1620.26 cm−1) and C–H bond stretching of alkanes (2926.11 cm−1) were also observed. IR fingerprint regions of the bacterial biomass (after dye decolorization) and metabolites (extracted) depicted significant variations in the distribution of functional groups, chemical compounds and band intensities in contrast to the control dye (prior to any bacterial treatment). Additionally, formation of the newer peaks and complete disappearance of the existing ones represented substantial changes in Mordant Black 11 dye structure, which might be attributed to the alteration and/or degradation after bacterial processing (Fig. 5; Table I provided as supplementary data). These results have ascertained the biotransfomation of the dye into respective metabolites including aromatic compounds, alcohols, alkyl halides and alkynes as suggested earlier by Senthilkumar and colleagues [32] for Congo red and Reactive Red 195 dyes using Pseudomonas sp.; and reactive textile dyes using anaerobic rumen consortium [33].
High-performance liquid chromatography for analyzing the dye metabolites after decolorization
Rate of transformation and degradation of Mordant Black 11 dye into various metabolites was assessed via HPLCLC-20AT. Elution peak of Mordant Black 11 dye before any bacterial processing was obtained at 5.045 min. New peak (in bacterial treated samples) appeared at 4.556 min accompanied by different peaks with various retention times, which indicated the formation of different metabolites of Mordant Black 11 dye (Fig. 6). Results of HPLC analysis ascertained the biodegradation and/or biotransformation of Mordant Black 11 dye by Klebsiella pneumoniae MB398 as reported in multiple research studies for Malachite Green by Saccharomyces cerevisiae [34], Congo Red by Hafnia alvei and Enterobacter cloaca [35], Malachite Green by Exiguobacterium sp. [36], textile azo dyes using Pseudomonas sp. [32], azo dyes (Congo Red, Amido Black and Ponceau BS) by Sphingomonas sp. [12], and Congo Red by Dietzia sp. [37]. Furthermore, HPLC analysis revealed 97.06% biodegradation of dye molecules by Klebsiella pneumoniae MB398 with approximately 2.52 mg L−1 of residual dye.
HPLC analysis of Mordant Black 11 dye a before bacterial treatment (control) and b after treatment with Klebsiella pneumoniae MB398. In comparison with the peak for parent compound Mordant Black 11 (RT = 5.045 min), presence of different peaks with various retention times in bacterial treated sample is prominently indicating the formation of different metabolites
GC-MS analysis of metabolites produced by Klebsiella pneumoniae MB398 and proposed pathway for biodegradation of Mordant Black 11 dye
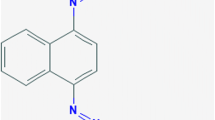
In samples treated with Klebsiella pneumoniae MB398 nine distinct peaks were detected during GC-MS spectroscopic analysis. Following intermediates/degradation compounds were identified using NIST1.10 library: phenol,2,4-bis(1,1-dimethylethyl), (b) 9-Eicosene, (E), (c) Ethanol,2,2-(2-propenyloxy), (d) acetic acid, (e) benzene, (f) 1-naphthol, (g) methyl formate, (h) valeraldehyde,2,4-dimethyl, and (i) 7-hexadecene (Z) as represented in Figure I and Table II (Supplementary Data) with their corresponding retention times (RT).
Consideration of the results of mineralized/degradation products of Mordant Black 11 dye during GC-MS analysis projected the formation of simpler metabolites and/or compounds having lower molecular weights thus validated the biotransformation of Mordant Black 11 dye by Klebsiella pneumoniae MB398. The chemical pathway for biotransformation of Mordant Black 11 dye (Fig. 7) by Klebsiella pneumoniae MB398 was proposed based on the retention time (RT), molecular weight (MW) and mass to charge ratios (m/z) of the products obtained through GC-MS spectroscopy and comparison with the NIST library. Klebsiella pneumoniae MB398 accomplished the cleavage of azo bond in Mordant Black 11 dye via catalytic reduction to form two intermediates (intermediate I and II). The degradation strategy followed deamination process to yield phenolsulfonic acid, nitrobenzene by intermediate I and ultimately phenol,2,4-bis(1,1-dimethylethyl) (RT: 10.3, MW: 206, m/z: 191), benzene (RT: 15.3, MW: 78.05, m/z: 78), acetic acid (RT: 8.3, MW:, m/z: 70), ethanol,2,2-(2-propenyloxy) (RT: 13.2, MW: 102, m/z: 58), and methyl formate (RT: 11.2, MW: 60, m/z: 31). While, intermediate II formed 1-naphthol (RT: 13.5, MW: 144, m/z: 144) followed by formation of 9-eicosene (E), (RT: 10.8, MW: 280, m/z: 55), valeraldehyde,2,4-dimethyl (RT: 7.0, MW: 114, m/z: 57), and (i) 7-hexadecene (Z), (RT: 17.2, MW: 224, m/z: 55) through deamination, desulfonation, denitrification, dehydroxylation, and ring fission processes.
Conclusion
This study highlighted the bioremediation potential of Klebsiella pneumoniae MB398 isolated from biofilm samples. The strain was acidophilic in nature and discerned the capability of optimally growing in pH 6.0. Klebsiella pneumoniae MB398 efficiently discolored and transformed more than 60–75% of Mordant Black 11 dye over a period of 72 h under the acidic conditions (pH 6.0). Quantitative analysis of optimization of various environmental parameters revealed the potential of Klebsiella pneumoniae MB398 to decolorize 74.58% of the dye at the initial dye concentration of 100 mg L−1 with inoculum size of 2% at 37 °C and pH 6.0 over a period of 24 h. The dye decolorization efficacy of the bacterial strain was also assessed against various metals added in the medium with maximum dye decolorization (63.71%) observed in the presence of cadmium (50 mg L−1). Klebsiella pneumoniae MB398 demonstrated the capability of growing and degrading the dye under variable environmental conditions, which may be attributed to its quick adaptive responses and genetic variations and characteristics, and resistance to the dye molecules. UV-VIS absorption spectra, FTIR and HPLC analysis discerned the decolorization of dye molecules and its transformation into the respective metabolites by Klebsiella pneumoniae MB398.
GC-MS analysis confirmed the effective biotransformation of Mordant Black 11 dye by Klebsiella pneumoniae MB398 projected via formation of unique compounds. Overall results projected the involvement of enzymatic activities of Klebsiella pneumoniae MB398, adsorption mechanisms and/or both in the biodegradation and biotransformation of Mordant Black 11 dye. These findings will serve as a baseline for application of Klebsiella pneumoniae MB398 in exploration of biotransformation approaches at larger scale for azo dyes comprising of recalcitrant compounds and heavy metals for retaining environmental sustainability.
References
Kansal SK, Sood S, Umar A, Mehta SK (2013) Photocatalytic degradation of Eriochrome Black T dye using well-crystalline anatase TiO2 nanoparticles. J Alloys Compd 581:392–397. https://doi.org/10.1016/j.jallcom.2013.07.069
Miao J, Jia Z, Lu H-B, Habibi D, Zhang L-C (2014) Heterogeneous photocatalytic degradation of mordant black 11 with ZnO nanoparticles under UV–Vis light. J Taiwan Inst Chem Eng 45:1636–1641
Ashraf SS, Rauf MA, Alhadrami S (2006) Degradation of Methyl Red using Fenton’s reagent and the effect of various salts. Dyes Pigments 69:74–78
El-Dars FMSE, Ibrahim HM, Farag HAB, Abdelwahhab MZ, Shalabi MEH (2015) Adsorption kinetics of Bromophenol Blue and Mordant Black 11 using bentonite carbon composite material. Int J Sci Eng Res 6(5):679–688
Jassal V, Shanker U, Kaith BS, Shankar S (2015) Green synthesis of potassium zinc hexacyanoferrate nanocubes and its potential application in photocatalytic degradation of organic dyes. RSC Adv 5:26141–26149
Jassal V, Shanker U, Kaith BS (2016) Aegle marmelos mediated green synthesis of different nanostructured metal hexacyanoferrates: activity against photodegradation of harmful organic dyes. Scientifica (Cairo) 2016:2715026–2715013. https://doi.org/10.1155/2016/2715026
Khalid A, Zubair M, Ihsanullah (2018) A comparative study on the adsorption of Mordant Black 11 dye from aqueous solution on graphene and acid-modified graphene. Arab J Sci Eng 43:2167–2179
Gobara HM, Elsalamony RA, Hassan SA (2016) Sonophotocatalytic degradation of eriochrome black-T dye in water using Ti grafted SBA-15. J Porous Mater 23:1311–1318. https://doi.org/10.1007/s10934-016-0190-3
Saritha B, Chockalingam P (2017) Photodegradation of eriochrome black-t dye from aqueous medium by photocatalysis. Intl J Pure Appl Maths 116(13):183–187
Chaieb K, Hagar M, Radwan NRE (2016) Biodegradation and decolorization of Azo dyes by adherent Staphylococcus lentus strain. Appl Biol Chem 59:1–10. https://doi.org/10.1007/s13765-016-0169-4
Intwala SM, Disha D (2017) Bioremediation of Azo Dye: Eriochrome Black T by the novel organism Bacillus lentus. Biosci Discov 8(4):771–775
Ali L, Alhassani H, Karuvantevida N, Rauf MA, Ashraf SS (2014) Efficient aerobic degradation of various Azo dyes by a Sphingomonas sp. isolated from petroleum sludge. J Bioremed Biodegrad 5(3):1–10
Cui D, Li G, Zhao M, Han S (2014) Decolorization of Azo dyes by a newly isolated Klebsiella sp. strain Y3, and effects of various factors on biodegradation. Biotechnol Biotechnol Equip 28(3):478–486
Kilany M (2017) Isolation, screening and molecular identification of novel bacterial strain removing methylene blue from water solutions. Appl Water Sci 7:4091–4098. https://doi.org/10.1007/s13201-017-0565-x
Zhang C, Hao Q, Zhang Z, Zhang X, Pan H, Zhang J, Zhang H, Sun F (2019) Whole genome sequencing and analysis of chlorimuron-ethyl degrading bacteria Klebsiella pneumoniae 2N3. Int J Mol Sci 20:3053–3066. https://doi.org/10.3390/ijms20123053
Nwaguma IV, Chikere CB, Okpokwasili GC (2016) Isolation, characterization, and application of biosurfactant by Klebsiella pneumoniae strain IVN51 isolated from hydrocarbon-polluted soil in Ogoniland, Nigeria. Bioresour Bioprocess 3:40–53. https://doi.org/10.1186/s40643-016-0118-4
Afzal AM, Rasool MH, Waseem M, Aslam B (2017) Assessment of heavy metal tolerance and biosorptive potential of Klebsiella variicola isolated from industrial effluents. AMB Express 7:184–192. https://doi.org/10.1186/s13568-017-0482-2
Aransiola EF, Ige OA, Ehinmitola EO, Layokun SK (2017) Heavy metals bioremediation potential of Klebsiella species isolated from diesel polluted soil. Afr J Biotech 16(19):1098–1105. https://doi.org/10.5897/AJB2016.15823
Wokem VC, Odokuma LO, Ariole CN (2017) Isolation and characterization of hydrocarbon-utilizing bacteria from petroleum sludge samples obtained from crude oil processing facility in Nigeria. J Appl Sci Environ Manag 21(2):355–359. https://doi.org/10.4314/jasem.v21i2.17
Ozyurek SB, Bilkay IS (2018) Biodegradation of petroleum by Klebsiella pneumoniae isolated from drilling fluid. Int J Environ Sci Technol 15:2107–2116. https://doi.org/10.1007/s13762-017-1581-y
Tarekegn MM, Salilih FZ, Ishetu AI (2020) Microbes used as a tool for bioremediation of heavy metal from the environment. Cogent Food Agri 6:1783174. https://doi.org/10.1080/23311932.2020.1783174
Ajaz M, Rehman A, Khan Z, Nisar MA, Hussain S (2019) Degradation of azo dyes by Alcaligenes aquatilis 3c and its potential use in the wastewater treatment. AMB Express 9:64. https://doi.org/10.1186/s13568-019-0788-3
Ewida AYI, El-Sesy ME, Zeid AA (2019) Complete degradation of azo dye acid red 337 by Bacillus megaterium KY848339.1 isolated from textile wastewater. Water Sci 33(1):154–161. https://doi.org/10.1080/11104929.2019.1688996
Sari IP, Simarani K (2019) Decolorization of selected azo dye by Lysinibacillus fusiformis W1B6: biodegradation optimization, isotherm, and kinetic study biosorption mechanism. Adsorpt Sci Technol 37:492–508. https://doi.org/10.1177/0263617419848897
Jain K, Shah V, Chapla D, Madamwar D (2012) Decolorization and degradation of azo dyes-Reactive Violet 5R by an acclimatized indigenous bacterial mixed cultures-SB4 isolated from anthropogenic dye contaminated soil. J Hazard Mater 213:378–386
Saratale RG, Saratale GD, Chang JS, Govindwar SP (2009) Ecofriendly degradation of sulfonated diAzo dye C.I. Reactive Green 19A using Micrococcus glutamicus NCIM-2168. Bioresour Technol 100:3897–8905
Shah K (2014) Biodegradation of Azo dye compounds. Int Res J Biochem Biotechnol 1(2):5–13
Karunya A, Rose C, Nachiyar VC (2013) Studies on degradation of textile azo dyes mordant black 17 using Pseudomanas aeruginosa sub 7, isolated from textile effluent. Intl J Appl Bio-Eng 7:9–14
Patel V, Cheturvedula S, Madamwar D (2012) Phenanthrene degradation by Pseudoxanthomonas sp. DMVP2 isolated from hydrocarbon contaminated sediment of Amlakhadii canal, Gujarat, India. J Hazard Mater 201:43–51
Chang J, Chou C, Lin Y, Lin P, Ho J, Hu T (2001) Kinetic characteristics of bacterial Azo dye decolorization by Pseudomonas luteola. Water Res 35(12):1114–1118
Asad S, Amoozegar MA, Pourbabee AA, Sarbolouki MN, Dastgheib SMM (2007) Decolorization of textile Azo dyes by newly isolated halophilic and halotolerant bacteria. Bioresour Technol 98:2082–2088
Senthilkumar S, Prabhu HJ, Perumalsamy M (2013) Response surface optimization for biodegradation of textile azo dyes using isolated bacterial strain Pseudomonas sp. Arab J Sci Eng 38(9):2279–2291. https://doi.org/10.1007/s13369-012-0507-8
Srinivas DM, Niraj ST, Nikhil BS, Unnati VN (2013) Remediation of textile reactive dyes using anaerobic rumen consortium. Int J Recent Sci Res 4(9):1400–1405
Jadhav JP, Govindwar SP (2006) Biotransformation of Malachite green by Saccharomyces cerevisiae MTCC 463. Yeast 23:315–323
Raj DS, Prabha RJ, Leena R (2012) Analysis of bacterial degradation of Azo dye Congo red using HPLC. J Indus Poll Cont 28(1):57–62
Wang J, Gao F, Liu Z, Qiao M, Niu X, Zhang K, Huang X (2012) Pathway and molecular mechanisms for Malachite green biodegradation in Exiguobacterium sp. MG2. PLoS One 7(12):1–10. https://doi.org/10.1371/journal.pone.0051808
Babu SS, Mohandass C, Vijayaraj AS, Dhale MA (2015) Detoxification and color removal of Congo red by a novel Dietzia sp. (DTS26)-a microcosm approach. Ecotoxicol Environ Saf 114:52–60
Acknowledgements
The authors are highly obliged and gratified to all those research labs and universities, which helped for accomplishing this research study.
Author information
Authors and Affiliations
Contributions
Conceived of or designed study: Uruj Tahir and Azra Yasmin; provision of lab facilities, chemicals and equipments: Azra Yasmin; performed research: Uruj Tahir; analyzed data: Uruj Tahir; contributed new methods or models: Uruj Tahir; wrote the paper: Uruj Tahir.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Responsible Editor: LUCY SELDIN
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 127 kb)
Rights and permissions
About this article
Cite this article
Tahir, U., Yasmin, A. Decolorization and discovery of metabolic pathway for the degradation of Mordant Black 11 dye by Klebsiella sp. MB398. Braz J Microbiol 52, 761–771 (2021). https://doi.org/10.1007/s42770-021-00470-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-021-00470-x