Abstract
The present research determines the effect of training data sets, correlation, and multicollinearity on the performance and overfitting of gene expression programming (GEP) and relevance vector machine (RVM) models in predicting the soaked CBR of fine-grained soil. For this purpose, one hundred and 82 training data sets have been compiled and subdivided into 50%, 60%, 70%, 80%, 90%, and 100%. In addition, 15 testing, 36 validation, and 12 laboratory-tested data sets have been compiled for trained models. The linear, polynomial, gaussian, and Laplacian kernels have been used to develop each GA and PSO optimized relevance vector machine (RVM) model, which have been trained by 50–100% training data sets. Thus, SRVM (single kernel-based) and HRVM (dual kernel-based) models have been developed and trained. The performance of models has been measured by RMSE, MAE, and R performance indicators. Based on the performance comparison, Model 21 (R = 0.9874) & Model 39 (R = 0.9748) of SRVM, and Model 51 (R = 0.9606) and Model 57 (R = 0.9701) of HRVM have been identified as better performing RVM models. However, GEP model 62 has performed (R = 0.8847) less than RVM models. The test performance comparison shows that model 21 has outperformed models 39, 51, 57, and 62 in predicting the soaked CBR of fine-grained soil. In addition, model 21 (performance = 0.8631) has predicted soaked CBR for the validation data set better than published models. Finally, the present research concludes that model 21 (GA optimized Laplacian kernel-based SRVM model) is a robust model that can predict the soaked CBR of fine-grained soil with the least prediction error and high performance.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
The transportation system is an essential component for the socio-economic development of a nation (Alam et al. 2020). The highway is an infrastructure facility to connect the Greenfield development. The most important element of highway construction is the mechanical properties of soil and design loads (Reddy et al. 2019). The California bearing ratio is a soil parameter widely used in designing flexible pavements in India. The CBR of soil is experimentally determined as per IS 2720 (P-16): 1979 to design the sub-base and base course. The CBR test determines an empirical value, which indicates the shear stress in subgrade, subbase, and base. Finally, the CBR test gives the soil’s mechanical strength and bearing value. The soaked and unsoaked are the types of CBR. The soaked CBR test involves 96 h of soaking of prepared soil sample on optimum moisture content, and a 50 mm diameter plunger penetrates the soil at the rate of 1.25 mm/min (IS 2720: 1979, P-16 2022). Determining unsoaked CBR value is easy and less time-consuming than soaked CBR. Still, both procedures are time-consuming, and sometimes, not accurate results are obtained due to improper maintenance of CBR equipment and sample disturbance (Taskiran, 2010). Therefore, the prediction models or AI approaches are applied to compute the CBR value for fine-grained soils because of their complex behaviour (González et al. 2018). Correlation equations are the most straightforward approach for determining the CBR of sub-grade materials (Taskiran, 2010). Several attempts have been made to establish a soaked CBR relationship with index parameters (gravel, sand, fine content, D10, D30, D50, D60), consistency limits (liquid limit, plastic limit, plasticity index), compaction parameters (optimum moisture content, maximum dry density), specific gravity, and compaction energy by numerous researchers. The researchers have derived the regression equations and concluded that the soil parameters LL, PI, PL, OMC, MDD, FC, MDD, G, S, M, and C affect the prediction of the CBR (Taskiran, 2010; Venkatasubramanian et al. 2011; Datta et al. 2011; Harini HN et al. 2014; Phani Kumar et al. 2015; Janjua et al. 2016; Pradeep Kumar et al. 2016; Chandrakar et al. 2016; Bourouis et al. 2016; Reddy et al. 2019). These parameters have been used as input parameters to construct the high-performance ANN models and compared with MLR, GEP, and OLS models.
Taskiran (2010) has compared GEP and ANN models and has reported that the GEP predicts CBR better than ANN with COD of 0.918. On the other hand, Ajalloeian et al. (2015) have concluded that the ANN model has performed better than the OLS approach. Vekatasubramanian et al. (2011) have compared ANN & MLR in predicting soil CBR and observed that MLR has predicted better than ANN. In addition, the authors have concluded the large data set can improve the performance of ANN models. Therefore, many researchers have used large data sets to develop ANN models and compare them with MLR.
Furthermore, Harini et al. (2014) and Pradeep Kumar et al. (2016) have concluded that the ANN model has outperformed the MLR model in predicting CBR. Rehman et al. (2017a; b) has reported that D50 and CU are the most influencing parameters for CBR of granular soil. Rehmanet al. (2017a; b) have concluded that the combined LL and PI predict soaked CBR of soil with a performance (R2) of 0.9. González et al. (2018) have estimated CBR using MLR, polynomial regression, and radial basis network models. The authors have suggested that soil can be divided into two groups based on the gravel content to predict CBR better. Ravichandra et al. (2019) have concluded that MDD is the most influencing parameter in computing the soaked CBR. The GMDH-NN model predicts CBR of soil better than the ANN and MLR models (Kurnaz et al. 2019). Alam et al. (2020) have reported that the Krigging method predicts CBR of soil better than ANN and GEP approaches. Vu et al. (2021) has concluded that the random forest model can predict the CBR of soil with a testing performance (R2) of 0.92.
In addition, many authors have used the field test data to predict the CBR of soil. For example, Al-Refeai et al. (1997) have successfully computed the CBR of fine-grained soil using gravel content and DCP parameters. Gül et al. (2021) have estimated the CBR from field measurements, such as laboratory soil and SPT-N parameters. The study of statistical relationships has revealed that the grain size, stiffness–looseness, DUW, and OMC influence the CBR of soil. In addition, it has also been observed that the ground vibration test and SPT predict CBR with high performance.
The literature study shows that the simple linear/ polynomial/ logistic regression analysis, multiple regression analysis, and artificial neural network approaches can predict the soaked CBR of fine-grained soil. Furthermore, many researchers have concluded that the G, S, FC, LL, PL, PI, OMC, and MDD of fine-grained soil play an important role in predicting soaked CBR. However, the effect of the correlation coefficient and training data set has not been studied in predicting soaked CBR of fine-grained soil. In addition, the hybrid learning approaches (relevance vector machine, least-square support vector machine, etc.) have not been applied to predict soaked CBR. Therefore, the following objectives are mapped for the present research work based on the literature study.
-
To develop and train the gene expression programming and GA/PSO optimized RVM models with 50%, 60%, 70%, 80%, 90%, and 100% of 182 training data sets.
-
To study the effect of optimization techniques (GA and PSO) on the performance of single-kernel-based RVM (SRVM) models in predicting soaked CBR of fine-grained soils.
-
To evolve dual kernel-based RVM (HRVM) models using a better-performing kernel (determined from each GA and PSO optimized SRVM model) and train the proposed HRVM models using 50%, 60%, 70%, 80%, 90%, and 100% of 182 training data sets.
-
To identify the best architectural model for predicting soaked CBR by mapping a comparative study between better-performing GA optimized SRVM, PSO optimized SRVM, GA optimized HRVM, PSO optimized HRVM, and GEP.
-
To determine the effect of the correlation coefficient and training data set on performance and overfitting of the developed RVM and GEP models.
-
To study the effect of multicollinearity in the training data sets on the performance and overfitting of the developed RVM and GEP models.
-
To determine the sensitivity of input parameters for soaked CBR using the cosine amplitude method.
2 Data analysis
This section briefly describes data sources, frequency distribution and histogram, Pearson's product-moment correlation coefficient, and statistics of collected data sets.
2.1 Data sources
The collected data set includes gravel, sand, fine content, liquid limit, plastic limit, plasticity index, optimum moisture content, maximum dry density, and soaked CBR of fine-grained soil. The soil data sets of soaked CBR have been collected from various sources, as given in Table 1.
Table 1 shows that the one hundred and eighty-two training data sets have been collected from different published research articles to train the RVM and GEP models. In addition, 15 and 36 data sets have been collected from the published articles to test and validate the trained AI models, respectively. Based on the performance comparison, the best architecture model has been determined to predict the soaked CBR. Furthermore, the 12 fine-grained soil samples have been collected from and around Kota, Rajasthan, and tested in the laboratory to cross-validate the performance and prediction of the best architecture model.
2.2 Frequency distribution and histogram of data sets
The gradational parameters, LL, PL, PI, OMC, and MDD, are the input parameters of models to predict soaked CBR. The frequency distribution of input parameters of soaked CBR data sets is shown in Fig. 1a–i.
2.3 Descriptive statistics
The present study has 182 training, 15 testing, 36 validation, and 12 cross-validation data sets of fine-grained soil. In addition, the training data sets have been subdivided into 50%, 60%, 70%, 80%, 90%, and 100% of 182 training data sets. The statistical parameters, minimum, maximum, mean (average), standard deviation (St. Dev), and confidence level (CL) at 95% are calculated for 50%, 60%, 70%, 80%, 90%, and 100% training data sets, as given in Table 2.
2.4 Pearson's product moment correlation coefficient
The Pearson's product-moment correlation coefficient has calculated the relationship between input and output parameters in the present study. The level of relationship in terms of the correlation coefficient is given in Table 3 (Hair et al. 2010).
Furthermore, the correlation coefficients are calculated for 50%, 60%, 70%, 80%, 90%, and 100% of 182 training data sets, as shown in Fig. 2a–f.
Figure 2a–f depicts the relationship between input and output parameters of 50%, 60%, 70%, 80%, 90%, and 100% of 182 training data sets. Figure 2 demonstrates the gravel content very strongly correlates with soaked CBR. On the other hand, the fine content very strongly correlates with maximum dry density. The liquid limit of fine-grained soil very strongly correlates with PL, PI, and OMC. In addition, it has also been observed that the sand content is strongly correlated with consistency limits. Still, the compaction parameters (OMC, MDD, and CBRs) are strongly influenced by gradational parameters and consistency limits of fine-grained soils. The plasticity index strongly influences the optimum moisture content and maximum dry density. In addition, fine content strongly influences consistency limits, and liquid limit strongly influences soaked CBR. In addition, the gravel content moderately correlates with liquid limit, plasticity index, and optimum moisture content of fine-grained soil. In addition, the plastic limit moderately influences soaked CBR. The correlation coefficient for soaked CBR shows that the sand content has no relationship with soaked CBR of fine-grained soil.
Figure 2a demonstrates that the sand content has multicollinearity of − 0.6644 and − 0.6700 with fine content and plastic limit, respectively. On the other hand, the OMC has multicollinearity with a plastic limit (0.7751) and a plasticity index (0.7410) of soil. Moreover, the liquid limit and PI have multicollinearity with soaked CBR. Figure 2b presents that the gravel content has multicollinearity with PL (− 0.4525) and PI (− 0.4830). On the other hand, sand content has multicollinearity with fine content (− 0.6684), LL (− 0.6856), and PL (− 0.4535) & PI (− 0.6052). Furthermore, FC has multicollinearity with PL (0.7461), PI (0.7511) and CBRs (− 0.7788). In addition, the liquid limit has multicollinearity with PL (0.8111) and OMC (0.8321). Moreover, the soaked CBR has multicollinearity with liquid limit and OMC.
Figure 2c shows that multicollinearity is present between sand content and liquid limit (− 0.6892), PL (− 0.6574), and PI (− 0.5346), and OMC (− 0.5543). Furthermore, maximum dry density has multicollinearity of − 0.8192 for FC and − 0.7949 for LL. In addition, optimum moisture content has multicollinearity with PL (0.6933) and PI (0.7198) of fine-grained soil. The correlation coefficient for CBRs shows that CBRs influence because of the multicollinearity between CBRs and liquid limit (− 0.5690) and OMC (− 0.5978) of fine-grained soil. Figure 2d illustrates that fine content has multicollinearity with gravel (− 0.7336) and sand (− 0.7059) content. The plastic limit and plasticity index have multicollinearity of − 0.4407 and − 0.4614, respectively, with gravel content. On the other hand, the liquid limit and plastic limit has multicollinearity of − 0.6440 and − 0.6230, respectively, with sand content. In addition, the plasticity index (− 0.5032) and OMC (− 0.5335) have multicollinearity with sand content. However, the liquid limit and plastic limit determine the plasticity index of soil, which develops a relationship (PI = LL−PL) between plastic limit and plasticity index. Thus, Fig. 2d shows that the plastic limit and plasticity index has multicollinearity of 0.7020 and 0.7259, respectively, with the optimum moisture content of fine-grained soils. In addition, the liquid limit (− 0.5751) and plasticity index (− 0.5343) contain the multicollinearity for soaked CBR of fine-grained soil.
Figure 2e depicts that the gravel content has multicollinearity with a plastic limit (− 0.4236) and plasticity index (− 0.4485). Furthermore, liquid limit (− 0.6719), plastic limit (− 0.6444) and, plasticity index (− 0.5359) and OMC (− 0.5478) have multicollinearity with sand content present in the fine-grained soils. Figure 2e shows that soaked CBR of fine-grained soils has multicollinearity with liquid limit and plasticity index. Figure 2f demonstrates that the fine content has multicollinearity with gravel (− 0.7067) and sand (− 0.7222) content. In addition, gravel content has multicollinearity with a plastic limit (− 0.4128) and a plasticity index (− 0.4332). On the other hand, the sand content has multicollinearity with a plastic limit (− 0.5119) and optimum moisture content (− 0.5271). Moreover, the OMC has multicollinearity of 0.6959 and 0.7308 with plastic limit and plasticity index, respectively. In addition, it has been found that the liquid limit and plasticity index influence the prediction of soaked CBR.
3 Methodology used in the present study
The relevance vector machine and gene expression programming artificial intelligence approaches have been used in the present research work to predict soaked CBR of fine-grained soils.
3.1 Relevance vector machine
A relevance vector machine is a hybrid artificial intelligence approach, and this approach is an advanced version of the support vector machine. RVM gives the most promising results, because it uses a Bayesian inference system (Tipping 2001):
The relevance vector machine models use Linear, Gaussian, Polynomial, and Laplacian kernels in the present study. The mathematical expression of kernels is
Linear Kernel
Gaussian Kernel
Polynomial Kernel
Laplacian Kernel
Sixty models are developed in the present study to predict the soaked CBR of fine-grained soil. The model designation with different percentages of training data sets is given in Table 4.
Six RVM models have been constructed by 50%, 60%, 70%, 80%, 90%, and 100% for each Linear, Polynomial, Gaussian, and Laplacian kernel. Six RVM models of single kernel function-based (SRVM) are optimized by GA and PSO techniques. In addition, the two kernel function-based (HRVM) models are developed by recognizing the maximum performance SRVM model in GA and PSO optimization. In this study, Laplacian and Gaussian kernel function-based SRVM models have performed better than the other RVM models. Therefore, HRVM models have been developed using Laplacian and Gaussian kernels as Kernel1 and Kernel2, respectively. The proposed HRVM models are also optimized by GA and PSO techniques. The hyperparameters of the proposed RVM models are given in Table 5.
3.2 Gene expression programming
Gene expression programming is an evolutionary algorithm developed to solve user-defined problems using automatic generation computer programs (Zhong, 2017), and it consists of mutation and crossover operators (Tenpe, 2020). It is a genotype–phenotype system encoded in simple linear chromosomes of fixed length. The general gene structure is shown in Fig. 3.
where “–” is the root node, “Rt, + , −” is the function node (Head), and “X1, X2, X3, 7” are terminals (tail). The head length (h) and tail length (t) are computed using Eq. 3.52:
The present study has developed six gene expression programming models to predict the soaked CBR of fine-grained soil. The model designation with different percentages of training data sets is given in Table 6.
The gene expression programming models have been trained by 50%, 60%, 70%, 80%, 90%, and 100% training data sets. The hyperparameters of the proposed GEP models are given in Table 7.
The following equations are derived while training the GEP models:
3.3 Methodology
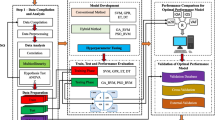
The present research predicts soaked CBR of fine-grained soil using the best architectural AI model. Therefore, a comparative study is mapped between adopted AI approaches, such as relevance vector machines and gene expression programming. The relevance vector machine models are developed using single and dual kernel functions, denoted by SRVM and HRVM. These kernel functions are Linear, Polynomial, Gaussian, and Laplacian. Each kernel function develops six SRVM models using 50%, 60%, 70%, 80%, 90% and 100% of 182 training data sets. The genetic algorithms and particle swarm optimization techniques optimize each SRVM model. The optimized SRVM models have predicted soaked CBR of 15 testing fine-grained soil data sets. Thus, four models (one from each kernel function) are the better-performing models from each GA and PSO identified and compared to obtain the best kernel function. Two kernel functions (one from GA and one from PSO) have been identified as the best kernel function and developed the dual kernel-function-based HRVM models. In the present study, the Laplacian and Gaussian kernel functions are the best kernel function identified from SRVM models. Therefore, six HRVM models are developed for 50%, 60%, 70%, 80%, 90%, and 100% of 182 training data sets and optimized by each GA and PSO technique. Based on the comparative study of the performance of HRVM models, one model from each GA and PSO technique is a better-performing model recognized. Finally, the present study has four better-performing models: one from SRVM optimized by GA, one from SRVM optimized by PSO, one from HRVM optimized by GA, and one from HRVM optimized by GA PSO.
On the other hand, 50%, 60%, 70%, 80%, 90%, and 100% of 182 training data sets have developed six GEP models. In addition, a GEP model having maximum performance is recognized as a better-performing model and compared with four models obtained from a comparison of RVM models. Thus, comparing one GEP and four RVM models gives the best architectural model to predict the soaked CBR of fine-grained soil. In addition, geotechnical properties of 36 fine-grained soils have been collected from published articles to compile a validation data set to validate the best architectural models. Moreover, the published models have predicted the soaked CBR of fine-grained soil in validation data sets and compared it with the best architectural model. After validating the best architectural model, the best architectural model has predicted the soaked CBR of twelve fine-grained soil samples tested in the laboratory.
4 Results and discussion
Numerous researchers have applied different AI approaches to predict the soaked CBR of soil for fine-grained soil. The present study uses the genetic algorithm and particle swarm optimized relevance vector machine AI approach to predict soaked CBR of fine-grained soils. In addition, the gene expression programming AI approach is also applied to predict the soaked CBR of fine-grained soil. The best architectural AI model has been determined by comparing the performance of developed models. Furthermore, the performance of models has been determined in terms of RMSE, MAE, and R.
4.1 Results of single kernel-based RVM (SRVM) models optimized by GA
The single kernel-based RVM models have been developed using Linear, Polynomial, Gaussian, and Laplacian kernel functions and optimized by a genetic algorithm. RVM models have been trained by 50%, 60%, 70%, 80%, 90%, and 100% of 182 data sets. Fifteen data sets have tested the capabilities of developed RVM models. The training and testing performance of the SRVM models is mapped for different percentages of training data sets, as shown in Fig. 4a–h.
Figure 4 depicts the performance of the developed Linear, Polynomial, Gaussian, and Laplacian SRVM models (GA optimized) using 50–100% training data sets. The following observations have been mapped from Fig. 4i Model 3 has predicted soaked CBR with the test RMSE of 1.0390, MAE of 0.8596, and R of 0.8974, (ii) Model 9 has predicted soaked CBR with test RMSE of 0.5116, MAE of 0.4169, and R of 0.9701, (iii) Model 15 has predicted soaked CBR with test RMSE of 0.8909, MAE of 0.7332, and R of 0.9260, (iv) Model 21 has predicted soaked CBR with test RMSE of 0.3140, MAE of 0.2418, and R of 0.9874. Models 3, 9, 15, and 21 are developed using Linear, Polynomial, Gaussian, and Laplacian kernels and trained by a 70% training data set. In the 70% training data set, gravel content is very strongly correlated, and fine content and MDD are strongly correlated with soaked CBR of fine-grained soils. The soaked CBR of fine-grained soil is highly influenced by gravel content (Al-Refeai et al. 1997), fine content (Reddy et al. 2019), and MDD (Ravichandra et al. 2019). The correlation coefficient of sand content shows no relationship, and the rest of the input parameters have a moderate relationship with the soaked CBR of fine-grained soil. The performance of Models 3, 9, 15, and 21 has been compared, and it has been observed that Model 21 (Laplacian kernel-based) has predicted soaked CBR with high performance and has been identified as a better performance model. Furthermore, the overfitting for single kernel-based RVM models optimized by GA has been calculated, as shown in Fig. 5.
The ratio of test RMSE to training RMSE (overfitting = \({\text{Test RMSE}}/{\text{Training RMSE}}\)) is known for the overfitting or overfitting ratio of the model (Tenpe et al. 2020). Figure 5 illustrates the comparison of overfitting of the model in predicting soaked CBR of fine-grained soil. Figure 5 shows that Models 3, 9, and 21 have the overfitting of 0.1279, 0.1319, and 0.2155, respectively, which is comparatively less than other models. On the other hand, the Gaussian SRVM (Model 18) model has an overfitting of 0.1686, which is less than other Gaussian SRVM (optimized by GA). The comparison of overfitting for models 3, 9, 18, and 21 demonstrates that model 3 (linear SRVM) has the least overfitting than other models. Therefore, it can be stated that the linear SRVM model optimized by the GA technique achieves minimum overfitting in predicting soaked CBR of fine-grained soil than nonlinear SRVM models optimized by the GA technique.
4.2 Results of single kernel-based RVM models optimized by PSO
Similarly, single kernel-based RVM models have been developed using Linear, Polynomial, Gaussian, and Laplacian kernel functions and optimized by the particle swarm optimization technique. Fifteen data sets have tested the capabilities of developed RVM models. The training and testing performance of the SRVM models is mapped for different percentages of training data sets, as shown in Fig. 6a–h.
Figure 6 depicts the performance of the developed Linear, Polynomial, Gaussian, and Laplacian SRVM models (PSO optimized) using 50–100% training data sets. Models 27, 33, 39, and 45 are developed using Linear, Polynomial, Gaussian, and Laplacian kernels and trained by a 70% training data set. The following observations have been mapped from Fig. 6, (i) Model 27 has predicted soaked CBR with the test RMSE of 0.8447, MAE of 0.7183, and R of 0.9315, (ii) Model 33 has predicted soaked CBR with test RMSE of 0.9350, MAE of 0.7585, and R of 0.9026, (iii) Model 39 has predicted soaked CBR with test RMSE of 0.4747, MAE of 0.3930, and R of 0.9748, (iv) Model 45 has predicted soaked CBR with test RMSE of 0.4788, MAE of 0.3553, and R of 0.9696. Models 27, 33, 39, and 45 are developed using Linear, Polynomial, Gaussian, and Laplacian kernels and trained by a 70% training data set. The input parameters gravel content, fine content, and maximum dry density very strongly correlate with the soaked CBR of fine-grained soil. In addition, it is found that the fine content strongly correlates with consistency limits, which indirectly enhances the prediction of soaked CBR of fine-grained soil. Furthermore, the overfitting for single kernel-based RVM models optimized by the PSO technique has been calculated, as shown in Fig. 7.
Figure 7 demonstrates the comparison of overfitting of SRVM models optimized by the PSO technique. Figure 7 shows that Models 27, 33, 39, and 45 have overfitting of 0.104, 0.2518, 0.1631, and 0.3134, respectively, which is comparatively the least than other SRVM models optimized by the PSO technique. The overfitting comparison of models 27, 33, 39, and 45 shows that Model 27 (linear model) has minimum overfitting in predicting the soaked CBR of fine-grained soil than nonlinear models (Models 33, 39, and 45).
On the other hand, the performance comparison has been mapped between GA and PSO optimized SRVM models. The particle swarm optimization technique has improved the performance (R) of Model 3 from 0.8974 to 0.9315 (Model 15) and Model 15 from 0.9260 to 0.9784 (Model 39). Model 3 and 15 are linear and Gaussian kernel-based SRVM models. Therefore, it can be stated that the PSO optimization technique boosts the performance of linear and Gaussian kernel-based SRVM models of soaked CBR. However, the performance of Model 9 has dropped from 0.9701 to 0.9026, and Model 21 has fallen from 0.9874 to 0.9696 using the PSO technique. Models 9 and 21 are polynomial and Laplacian kernel-based SRVM models. Therefore, it can be stated that the PSO optimization technique decreases the performance of polynomial and Laplacian kernel-based SRVM models of soaked CBR, predicting soaked CBR of fine-grained soil.
Furthermore, genetic algorithm optimized SRVM Model 21 has outperformed the other GA optimized SRVM models, and PSO optimized SRVM Model 39 has outperformed the other PSO optimized SRVM models. However, Model 21 and Model 39 has constructed by Laplacian and Gaussian kernel functions. Therefore, the hybrid relevance vector machine (HRVM) models are developed using Laplacian and Gaussian kernel as kernel1 and kernel2, respectively, and optimized by GA and PSO techniques.
4.3 Results of two kernel-based RVM models optimized by GA
HRVM models have also been trained by 50%, 60%, 70%, 80%, 90%, and 100% of 182 data sets and optimized by GA. Fifteen data sets have tested the ability of developed HRVM models. The training and testing performance of the HRVM models is mapped for different percentages of training data sets, as shown in Fig. 8a, b.
Figure 8 depicts the performance of the developed GA optimized HRVM models using 50–100% training data sets. From Fig. 8, it has been observed that Model 51 has predicted soaked CBR with test RMSE of 0.7381, MAE 0.6094, and R of 0.9606. Model 51 has been trained by 70% training data sets. Therefore, Model 51 has outperformed the GA-optimized Models 3 and 15 and PSO-optimized Models 27 and 33. Still, Models 9, 21, 39, and 45 have predicted soaked CBR better than Model 51. Furthermore, the HRVM (GA Optimized) overfitting has been calculated, as shown in Fig. 9.
Figure 9 illustrates that Model 51 has an overfitting of 0.2805 in predicting soaked CBR of fine-grained soil, which is comparatively less than other GA-optimized HRVM models. However, model 51 has been trained by 70% of 182 training data sets. As the result of the relationship between soaked CBR and input parameters (gravel content, fine content, and maximum dry density), model 51 has predicted soaked CBR of fine-grained soil with the least overfitting.
4.4 Results of two kernel-based RVM models optimized by PSO
Similarly, HRVM models (PSO optimized) have been trained by 50%, 60%, 70%, 80%, 90%, and 100% of 182 data sets and optimized by the PSO technique. Fifteen data sets have tested the capabilities of developed HRVM models. The training and testing performance of the HRVM models is mapped for different percentages of training data sets, as shown in Fig. 10a, b.
Figure 10 depicts the performance of the developed PSO optimized HRVM models using 50–100% training data sets. From Fig. 10, it has been observed that Model 57 has predicted soaked CBR with test RMSE of 0.4761, MAE 0.3895, and R of 0.9701. On the other hand, model 51 has been trained by 70% training data sets. Therefore, Model 57 has outperformed the GA optimized Models 3 & 15 and PSO optimized Models 27, 33 and 45. Still, Model 9, 21, and 39 have predicted soaked CBR better than Model 57. On the other hand, the comparison of overfitting of the model, as shown in Fig. 11, demonstrates that model 57 has predicted the soaked CBR of fine-grained soil with the least overfitting than other PSO optimized HRVM models, i.e., 0.2812.
Finally, from the comparison of single and dual kernel RVM models, the single kernel-based Models 21 (GA optimized), 39 (PSO optimized), and two kernel-based Models 51 (GA optimized) and 57 (PSO optimized) have been identified as better performance RVM models in predicting soaked CBR of fine-grained soils. The results of RVM models show that the high performance and accuracy in predicting soaked CBR can be achieved by a 70% training data set. The data set has a very strong correlation with gravel content and a strong correlation with FC and MDD of soil.
On the other hand, multicollinearity has been observed in the 70% of 182 training data sets. The multicollinearity is a statistical parameter affecting the models' prediction and performance. 70% of the 182 training data set shows that the fine content has multicollinearity with gravel and sand content. However, the sand content is the least influencing input parameter in predicting soaked CBR of fine-grained soil. Moreover, the sand content has multicollinearity with consistency limits and optimum moisture content. Thus, the effect of consistency limits and optimum moisture content becomes less in predicting soaked CBR. In addition, it is found that the maximum dry density has multicollinearity with fine content and liquid limit. In addition, the optimum moisture content has multicollinearity with plastic limit and plasticity index. Instead of the presence of multicollinearity in the 70% training data set, nonlinear RVM models have predicted the soaked CBR with a performance of more than 0.95, which shows that the RVM approach is a robust approach and it is not getting affected by the presence of multicollinearity during the prediction of soaked CBR of fine-grained soil.
4.5 Results of gene expression programming
The gene expression programming models have been trained by 50%, 60%, 70%, 80%, 90%, and 100% of 182 data sets. Fifteen data sets have tested the capabilities of developed GEP models. The training and testing performance of the GEP models is mapped for different percentages of training data sets, as shown in Fig. 12a, b.
Figure 12 depicts the performance comparison of the trained and tested GEP models using 50–100% training data sets. From Fig. 12, it has been observed that Model 62 has predicted soaked CBR with test RMSE of 1.2148%, MAE 0.9902%, and R of 0.8847. Model 62 has been trained by 60% training data sets. Therefore, Model 62 has outperformed the other GEP models in predicting the soaked CBR of soil. Furthermore, the overfitting is also calculated for the GEP models, as shown in Fig. 13.
Figure 13 demonstrates that Model 62 has achieved an overfitting of 0.3063 in predicting soaked CBR of fine-grained soil, which is comparatively less than other GEP models. The expression tree of model 62 is shown in Fig. 14.
Figure 14 shows that Model 62 has three genes, i.e., Sub-ET1, Sub-ET2, and Sub-ET3, which consists of three constant values viz. c0 = 2.07, c6 = 6.63, and c7 = − 4.31 and input parameters: d0 = G, d1 = S, d3 = LL, d4 = PL, d6 = OMC. The fine content and maximum dry density play a vital role in predicting the soaked CBR of fine-grained soil (Taskiran 2010; Chandrakar et al. 2016; Bourouis et al. 2016; Reddy et al. 2019). However, both input parameters are missing in the expression tree, indicating the less effective models for predicting the soaked CBR of fine-grained soil. In addition, plasticity index and C + S content are absent in Model 62. The GEP models have been tested by the data set of inorganic clays of high plasticity soils. The high plasticity clay requires the plasticity index to predict soaked CBR (Datta et al. 2011). Therefore, Model 62 has predicted soaked CBR lower than experimental values of CH soil.
On the other hand, 60% of 182 training data sets have trained the model 62. 60% of 182 training data sets have multicollinearity between gradational parameters and consistency limits. The gravel content has moderate multicollinearity (0.41 < R < 0.60) with plastic limit and plasticity index. The sand content has multicollinearity with FC and liquid limits, and the expression tree does not have FC as the input parameter. Therefore, the present multicollinearity between sand and fine content does not affect the prediction. Still, the liquid limit has very strong multicollinearity (R > 0.8) with plastic limit and optimum moisture content. Therefore, model 62 did not perform well compared to the RVM models in the presence of multicollinearity.
4.6 Best architecture model
The test performance of Models 21, 39, 51, 57, and 62 has been compared to determine the best architecture model for predicting soaked CBR of soil, as shown in Fig. 15.
Figure 15 illustrates that Model 21 (SRVM optimized by GA) has predicted soaked CBR of fine-grained soil with RMSE of 0.3140%, MAE of 0.2418, and R of 0.9874, which is comparatively less than other better-performing models. Therefore, the best architectural model for predicting the soaked CBR of fine-grained soil is Model 21.
Furthermore, the data sets of 36 fine-grained soil specimens have been collected from the published research articles to validate Model 21. In addition, the published equations/ models have also been used to compute the soaked CBR of 36 fine-grained soil and compared with Model 21. Patel et al. (2010) derived a multilinear regression equation to predict soaked CBR of soil using PI, MDD, and OMC:
Harini et al. (2014) also developed a regression model to predict soaked CBR of soil using liquid limit and fine content:
Pradeep Kumar et al. (2016) proposed a multiple linear regression equation to predict the soaked CBR of soil using MDD, OMC, PL, LL, PI, G, S, and FC:
Reddy et al. (2019) also suggested a simple regression equation to predict the CBR using the plasticity index of soil:
Equations 13–16 and model 21 have been used to predict the soaked CBR of 36 soil specimens collected from published articles and results, as shown in Fig. 16.
Figure 16 depicts the actual vs predicted plot of soaked CBR of fine-grained soil using Model 21 and Eqs. 13–16. Figure 16 shows that Model 21, Eqs. 13, 14, 15, and 16 have predicted soaked CBR with a performance (R) of 0.8631, 0.7743, 0.8437, 0.8133, and 0.7919, respectively. The performance comparison shows that Model 21 has performed better than published equations/models. Furthermore, 12 fine-grained soil samples have been collected from and around Kota, Rajasthan, for the laboratory validation of model 21. The gravel, sand, fine content, liquid limit, plastic limit, plasticity index, OMC, MDD and soaked CBR have been determined in the geotechnical laboratory. The prediction vs actual plot for laboratory-tested soil data sets is shown in Fig. 17.
Figure 17 depicts the actual vs predicted plot of soaked CBR of 12 fine-grained soil using Model 21. Figure 17 illustrates that Model 21 predicts the soaked CBR for laboratory-tested soil with RMSE of 0.4237%, MAE of 0.32% and R of 0.8139. The correlation coefficient (R) of 0.8 or more than 0.8 shows a strong relationship between actual and predicted values (Smith 1986). Therefore, Model 21 can be used to predict the soaked CBR of fine-grained soil.
5 Sensitivity analysis
In the present study, the non-linear sensitivity analysis has been performed using the cosine amplitude method. The sensitivity analysis is performed by Eq. 17 (Ardakani et al. 2019):
where \(X_{ic}\) is input parameters G, S, FC, LL, PL, PI, OMC, and MDD, and \(X_{jk}\) is output parameter soaked CBR of fine-grained soil. The value of SS equal to one shows that the input parameter highly influences the output parameter. The sensitivity analysis has been performed for 50%, 60%, 70%, 80%, 90%, and 100% of 182 training data sets in this study, as shown in Fig. 18.
Figure 18 illustrates the sensitivity analysis for different percentages of training data sets. However, Model 21 has been identified as the best architectural model by considering the test performance in the present study, which is trained by 70% of 182 training data sets. In addition, model 21 has outperformed the published models and equation in predicting soaked CBR of fine-grained soil. Furthermore, Fig. 18 shows that the 70% training data sets including G, S, FC, LL, PL, PI, OMC and MDD has sensitivity of 0.9209, 0.5139, 0.2281, 0.3079, 0.3885, 0.1970, 0.2673, and 0.6265, respectively, with soaked CBR of fine-grained soil. In addition, the sensitivity for consistency limits and compaction parameters has increased with training data sets. The soaked CBR of fine-grained soil is highly affected by index properties (Taskiran 2010). Therefore, the proposed AI models in the present study have achieved high performance and prediction for Model 21 trained by the 70% of the 182 training data sets.
6 Conclusions
The present study has successfully employed gene expression programming (GEP) and relevance vector machine (optimized by GA and PSO technique) models to predict the soaked CBR of fine-grained soils. Based on the outcomes of the study, the following conclusions are mapped.
-
The performance comparison of GA and PSO optimized SRVM models demonstrates that the performance of the linear and Gaussian SRVM models increases after optimizing the model by the PSO optimization technique. On the other hand, the performance of polynomial and Laplacian SRVM models decrease after optimizing the models by the PSO optimization technique.
-
From the performance comparison of the SRVM model, the Laplacian SRVM model (Model 21) and Gaussian SRVM model (Model 39) have been identified as better-performing models with a performance of 0.9874 and 0.9748, respectively. Therefore, the dual-kernel-based HRVM (optimized by GA and PSO) models are developed using the Laplacian kernel (as kernel1) and Gaussian kernel (as kernel2). The test performance of HRVM models illustrates that the PSO optimization technique enhances the performance and predicts the soaked CBR better than GA-optimized HRVM models.
-
Model 21 has been identified as the best architectural model for predicting soaked CBR of fine-grained soils by comparing the performance of Model 21 (0.9874), Model 39 (0.9748), Model 51 (0.9606), Model 57 (0.9701), and Model 62 (0.8847). In addition, the validation and cross-validation (laboratory validation) results have confirmed that Model 21 can predict the soaked CBR better than published models in the literature.
-
The present study demonstrates that the RVM model predicts soaked CBR better if the input and output parameters of training data sets have a strong (0.61 < R < 0.80) to very strong (0.81 < R < 1.0) correlation. In addition, the developed RVM model predicts the soaked CBR with the least prediction error (RMSE and MAE) and overfitting.
-
Based on the performance comparison of GEP models, Model 62 outperformed the other GEP models with a performance of 0.8847. Model 62 was trained using input parameters gravel, sand content, OMC, and PL to predict the soaked CBR. Model 62 did not perform well in the absence of the maximum dry density and the presence of multicollinearity in 60% of 182 training data sets.
-
Multicollinearity in the training data set affects the performance and overfitting of the GEP model. On the other hand, the optimized SRVM and HRVM models are less influenced by multicollinearity.
-
The sensitivity analysis confirms that the soaked CBR is highly influenced by the gravel, sand content, and maximum dry density of fine-grained soils.
Finally, the present research introduces a robust SRVM (Laplacian SRVM model optimized by GA optimization) model to predict the soaked CBR of fine-grained soil.
Data availability
All data, models, and code generated or used during the study appear in the submitted article. The laboratory-test database can be provided on the request.
Abbreviations
- AI:
-
Artificial intelligence
- ANN:
-
Artificial neural networks
- C:
-
Clay content
- CBR:
-
California bearing ratio
- CC :
-
Coefficient of curvature
- CL:
-
Confidence level
- CL:
-
Inorganic clays of low plasticity
- CS:
-
Coarse sand
- C U :
-
Coefficient of uniformity
- D 10 :
-
Particle size at 10% fine
- D 30 :
-
Particle size at 30% fine
- D 50 :
-
Particle size at 50% fine
- D 60 :
-
Particle size at 60% fine
- DCP:
-
Dynamic cone penetration
- DUW:
-
Dry unit weight of soil
- FC:
-
Fine content
- FD:
-
Frequency distribution
- FS:
-
Fine sand
- FSI:
-
Free swell index
- G:
-
Gravel content
- GA:
-
Genetic algorithm
- GEP:
-
Gene expression programming
- GMDH-NN:
-
Group method of data handling neural network
- HRVM:
-
Hybrid/two kernel-based relevance vector machine
- IS:
-
Indian Standards
- LL:
-
Liquid limit
- M:
-
Silt content
- MAE:
-
Mean absolute error
- MDD:
-
Maximum dry density
- MLR:
-
Multiple linear regression
- NUW:
-
Natural unit weight of soil
- OC:
-
Organic content
- OMC:
-
Optimum moisture content
- OWC:
-
Optimum water content
- PI:
-
Plasticity index
- PL:
-
Plastic limit
- PSO:
-
Particle swarm optimization
- R :
-
Correlation coefficient
- R 2 :
-
Coefficient of determination
- RMSE:
-
Root mean square error
- RVM:
-
Relevance vector machine
- S :
-
Sand content
- SPSS:
-
Statistical package for the social sciences
- SPT-N:
-
Standard penetration test number
- SRVM:
-
Single kernel-based relevance vector machine
- St. Dev:
-
Standard deviation
References
Abdelkader HA, Hussein M, Ye H (2021) Influence of waste marble dust on the improvement of expansive clay soils. Adv Civil Eng. https://doi.org/10.1155/2021/3192122
Ajalloeian R, Mojtaba K (2015) Predicting maximum dry density, optimum moisture content and California bearing ratio (CBR) through soil index using ordinary least squares (OLS) and artificial neural networks (ANNS). Int J Innov Technol Explor Eng 3(3):1–5
Akanbi DO (2021) Performance evaluation of crushed glass in stabilizing lateritic soil for road pavement layers. Saudi J Eng Technol 6(4):77–83. https://doi.org/10.36348/sjet.2021.v06i04.006
Alam SK, Mondal A, Shiuly A (2020) Prediction of CBR value of fine grained soils of Bengal Basin by genetic expression programming, artificial neural network and krigging method. J Geol Soc India 95(2):190–196. https://doi.org/10.1007/s12594-020-1409-0
Alhassan M (2008) Potentials of rice husk ash for soil stabilization. AU J Technol 11(4):246–250
Al-Refeai T, Al-Suhaibani A (1997) Prediction of CBR using dynamic cone penetrometer. J King Saud Univ Eng Sci 9(2):191–203. https://doi.org/10.1016/S1018-3639(18)30676-7
Al-Soudany K (2018) Remediation of clayey soil using silica fume. In: MATEC Web of Conferences (Vol. 162, p. 01017). EDP Sciences. https://doi.org/10.1051/matecconf/201816201017.
Amena S (2021) Experimental study on the effect of plastic waste strips and waste brick powder on strength parameters of expansive soils. Heliyon 7(11):e08278. https://doi.org/10.1016/j.heliyon.2021.e08278
Araujo W, Ruiz G (2016) Correlation equations of CBR with index properties of soil in the city of Piura. In: Proceeding of 14th LACCEI International Multi-conference for Engineering, Education and Technology (pp. 1–7).
Ardakani A, Kordnaeij A (2019) Soil compaction parameters prediction using GMDH-type neural network and genetic algorithm. Eur J Environ Civ Eng 23(4):449–462. https://doi.org/10.1080/19648189.2017.1304269
Azim MAM, Azhar ATS, Tarmizi AKA, Shahidan S, Nabila ATA (2017) Enhancing the compressive strength of landfill soil using cement and bagasse ash. In: IOP Conference Series: Materials Science and Engineering (Vol. 271, No. 1, p. 012053). IOP Publishing, https://doi.org/10.1088/1757-899X/271/1/012053.
Bandyopadhyay TS, Singh AA, Pandey V, Singh JP (2016) Stabilization of soil using GGBS and calcium carbide residue. Int J Innov Res Sci Eng Technol 5(9):17023–17030
Blayi RA, Sherwani AFH, Ibrahim HH, Abdullah SJ (2020a) Stabilization of high-plasticity silt using waste brick powder. SN Appl Sci 2(12):1–12. https://doi.org/10.1007/s42452-020-03814-8
Blayi RA, Sherwani AFH, Ibrahim HH, Faraj RH, Daraei A (2020b) Strength improvement of expansive soil by utilizing waste glass powder. Case Stud Construct Mater 13:e00427. https://doi.org/10.1016/j.cscm.2020.e00427
Bourouis MA, Zadjaoui A, Djedid A (2016) Neuro-genetic approach for immersed CBR index prediction. In: Proceedings of the First Southern African Geotechnical Conference, Sun City (pp. 143–148). https://doi.org/10.3217/978-3-85125-564-5-046.
Chandrakar V, Yadav RK (2016) Study of correlation of CBR value with engineering properties and index properties of coarse grained soil. Internat Res J Eng Tech (IRJET) 3: 772–778.
Dabou B, Kanali C (2021) Structural performance of laterite soil stabilised with cement and blue gum (Eucalyptus Globulus) wood ash for use as a road base material. arXiv preprint https://arxiv.org/abs/2109.08507, https://doi.org/10.14445/22315381/IJETT-V69I9P231.
Datta T, Chottopadhyay BC (2011) Correlation between CBR and index properties of soil. In: Proceedings of Indian Geotechnical Conference, Kochi (pp. 131–133).
Edil TB, Acosta HA, Benson CH (2006) Stabilizing soft fine-grained soils with fly ash. J Mater Civ Eng 18(2):283–294. https://doi.org/10.1061/ASCE0899-1561200618:2(283)
Eltwati AS, Tarhuni F, Elkaseh A (2020) Engineering properties of clayey soil stabilized with waste granite dust. J Criti Rev 7(16):794–802
González Farias I, Araujo W, Ruiz G (2018) Prediction of California bearing ratio from index properties of soils using parametric and non-parametric models. Geotech Geol Eng 36(6):3485–3498. https://doi.org/10.1007/s10706-018-0548-1
Gül Y, Çayir HM (2021) Prediction of the California bearing ratio from some field measurements of soils. In: Proceedings of the Institution of Civil Engineers-Municipal Engineer (Vol. 174, No. 4, pp. 241–250). Thomas Telford Ltd, https://doi.org/10.1680/jmuen.19.00020.
Hair JF, Ortinau DJ, Harrison DE (2010) Essentials of marketing research (vol. 2). McGraw-Hill/Irwin, New York
Harini H, Naagesh S (2014) Predicting CBR of fine grained soils by artificial neural network and multiple linear regression. Int J Civil Eng 5(2):119–126
Indian Standard: 2720: (P-16) 1979, Method of test for soil: Laboratory determination of CBR.
Janjua ZS, Chand J (2016) Correlation of CBR with index properties of soil. Int J Civil Eng Technol 7(5):57–62
Ji-ru Z, Xing C (2002) Stabilization of expansive soil by lime and fly ash. J Wuhan Univ Technol Mater Sci Ed 17(4): 73–77. https://doi.org/10.1007/BF02838423.
Kadhim YM, Al-Adhamii RA, Fattah MY (2022) Geotechnical properties of clayey soil improved by sewage sludge ash. J Air Waste Manag Assoc 72(1):34–47. https://doi.org/10.1080/10962247.2020.1862939
Keshav K, Mishra MK, Singh JP (2018) Stabilization of soil using sodium hydroxide additive. Int J Res Eng Appl Manag 4(2):328–332. https://doi.org/10.18231/2454-9150.2018.0166
Kulkarni PP, Mandal JN (2017) Performance assessment of stabilized soil with fly ash-nano material mixes. J Geotech Transp Eng 3(2):35–46
Kumar S, Chouksey A, Priyanka (2017) Stabilization of sub-grade using fly ash and geosynthetic material. Int J Adv Technol Eng Sci 5(06).
Kurnaz TF, Kaya Y (2019) Prediction of the California bearing ratio (CBR) of compacted soils by using GMDH-type neural network. Eur Phys J plus 134(7):326. https://doi.org/10.1140/epjp/i2019-12692-0
Mahdi Z, Alhassnawi N (2018) Assessment of subgrade soil improvement by waste glass powder. Int J Civil Eng Technol 9:12–21
Mittal A (2021) Effect of rice husk ash and stone dust on selecting engineering properties of poor subgrade soil. Jordan J Civil Eng 15(1):133–141
Murmu AL, Dhole N, Patel A (2020) Stabilisation of black cotton soil for subgrade application using fly ash geopolymer. Road Mater Pavem Design 21(3):867–885. https://doi.org/10.1080/14680629.2018.1530131
Negi C, Yadav RK, Singhai AK (2013) Effect of silica fume on engineering properties of black cotton soil. Int J Comput Eng Res(IJCER): 83.
Patel RS, Desai MD (2010) CBR predicted by index properties for alluvial soils of South Gujarat. In: Proceedings of the Indian geotechnical conference, Mumbai (pp. 79–82).
Phani Kumar V, Manjula Ch, Poornima P (2015) Artificial neural networks (ANNS) for predicting of California bearing ratio of soils. Int J Mod Eng Res 5(1):15–21
Pradeep Kumar KJ, Harish Patel YM (2016) Soft computing technique for prediction of CBR form index properties of subgrade soil. Int J Innov Res Sci Eng Technol 5(7):13852–13860. https://doi.org/10.15680/IJIRSET.2016.0507214
Ramasubbarao G, Sankar SG (2013) Predicting soaked CBR value of fine grained soils using index and compaction characteristics. Jordan J Civil Eng 7(3):354–360
Ravichandra AH, Shivakumar K, Vinaykumar H, Md. Khalid S, Basavaraj B (2019) Prediction of CBR value by using index properties of soil. Int Res J Eng Technol 6(7):3740–3747
Reddy S, Ruchita N, Sharma P, Satyanarayana SV (2019) Prediction of California bearing ratio through empirical correlations of index properties for tropical indian soils. Int J Innov Eng Technol 15(1): 67–77. https://doi.org/10.21172/ijiet.151.09.
Rehman AU, Farooq K, Mujtaba H (2017a) Prediction of California bearing ratio (CBR) and compaction characteristics of granular soils. Acta Geotechnica Slovenica 14(1):63–72
Rehman ZU, Khalid U, Farooq K, Mujtaba H (2017b) Prediction of CBR value from index properties of different soils. Technol J Univ Eng Technol (UET) 22:17–26
Rohith MS, Kumar DRS, Paul W, KumaraSwamy N (2018) A study on the effect of stabilizers (zycobond and terrasil) on strength of subgrade on BC soil. Indian J Sci Res 17(2):86–92
Senol A, Bin-Shafique MS, Edil TB, Benson CH (2002) Use of class C fly ash for stabilization of soft subgrade. In: Fifth International Congress on Advances in Civil Engineering (pp. 25–27). Istanbul, Turkey: Istanbul Technical University.
Shirur NB, Hiremath SG (2014) Establishing relationship between CBR value and physical properties of soil. IOSR J Mech Civil Eng 11(5):26–30
Shivaramaiah A, Ravi Shankar AU, Singh A, Pammar KH (2020) Utilization of lateritic soil stabilized with alkali solution and ground granulated blast furnace slag as a base course in flexible pavement construction. Int J Pavem Res Technol 13(5):478–488. https://doi.org/10.1007/s42947-020-0251-5
Smith GN (1986) Probability and statistics in civil engineering. Collins professional and technical books, pp 244.
Talukdar DK (2014) A study of correlation between California bearing ratio (CBR) value with other properties of soil. Int J Emerg Technol Adv Eng 4(1):559–562
Taskiran T (2010) Prediction of California bearing ratio (CBR) of fine grained soils by AI methods. Adv Eng Softw 41(6):886–892. https://doi.org/10.1016/j.advengsoft.2010.01.003
Teklehaymanot FG, Alene E (2021) CBR predictive model development from soil index and compaction properties in case of fine-grained soils of Debre-Tabor City, Ethiopia. Int J Adv Sci Eng 8(2):2224–2234
Tenpe AR, Patel A (2020) Utilization of support vector models and gene expression programming for soil strength modeling. Arab J Sci Eng 45(5):4301–4319. https://doi.org/10.1007/s13369-020-04441-6
Tipping ME (2001) Sparse Bayesian learning and the relevance vector machine. J Mach Learn Res 1(Jun):211–244.
Vandana Rao P, Sudheer Kumar G, Prasanthi B (2018) A parametric study on black cotton soil stabilized using rice husk ash. Int J Adv Manag Technol Eng Sci 8(4):623–632
Venkatasubramanian C, Dhinakaran G (2011) ANN model for predicting CBR from index properties of soils. Int J Civ Struct Eng 2(2):614–620
Vu DQ, Nguyen DD, Bui QAT, Trong DK, Prakash I, Pham BT (2021) Estimation of California bearing ratio of soils using random forest based machine learning. J Sci Transp Technol:48–61
Vukićević M, Maraš-Dragojević S, Jocković S, Marjanović M, Pujević V (2013) Research results of fine-grained soil stabilization using fly ash from Serbian electric power plants. In: 18th International Conference on Soil Mechanics and Geotechnical Engineering (pp. 2–6).
Vukićević M, Pujević V, Marjanović M, Jocković S, Maraš-Dragojević S (2015) Fine grained soil stabilization using class F fly ash with and without cement. In: Proceedings of the XVI ECSMGE Geotechnical Engineering for Infrastructure and Development, ISBN 978–0–7277–6067–8, pp. 2671–2676.
Wilches FJ, Burbano JLA, Sierra EEC (2020) Subgrade soils characterization data, for correlation of geotechnical variables on urban roads in northern Colombia. Data Brief 32:106095. https://doi.org/10.1016/j.dib.2020.106095
Yin Z, Lekalpure RL, Ndiema KM (2022) Experimental study of black cotton soil stabilization with natural lime and pozzolans in pavement subgrade construction. Coatings 12(1):103. https://doi.org/10.3390/coatings12010103
Zhong J, Feng L, Ong YS (2017) Gene expression programming: A survey. IEEE Comput Intell Mag 12(3):54–72. https://doi.org/10.1109/MCI.2017.2708618
Funding
No funding was received in assisting the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
JK: main author, conceptualization, literature review, manuscript preparation, application of AI models, methodological development, statistical analysis, detailing, and overall analysis; KSG: conceptualization, manuscript finalization, detailed review and editing.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Khatti, J., Grover, K.S. Prediction of soaked CBR of fine-grained soils using soft computing techniques. Multiscale and Multidiscip. Model. Exp. and Des. 6, 97–121 (2023). https://doi.org/10.1007/s41939-022-00131-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41939-022-00131-y