Abstract
Biomedical relation extraction (RE) is critical in constructing high-quality knowledge graphs and databases as well as supporting many downstream text mining applications. This paper explores prompt tuning on biomedical RE and its few-shot scenarios, aiming to propose a simple yet effective model for this specific task. Prompt tuning reformulates natural language processing (NLP) downstream tasks into masked language problems by embedding specific text prompts into the original input, facilitating the adaption of pre-trained language models (PLMs) to better address these tasks. This study presents a customized prompt tuning model designed explicitly for biomedical RE, including its applicability in few-shot learning contexts. The model’s performance was rigorously assessed using the chemical-protein relation (CHEMPROT) dataset from BioCreative VI and the drug-drug interaction (DDI) dataset from SemEval-2013, showcasing its superior performance over conventional fine-tuned PLMs across both datasets, encompassing few-shot scenarios. This observation underscores the effectiveness of prompt tuning in enhancing the capabilities of conventional PLMs, though the extent of enhancement may vary by specific model. Additionally, the model demonstrated a harmonious balance between simplicity and efficiency, matching state-of-the-art performance without needing external knowledge or extra computational resources. The pivotal contribution of our study is the development of a suitably designed prompt tuning model, highlighting prompt tuning’s effectiveness in biomedical RE. It offers a robust, efficient approach to the field’s challenges and represents a significant advancement in extracting complex relations from biomedical texts.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Relation extraction (RE) is essential for fully utilizing unstructured data for biomedical research. Approximately 80% of biomedical data remains unstructured, making analysis challenging [1]. Information extraction techniques play a critical role in extracting valuable knowledge from unstructured text, which is a necessary step in automatically constructing high-quality biomedical databases and knowledge graphs to support downstream applications [2]. RE, which refers to extracting the semantic relations between the entities from unstructured documents, is one of the vital information extraction tasks in natural language processing (NLP) [3].
Although biomedical RE is an essential task, its current performance is not satisfactory due to the complexity of the task itself and lack of labeled data [4]. Error introduced from RE can affect downstream applications and hence reduce the reliability of the analysis. The emergence of machine learning methods significantly improves the performance of RE systems as compared with previous rules-based approaches. High-quality labeled corpora, however, are necessary for supervised machine learning models to achieve better RE performance [5]. Data annotation to build high-quality corpora, however, is a human labor-intensive task [4, 5]. Therefore, making the RE system perform better in few-shot scenarios is an important task [5].
Fine-tuning pre-trained language models (PLMs) to downstream tasks became a paradigm in the NLP field after the emergence of Bidirectional Encoder Representations from Transformers (BERT) [6]. Pre-training in the context of BERT variants refers to the initial training of these models from scratch using a large corpus. Conventionally, to harness the full potential of these pre-trained models, they should undergo fine-tuning with task-specific training data. This fine-tuned model is then deployed to address specific tasks. In this paper, we designate these models as “Conventional fine-tuned PLMs.” This terminology is used because, in our research, we did not pre-train the models from scratch; instead, we utilized models already fine-tuned and available on Hugging Face, and further fine-tuned them on our relation extraction training dataset. These models were then applied to our test sets. Hence, we refer to the baseline models in this study as conventional fine-tuned PLMs.
Recently, prompt tuning has tended to move conventional fine-tuned PLMs to a new paradigm in the NLP field [7]. Prompt tuning involves transforming NLP tasks into masked language problems by incorporating a specific text fragment, known as a prompt template, into the original input. This approach entails fine-tuning the PLMs in the format of masked language problems using task-specific training data. The fine-tuned PLMs are then utilized to solve these masked language problems by predicting the correct tokens from predefined label words that fit into the masked positions. This method of integrating a prompt template into the original input effectively adapts NLP tasks to be more amenable to solutions via PLMs [7]. Research has shown that prompt tuning can perform well in various NLP tasks [8,9,10,11] and their few-shot scenarios [12, 13].
This paper introduces a streamlined prompt tuning model, tailored for biomedical RE, including its application in few-shot scenarios. Additionally, this paper thoroughly evaluates the proposed model’s efficacy in this domain. To this end, the study conducts an extensive appraisal of models including BioBERT [14], BlueBERT [15], BioClinicalBERT [16], and PubMedBERT [17]. The chemical-protein relation (CHEMPROT) dataset of BioCreative VI [18] and the drug-drug interaction (DDI) dataset of SemEval-2013 [19] are utilized as evaluation benchmarks. Our results demonstrate that the proposed prompt tuning model outperformed the baseline, conventional fine-tuned PLMs across two datasets, encompassing few-shot scenarios. Moreover, the model demonstrated a harmonious balance between simplicity and efficiency, attaining comparable performance to the state-of-the-art models without the reliance on external knowledge resources and additional computational overhead.
2 Related Work
2.1 Biomedical RE
The methods used for biomedical RE have encompassed several stages, including rule-based methods, traditional machine learning methods, conventional deep learning methods, and fine-tuned PLMs. Starting from BERT [6], fine-tuned PLMs, in general, outperform the previous rule-based methods, traditional machine learning methods, and conventional deep learning methods in different NLP downstream tasks, including RE [20].
In terms of traditional machine learning methods, Warikoo et al. [21] integrated the linguistic patterns into the kernel methods and applied this model to the CHEMPROT dataset. Abacha et al. [22] integrated feature engineering into the kernel approaches and used this model on the DDI dataset. Overall, the traditional machine learning methods achieved reasonable performance (F1 score ~ 60%) on both datasets.
Considerable research has explored conventional deep learning methods to tackle biomedical RE, especially using the CHEMPROT dataset. Lim and Kang [2] proposed three recurrent neural networks (RNNs)—a tree-Long Short-Term Memory network (tree-LSTM) using additional features, a tree-LSTM with an extra preprocessing step, and a Stack-augmented Parser Interpreter Neural Network (SPINN). Corbett and Boyle [23] used an unlabeled corpus to pre-train the word embedding and multiple LSTM layers in the RE system. Peng et al. [24] ensembled a support vector machine (SVM), a convolutional neural network (CNN), and an RNN. Liu et al. [25] demonstrated that attention-based (ATT-) RNNs could outperform identical models without an attention mechanism. Mehryary et al. [26] proposed three systems—an SVM classifier; a shared task artificial neural network (ST-ANN) system, which consists of three LSTM chains; and an improved ANN (I-ANN) system, which adds a bidirectional LSTM layer. Zhang et al. [27] integrated deep context representation and multi-head attention with a bidirectional LSTM layer, which can extract more comprehensive features from a sentence and determine the important information. Antunes and Matos [28] integrated a relatively narrow representation of the relations into LSTM/CNN. Wang et al. [29] applied a Graph Convolutional Neural Network in biomedical RE. Notably, however, the performance of each of these methods is not optimal. The F1 score of all methods mentioned above is less than 70%.
The emerging transformer-based PLMs [30] have been used for many NLP tasks, including biomedical RE, and the studies obtained good performance [20]. For example, Sun et al. [31] proposed BERT-based attention-guided capsule (BERT-Att-Capsule) networks, which showed promising performance on the CHEMPROT dataset (F1 = 74.80%). They then integrated Gaussian probability into their model and achieved better performance (F1 = 76.56%) [32]. In addition, they applied the improved model to the DDI dataset and achieved promising performance (F1 = 82.04%) [32].
In terms of the joint model that can be used for extracting the entities and relations simultaneously, Zuo and Zhang [33] proposed a model on top of BERT, in which all spans are considered as candidate entities (F1 = 64.6% on BB—rel 2019 dataset [34]). Sun et al. [35] proposed a joint model based on BERT, which integrated a tagging strategy to address the overlapping triples in the dataset (F1 = 66.0% on CHEMPROT, F1 = 75.7% on DDI).
2.2 Prompt Tuning
Conventional fine-tuned PLMs contain BERT and BERT variants such as BioBERT [14], BlueBERT [15], BioClinicalBERT [16], and PubMedBERT [17] in the biomedical field. The model architectures are constructed by assembling transformer encoders. The models’ weights are initially initialized and subsequently pre-trained on a large text corpus. During the pre-training stage, two tasks are conducted: the masked language problem and the next sentence prediction [36]. The masked language problem task involves masking a portion of the input words, prompting the model to predict the masked tokens. Additionally, the next sentence prediction task entails determining whether a sentence logically follows another within a given context. The pre-training process involves iterative weight adjustments via backpropagation. The distinctiveness of each PLM predominantly arises from the specificities of the text corpora used for pre-training.
Prior to deploying PLMs for various NLP downstream tasks, such as sentiment analysis and named entity recognition, an additional step of fine-tuning is required. Fine-tuning entails the adaptation of PLMs to specific NLP tasks by training them on a task-specific corpus. This process refines the weights within the PLMs, rendering them more suitable for the intended downstream applications. Subsequently, these fine-tuned PLMs are employed to address the respective NLP downstream tasks.
Prompt tuning converts NLP downstream tasks into masked language problems through insertion of a prompt template, which refers to a piece of text that contains mask tokens, into the original input [7]. In this way, the format of the downstream tasks can become identical to the pre-training task, which can be better addressed by PLMs [7].
Emerging studies have yielded encouraging outcomes concerning the efficacy of prompt tuning across a spectrum of NLP tasks. This technique has been initially validated through its successful application in areas such as sentiment classification [8], dialog generation [9], factual probing [10], and text generation [37]. Furthermore, prompt tuning has demonstrated notable performance in RE within the general domain, as evidenced by multiple studies [38,39,40,41]. The utility of prompt tuning extends into few-shot scenarios, with research exploring its application in text classification [12, 13], natural language inference [12], dialog generation [9], and named entity recognition [42]. In the context of few-shot learning in RE, Sainz et al. [41] have implemented prompt tuning within the general field and substantiated its effectiveness in concurrent RE tasks and few-shot scenarios.
2.3 Prompt Tuning in Biomedical RE
Our previous work was one of the first attempts that presented a novel application of prompt tuning for biomedical RE utilizing the CHEMPROT dataset [43], showcasing its efficacy with BlueBERT [15] and PubMedBERT [17]. In a later study, Yeh et al. [44] furthered this exploration by applying prompt tuning to the same dataset using the RoBERTa-base and BioMed-RoBERTa-base models. However, these studies relied on a limited selection of benchmark datasets and models, which led to circumscribed performance outcomes. Specifically, our seminal research [43] yielded a macro-F1 score of 73.44, while Yeh et al. [44] recorded a score of 76.31. To enhance these outcomes, the current study introduces a refined methodology. This comprises a more suitable prompt template, informed by expert knowledge and the specific definitions of relation types. We also augment the tokenizers of PLMs with tokens absent from their native vocabularies. Furthermore, our approach encompasses a broader spectrum of benchmark datasets, specifically CHEMPROT and DDI, and a diverse array of PLMs including BioBERT, BlueBERT, BioClinicalBERT, and PubMedBERT.
Following these initial forays into the realm of prompt tuning for biomedical RE, Li et al. [45] introduced the BioKnowPrompt model. This innovative model merges knowledge injection with prompt tuning, manifesting noteworthy performance on both the CHEMPROT and DDI datasets. However, it’s pertinent to note that in real-world applications, expansive knowledge repositories and extensive computational resources might not always be accessible. Therefore, the introduction of a streamlined and efficient prompt tuning model, equipped with a well-designed prompt template, is essential. This approach aims to strike an optimal balance between simplicity and effectiveness, enabling comparable performance without the necessity for extensive knowledge resources and computational power.
3 Methods and Materials
As noted, this paper introduces a novel model, characterized by its simplicity and efficacy, for biomedical RE. It employs the CHEMPROT dataset of BioCreative VI [18] and the DDI dataset of SemEval-2013 [19] as the evaluation benchmark datasets. Detailed descriptions of these two datasets are provided in Sect. 3.1. Furthermore, the specifics of the proposed prompt tuning model are elucidated in Sect. 3.2.
3.1 Materials
3.1.1 CHEMPROT Dataset
One dataset used in this study is from the BioCreative VI Track 5—Text Mining Chemical-Protein Interactions (CHEMPROT) [18]. There are ten types of relations in this dataset (CPR:1 to CPR:10). As instructed by the BioCreative VI Track 5, five types of relations were used for evaluation purposes, including CPR:3, CPR:4, CPR:5, CPR:6, and CPR:9 (Table 1). Therefore, we focused only on these five types of relations to develop and evaluate our models. The dataset comprises a training set, development set, and test set, with 1020, 621, and 800 abstracts from the PubMed database, respectively.
3.1.2 DDI Dataset
The second dataset used in this study is from SemEval-2013 Track 9—Extraction of Drug-Drug Interactions from Biomedical Texts (DDI) [19]. As shown in Table 2, the relation classifications include DDI-advise, DDI-effect, DDI-int, and DDI-mechanism. This dataset consists of 792 narratives from the DrugBank database and 233 abstracts from Medline. The training set contains 624 files, and the test set contains 191 files. It should be noted that the development set needs to be separated from the training set.
3.2 Methods
3.2.1 Proposed Prompt Tuning Model
As explained in Sect. 2.2, conventional fine-tuned PLMs in our study specifically refer to BERT variants without prompt engineering. These models undergo pre-training on a large corpus and subsequent fine-tuning on specific NLP tasks. The primary difference between the proposed prompt tuning model and conventional fine-tuned PLMs lies in the prompt engineering, which consists of two components: a prompt template and label words, as depicted in Fig. 1. A prompt template is a piece of text inserted into the original input (illustrated as “w1 w2 w3 … wm.” in Fig. 1.), which has [MASK] tokens and can convert the initial NLP downstream tasks into masked language problems. The label words refer to the potential values inserted into the [MASK] tokens and should be pre-defined.
Han et al. [11] developed a prompt template and label words for RE in a general context, and our model was built upon the foundations of Han’s study. In this paper, we customized the prompt template and label words, guided by expert knowledge, to specifically match the unique features of our dataset and the definitions of the different relation types. For CHEMPROT, the prompt template was “@CHEMICAL$ [MASK] [MASK] [MASK] [MASK] @PROTEIN$.”; the label words were {is the upregulator of, is the downregulator of, is the modulator of, is the antagonist of, is the participant of, is not associated with}, each of which could be mapped to the corresponding relation classification in BioCreative VI Track 5 (i.e., CPR:3, CPR:4, CPR:5, CPR:6, CPR:9, and no_relation). Regarding DDI, the prompt template was “This describes [MASK] [MASK] [MASK] regarding drug-drug interactions.”; the label words were {a significant recommendation, a pharmacodynamic mechanism, a basic notification, a pharmacokinetic detail, an irrelevant subject}, which corresponded to the relation type {DDI-advise, DDI-effect, DDI-int, DDI-mechanism, no_relation}.
After prompt engineering, the fine-tuned PLMs can be utilized to pick the correct ones from the pre-defined label words and map them into the final relation classification to conduct the biomedical RE task. We utilized four PLMs, BioBERT (dmis-lab/biobert-large-cased-v1.1) [14], BlueBERT (bionlp/bluebert_pubmed_mimic_uncased_L-12_H-768_A-12) [15], BioClinicalBERT (emilyalsentzer/Bio_ClinicalBERT) [16], and PubMedBERT (microsoft/BiomedNLP-PubMedBERT-base-uncased-abstract-fulltext) [17], as the fine-tuned PLMs. These four PLMs have undergone pretraining on a biomedical corpus; consequently, they are well-suited for tasks within the domain of biomedical NLP. The corpus used for each of the four models varies slightly. Diverse PLMs employ tokenizers that operate with unique vocabulary lists. These tokenizers are utilized to convert sequences of characters into sequences of tokens. To guarantee that specific tokens in the label words remain intact and are not further split by the tokenizer, it is vital to add them directly to the tokenizer’s vocabulary list. This inclusion ensures that these tokens are acknowledged as discrete entities, thus preserving their intended meaning and representation. Table 3 provides details about the special tokens that have been appended to the vocabularies of various models.
Figure 2 presents the pipeline of the proposed prompt tuning model with a running example, which consists of pre-processing, prompt engineering, and RE.
The first step of the pipeline is pre-processing, which consists of splitting sentences in the corpus, identifying candidate entity pairs, and replacing the target entities with placeholders. First, each unit in the corpus was split into individual sentences. The sentences with more than one entity were assumed to potentially have candidate relations and, thus, were retained. We focused on intra-sentence relations due to limited coverage of cross-sentence relations from the datasets. Each pair of entities within a sentence was regarded as a relation-instance candidate. The original sentence that contained at least one chemical entity and one protein entity (for CHEMPROT) or two drug entities (for DDI) was included as one record in our dataset. The candidate relations that appeared in gold standard relations were labeled as positive relation instances, and the remaining ones were labeled as negative relation instances. In addition, chemical entities, protein entities, and drug entities tend to have complex names, which adds ambiguity to the model in predicting the relation between the entities using the context information in the sentences. To address this challenge, we replaced the entities of interest in the sentences with unified placeholder tags, such as @CHEMICAL$, @PROTEIN$, and @DRUG$. In the running example, the preprocessed text module shows one record in our dataset. The subject “15d-PGJ2” and the object “iNOS” between which candidate relations potentially existed were replaced by placeholders—“@CHEMICAL$” and “@PROTEIN$”.
The second step of the pipeline was prompt engineering. After inserting the prompt template into the preprocessed text, the RE task was converted into a masked language problem. As shown in the running example, the original input was converted into “In activated microglia, @CHEMICAL$ suppressed iNOS promoter activity, @PROTEIN$ mRNA, and protein levels. @ CHEMICAL$ [MASK] [MASK] [MASK] [MASK] @PROTEIN$.”, in which four [MASK] tokens represent the relation between the chemical and the protein. The corresponding label words that can be inserted into these four [MASK] tokens were pre-defined—{is the upregulator of, is the downregulator of, is the modulator of, is the antagonist of, is the participant of, is not associated with}.
The final step of the pipeline was relation classification—utilizing PLMs to predict the correct relation labels, which involve three sub-steps: fine-tuning, predicting, and label mapping. Fine-tuning refers to using the training set to adjust the weights in the PLMs to make them more appropriate for our task. Predicting refers to using fine-tuned PLMs to identify the correct label words that can fill in the [MASK] tokens from the pre-defined set, with an elaborate depiction provided in Fig. 3. The process retrieves the context vectors for the [MASK] tokens from the PLMs, the dimensions of which are the product of the number of [MASK] tokens and the hidden state dimension. Conversely, in accordance with the predefined label words, the possible insertions for each masked token varied: one for the first, two for the second, six for the third, and two for the fourth. These label words are converted into token IDs by the PLMs’ tokenizers and subsequently into embedding vectors by the word embedding layer of the PLMs. For each masked token within the framework, the dimensions of the resulting embedding vectors are computed as the product of the number of feasible insertions and the hidden state dimension. This is followed by a transposition of these vector dimensions. The subsequent process involves performing matrix multiplication between the context vectors for the [MASK] tokens and the embedding vectors of the label words, leading to the generation of logits that represent the choices of label words for each masked token position. This methodology is instrumental in determining the most suitable label word choice for each position. In the running example, the label words “is the downregulator of” were chosen to fill into the [MASK] tokens. Label mapping refers to mapping the label words to the final relation classification. In the running example, the label words “is the downregulator of” were mapped to the relation classification “CPR:4”. We used micro-F1, weighted-F1, and macro-F1 to evaluate the performance of the models.
3.2.2 Few-Shot Scenarios
In our paper, we examine four few-shot scenarios: 1-shot, 8-shot, and 16-shot. In a K-shot scenario, we sample “K” instances for each type of relation from the original complete training and validation sets. These sampled datasets then serve as the training and validation sets for the respective K-shot scenarios. For example, in the 1-shot scenario for the CHEMPROT dataset, we sample one instance from each category in both the training and validation sets. The sampled data then form the training and validation sets for the 1-shot scenario. For evaluation purposes, we retain the entire test set in all few-shot scenarios.
3.2.3 Experimental Settings
To ensure a fair comparison, identical hardware configurations were maintained for both baseline and proposed prompt tuning models. Furthermore, a standardized optimization process was adhered to across all models. The learning rate was selected from a predefined set: [1e-1, 5e-2, 1e-2, 5e-3, 1e-3, 5e-4, 1e-4, 5e-5, 1e-5, 5e-6, 1e-6, 5e-7]. The number of warmup steps was set to either 0 or 300, and the weight decay parameter was consistently maintained at zero. A fixed random seed of 42 was employed for reproducibility. For BlueBERT, BioClinicalBERT, and PubMedBERT, the training batch size was set at either 32 or 36, whereas for BioBERT, a smaller batch size of 8 or 12 was utilized. In the context of fully supervised learning, each model underwent training for 15 epochs. Conversely, in the few-shot learning scenarios, the number of training epochs for the models was extended to 20. The optimal epoch, as determined by the performance on the development set, was selected and used to evaluate the final performance on the test set. More detailed information regarding hyperparameters can be found in the supplementary_hyperparameter file.
4 Results
4.1 Data Preprocessing
For data preprocessing and partitioning, we adopted the methodology delineated in Peng’s study [46]. After preprocessing, one record in the dataset refers to an instance (either positive or negative) that contains at least one chemical entity and one protein entity, or two drug entities. For CHEMPROT, the training set consists of 19,460 relation instances, in which 4154 were positive and 15,306 were negative; the validation dataset consists of 11,820 relation instances, in which 2416 were positive and 9404 were negative; the test dataset consists of 16,943 relation instances, in which 3458 were positive and 13,485 were negative. For DDI, the training set consists of 18,779 relation instances, in which 2937 were positive and 15,842 were negative; the validation dataset consists of 7244 relation instances, in which 1004 were positive and 6240 were negative; the test dataset consists of 5761 relation instances, in which 979 were positive and 4782 were negative. Table 4 presents the statistical summarization of the CHEMPROT and DDI datasets for model development.
4.2 Performance of the Proposed Prompt Tuning Model in the Fully Supervised Setting
Table 5 presents a comparative analysis of various PLMs and their performance improvements when using proposed prompt tuning methodologies, specifically in the absence of external knowledge resources. Our evaluation spanned two distinct biomedical text mining tasks, namely CHEMPORT and DDI. We assessed the efficacy of each model using three established metrics: Micro-F1, Weighted F1, and Macro-F1 scores.
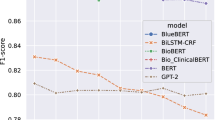
In the scope of previous research, Yeh et al.’s model is showcased, exhibiting a Micro-F1 score of 90.09 and a Macro-F1 score of 76.31 on the CHEMPORT task, with no reported scores for the DDI task [44]. The performance of conventional PLMs as reported in other studies is listed next, where only the Macro-F1 scores are provided. Among the models compared—BioBERT, BlueBERT, and PubMedBERT, PubMedBERT achieves the highest Macro-F1 score of 77.24 in the CHEMPROT task and 82.36 in the DDI task. To ensure a fair comparison, we conducted experiments with both the conventional PLMs and the proposed prompt tuning models. For our experimental results regarding conventional PLMs, a full spectrum of scores is presented. BioBERT displays strong performance with Macro-F1 score of 77.46 in the CHEMPORT task, and notably higher scores in the DDI task. BlueBERT and BioClinicalBERT show moderate performance, with BlueBERT achieving a Macro-F1 score of 71.47 and BioClinicalBERT a Macro-F1 score of 73.32 in the CHEMPORT task. PubMedBERT demonstrates robust performance with a Macro-F1 score of 77.22 in CHEMPORT and 82.26 in DDI, indicating a consistent high-level efficacy across both tasks.
The focus then shifts to our proposed prompt tuning models. These models apply a novel prompt tuning methodology without the use of external knowledge resources and exhibit enhancements in performance. For instance, the prompt-tuned BioBERT model demonstrates improvement by achieving a Macro-F1 score of 77.55 in CHEMPORT and a Macro-F1 score of 81.64 in DDI, thereby surpassing the scores of its conventional counterpart. Similarly, the prompt-tuned BlueBERT and BioClinicalBERT models demonstrate slight performance gains in certain metrics. The prompt-tuned PubMedBERT model not only surpasses its conventional version but also attains the highest Macro-F1 score of 78.02 in the CHEMPROT task and the highest Macro-F1 score of 82.54 in the DDI task among all the models presented.
4.3 Performance of the Proposed Prompt Tuning Model in the Few-Shot Scenarios
Table 6 illustrates the performance of our proposed prompt tuning models for few-shot scenarios. We observed notable variations in performance across different PLMs and the number of shots (K). Our results show that prompt tuning generally led to better performance compared to conventional PLMs. For instance, with BioBERT on the CHEMPORT task, the prompt tuning model exhibited a performance increase from 16.08 to 21.88 at K = 1, from 28.13 to 30.41 at K = 8, and from 34.91 to 39.77 at K = 16. These results affirm the potential of prompt tuning models in few-shot learning, highlighting their capacity to adapt and learn in data-constrained situations. For a detailed performance evaluation, please refer to the supplementary_hyperparameters file.
5 Discussion
The results demonstrate that the prompt tuned PubMedBERT model significantly outperforms conventional PLMs in both the CHEMPROT and DDI tasks, achieving the best results in these two tasks. Furthermore, in few-shot scenarios, the prompt-tuned BioBERT models emerged as the top performers. Collectively, these findings underscore the effectiveness of prompt tuning. This improvement may be attributed to the application of prompt engineering, which restructures the downstream biomedical RE task into a masked language problem format akin to the pre-training phase, thereby bridging the gap between the pre-training and fine-tuning stages. This alignment potentially explains the enhanced efficacy of PLMs when employing prompts for biomedical RE/classification. Notably, in most of the cases, as evidenced in Table 5 and 6, the proposed prompt tuning models demonstrate enhanced performance compared to their conventional fine-tuned PLM counterparts. This trend underlines the value of prompt tuning in augmenting the capabilities of conventional PLMs, though it’s important to acknowledge that its efficacy may vary depending on the specific model.
Our proposed prompt tuning models demonstrate exceptional efficiency, striking an optimal balance between simplicity and effectiveness, which is crucial in making our model both practical and accessible for a wider range of applications in biomedical RE. In contrast to rule-based methods, which demand substantial expert involvement and human resources, and traditional machine learning or conventional deep learning methods, which require extensive annotated corpora and thus significant expert input, our models achieve promising performance without necessitating expert-driven rule creation or corpus annotation. Furthermore, this study marks a significant advancement in the field of biomedical RE, achieving state-of-the-art results within prompt tuning models that do not rely on external knowledge resources. This positions our work at the forefront of current research employing prompt tuning in biomedical RE. Notably, our results remain competitive even when compared to the BioKnowPrompt model [45], which integrates prompt tuning with knowledge injection and incurs higher GPU costs for model development. This is a noteworthy achievement, considering the different data splitting methods and the larger training set used by BioKnowPrompt, in addition to their use of external resources.
This study underscores the efficiency and effectiveness of our streamlined model in biomedical RE, suggesting that prompt tuning is a viable strategy for optimizing performance in biomedical text mining applications. Such improvements have important implications for the rapid development of large-scale biomedical databases and knowledge graphs, enhancing their precision. Additionally, prompt tuning offers a practical alternative for NLP tasks, especially in situations where labeled datasets are limited.
Nonetheless, our study is not without its limitations. Our approach centered on four BERT model variants; future research will aim to integrate prompt tuning with more sophisticated RE models. For instance, Sun et al. [32] incorporated Gaussian probability distribution with BERT, resulting in substantial performance gains. Exploring such advanced models in conjunction with prompt tuning could potentially lead to further improvements. Our focus was also confined to CHEMPROT and DDI RE tasks, but we anticipate applying the model to a broader spectrum of biomedical RE benchmarks.
6 Conclusion
In our exploration of prompt tuning within biomedical RE, we utilized the CHEMPROT and DDI datasets as benchmarks for evaluation. Our findings suggest that prompt tuning can surpass the performance of baseline, conventionally fine-tuned PLMs in both full dataset and few-shot scenarios. We deduce that prompt engineering significantly enhances the efficacy of PLMs in biomedical RE tasks though this enhancement may vary depending on the specific model employed. This leads to the conclusion that with a well-designed prompt template, prompt tuning stands as an exceptionally effective approach for biomedical RE. Looking ahead, we plan to expand the application of our prompt tuning model to a broader range of biomedical RE tasks, thereby facilitating advancements in text mining, knowledge graph construction, and other related downstream applications.
References
SyTrue (2015) Why unstructured data holds the key to intelligent healthcare systems. Consultant HIT. https://hitconsultant.net/2015/03/31. Accessed 24 Jun 2023
Lim S, Kang J (2018) Chemical–gene relation extraction using recursive neural network. Database. https://doi.org/10.1093/database/bay060
Zelenko D, Aone C, Richardella A (2003) Kernel methods for relation extraction. J Mach Learn Res 3:1083–1106
Nasar Z, Jaffry SW, Malik MK (2021) Named entity recognition and relation extraction: state-of-the-art. ACM. Comput Surv. https://doi.org/10.1145/3445965
Shi Y, Xiao Y, Quan P, Lei M, Niu L (2021) Distant supervision relation extraction via adaptive dependency-path and additional knowledge graph supervision. Neural networks: the official journal of the International Neural Network Society. https://doi.org/10.1016/j.neunet.2020.10.012
Devlin J, Chang MW, Lee K, Toutanova K (2019) BERT: Pre-training of deep bidirectional transformers for language understanding. Proceedings of the 2019 conference of the North American chapter of the association for computational linguistics: human language technologies. https://doi.org/10.18653/v1/N19-1423
Liu P, Yuan W, Fu J, Jiang Z, Hayashi H, Neubig G (2023) Pre-train, prompt, and predict: a systematic survey of prompting methods in natural language processing. ACM Comput Surv. https://doi.org/10.1145/3560815
Li C, Gao F, Bu J, Xu L, Chen X, Gu Y, Shao Z, Zheng Q, Zhang N, Wang Y, Yu Z (2021) SentiPrompt: sentiment knowledge enhanced prompt-tuning for aspect-based sentiment analysis. arXiv. https://doi.org/10.48550/arXiv.2109.08306
Zheng C, Huang M (2021) Exploring prompt-based few-shot learning for grounded dialog generation. arXiv. https://doi.org/10.48550/arXiv.2109.06513
Zhong Z, Friedman D, Chen D (2021) Factual probing is [MASK]: learning vs. learning to recall. arXiv. https://doi.org/10.48550/arXiv.2104.05240
Han X, Zhao W, Ding N, Liu Z, Sun M (2021) PTR: prompt tuning with rules for text classification. arXiv. https://doi.org/10.1016/j.aiopen.2022.11.003
Schick T, Schütze H (2020) Exploiting cloze questions for few shot text classification and natural language inference. arXiv. https://doi.org/10.48550/arXiv.2001.07676
Schick T, Schmid H, Schütze H (2020) Automatically identifying words that can serve as labels for few-shot text classification. arXiv. https://doi.org/10.48550/arXiv.2010.13641
dmis-lab (2020) Biobert-large-cased-v1.1. Hugging face. https://huggingface.co/dmis-lab/biobert-large-cased-v1.1. Accessed 15 Oct 2023
bionlp (2020) Bluebert_pubmed_mimic_uncased_L-12_H-768_A-12. Hugging face. https://huggingface.co/bionlp/bluebert_pubmed_mimic_uncased_L-12_H-768_A-12. Accessed 15 Oct 2023
emilyalsentzer (2020) Bio_ClinicalBERT. Hugging face. https://huggingface.co/emilyalsentzer/Bio_ClinicalBERT. Accessed 15 Oct 2023
Microsoft (2021) BiomedNLP-BiomedBERT-base-uncased-abstract-fulltext. hugging face. https://huggingface.co/microsoft/BiomedNLP-BiomedBERT-base-uncased-abstract-fulltext. Accessed 19 Nov 2023
Krallinger M, Rabal O, Akhondi SA, Perez M, Santamaria J, Rodríguez GP, Tsatsaronis G, Intxaurrondo A, López JAB, Nandal U, Buel EV, Chandrasekhar A, Rodenburg M, Lægreid A, Doornenbal MA, Oyarzábal J, Lourenço A, Valencia A (2017) Overview of the BioCreative VI chemical-protein interaction track. Semantic Scholar. https://www.semanticscholar.org/paper/Overview-of-the-BioCreative-VI-chemical-protein-Krallinger-Rabal/eed781f498b563df5a9e8a241c67d63dd1d92ad5. Accessed 15 Oct 2021
Herrero-Zazo M, Segura-Bedmar I, Martínez P, Declerck T (2013) The DDI corpus: an annotated corpus with pharmacological substances and drug–drug interactions. J Biomed Inform. https://doi.org/10.1016/j.jbi.2013.07.011
Li Z, Lin H, Shen C, Zheng W, Yang Z, Wang J (2020) Cross2Self-attentive bidirectional recurrent neural network with BERT for biomedical semantic text similarity. 2020 IEEE International Conference on Bioinformatics and Biomedicine. https://doi.org/10.1109/BIBM49941.2020.9313452
Warikoo N, Chang YC, Hsu WL (2018) LPTK: a linguistic pattern-aware dependency tree kernel approach for the BioCreative VI CHEMPROT task. Database. https://doi.org/10.1093/database/bay108
Ben Abacha A, Chowdhury MFM, Karanasiou A, Mrabet Y, Lavelli A, Zweigenbaum P (2015) Text mining for pharmacovigilance: using machine learning for drug name recognition and drug-drug interaction extraction and classification. J Biomed Inform. https://doi.org/10.1016/j.jbi.2015.09.015
Corbett P, Boyle J (2018) Improving the learning of chemical-protein interactions from literature using transfer learning and specialized word embeddings. Database. https://doi.org/10.1093/database/bay066
Peng Y, Rios A, Kavuluru R, Lu Z (2018) Extracting chemical–protein relations with ensembles of SVM and deep learning models. Database. https://doi.org/10.1093/database/bay073
Liu S, Shen F, Komandur Elayavilli R, Wang Y, Rastegar-Mojarad M, Chaudhary V, Liu H (2018) Extracting chemical-protein relations using attention-based neural networks. Database: the journal of biological databases and curation. https://doi.org/10.1093/database/bay102
Mehryary F, Björne J, Salakoski T, Ginter F (2018) Potent pairing: ensemble of long short-term memory networks and support vector machine for chemical-protein relation extraction. Database: the journal of biological databases and curation. https://doi.org/10.1093/database/bay120
Zhang Y, Lin H, Yang Z, Wang J, Sun Y (2019) Chemical–protein interaction extraction via contextualized word representations and multihead attention. Database. https://doi.org/10.1093/database/baz054
Antunes R, Matos S (2019) Extraction of chemical–protein interactions from the literature using neural networks and narrow instance representation. Database. https://doi.org/10.1093/database/baz095
Wang E, Wang F, Yang Z, Wang L, Zhang Y, Lin H, Wang J (2020) A graph convolutional network-based method for chemical-protein interaction extraction: algorithm development. JMIR medical informatics. https://doi.org/10.2196/17643
Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser L, Polosukhin I (2017) Attention is all you need. arXiv. https://doi.org/10.48550/arXiv.1706.03762
Sun C, Yang Z, Wang L, Zhang Y, Lin H, Wang J (2020) Attention guided capsule networks for chemical-protein interaction extraction. J Biomed Inform. https://doi.org/10.1016/j.jbi.2020.103392
Sun C, Yang Z, Su L, Wang L, Zhang Y, Lin H, Wang J (2020) Chemical–protein interaction extraction via gaussian probability distribution and external biomedical knowledge. Bioinformatics. https://doi.org/10.1093/bioinformatics/btaa491
Zuo M, Zhang Y (2021) A span-based joint model for extracting entities and relations of bacteria biotopes. Bioinformatics. https://doi.org/10.1093/bioinformatics/btab593
Corpus Statistics (2019) BB 2019. https://sites.google.com/view/bb-2019/dataset/. Accessed 19 Jan 2024
Sun C, Yang Z, Wang L, Zhang Y, Lin H, Wang J (2022) MRC4BioER: joint extraction of biomedical entities and relations in the machine reading comprehension framework. J Biomed Inform. https://doi.org/10.1016/j.jbi.2021.103956
google research (2018) Bert: tensorFlow code and pre-trained models for BERT. Github. https://github.com/google-research/bert. Accessed 17 Sep 2022
Guo H, Tan B, Liu Z, Xing EP, Hu Z (2021) Text generation with efficient (soft) Q-learning. arXiv. https://doi.org/10.48550/arXiv.2106.07704
Chen X, Li L, Zhang N, Tan C, Huang F, Si L, Chen H (2022) Relation extraction as open-book examination: retrieval-enhanced prompt tuning. arXiv. https://doi.org/10.1145/3477495.3531746
Chen X, Zhang N, Li L, Yao Y, Deng S, Tan C, Huang F, Si L, Chen H (2022) Good visual guidance make a better extractor: hierarchical visual prefix for multimodal entity and relation extraction. Findings of the Association for Computational Linguistics. https://doi.org/10.18653/v1/2022.findings-naacl.121
Chen X, Zhang N, Xie X, Deng S, Yao Y, Tan C, Huang F, Si L, Chen H (2022) KnowPrompt: knowledge-aware prompt-tuning with synergistic optimization for relation extraction. Proceedings of the ACM Web Conference 2022. https://doi.org/10.1145/3485447.3511998
Sainz O, de Lacalle OL, Labaka G, Barrena A, Agirre E (2021) Label verbalization and entailment for effective zero and few-shot relation extraction. Proceedings of the 2021 Conference on Empirical Methods in Natural Language Processing. https://doi.org/10.18653/v1/2021.emnlp-main.92
Ma R, Zhou X, Gui T, Tan Y, Li L, Zhang Q, Huang X (2021) Template-free prompt tuning for few-shot NER. arXiv. https://doi.org/10.48550/arXiv.2109.13532
He J, Li F, Hu X, Li J, Nian Y, Wang J, Xiang Y, Wei Q, Xu H, Tao C (2022) Chemical-protein relation extraction with pre-trained prompt tuning. IEEE Int Conf Healthc Inform. https://doi.org/10.1109/ichi54592.2022.00120
Yeh HS, Lavergne T, Zweigenbaum P (2022) Decorate the examples: a simple method of prompt design for biomedical relation extraction. arXiv. https://doi.org/10.48550/arXiv.2204.10360
Li Q, Wang Y, You T, Lu Y (2022) BioKnowPrompt: incorporating imprecise knowledge into prompt-tuning verbalizer with biomedical text for relation extraction. Inf Sci. https://doi.org/10.1016/j.ins.2022.10.063
Peng Y, Yan S, Lu Z (2019) Transfer learning in biomedical natural language processing: an evaluation of BERT and ELMo on ten benchmarking datasets. Proceedings of the 18th BioNLP Workshop and Shared Task. https://doi.org/10.18653/v1/w19-5006
Lee J, Yoon W, Kim S, Kim D, Kim S, So CH, Kang J (2020) BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. https://doi.org/10.1093/bioinformatics/btz682
Gu Y, Tinn R, Cheng H, Lucas M, Usuyama N, Liu X, Naumann T, Gao J, Poon H (2022) Domain-specific language model pretraining for biomedical natural language processing. ACM Transactions on Computing for Healthcare. https://doi.org/10.1145/3458754
Funding
The study was partly supported by the National Institute on Aging under Project Nos. R01AG084236, R56AG074604, U24AI171008, and RF1AG072799.
Author information
Authors and Affiliations
Contributions
CT supervised the study. CT, FL, and YX contributed to the study conception and design. JH, JL, XH, YN, JW, QW, and YL contributed to data acquisition, processing, and model development. JH was responsible for drafting of the manuscript. CT, FL, and HX were responsible for critical revision of the manuscript. All authors revised and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical Approval
Not applicable.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
He, J., Li, F., Li, J. et al. Prompt Tuning in Biomedical Relation Extraction. J Healthc Inform Res 8, 206–224 (2024). https://doi.org/10.1007/s41666-024-00162-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41666-024-00162-9