Abstract
Banana bunchy top virus (BBTV), banana streak viruses (BSVs) and cucumber mosaic virus (CMV) are frequently reported infecting bananas globally. Effective control of their spread depends on robust detection of these viruses in propagation stock, planting material, infected nursery plants, and through strict quarantine. We developed single reverse transcription loop-mediated isothermal amplification (RT-LAMP) assays for BBTV, banana streak OL virus (BSV-OL) and CMV that were sensitive, specific, efficient, and completed in less than 60 min. RNA-based RT-LAMP minimized false positives that arose from banana genomes harboring endogenous viral genomes, such as BSVs. RT-LAMP was also more sensitive than RT-PCR in detecting the DNA viruses, BBTV and BSV-OL, in infected plants. We also developed a multiplex assay using three sets of primers specific for each virus to simultaneously detect BBTV, BSV-OL and CMV in a sample of RNA from the same plant. The reliability and convenience of this assay makes it useful for plant quarantine and indexing plants for propagation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bananas and plantains (Musa spp. L., Musaceae, Zingiberales) are large perennial herbs vital to food security in many tropical and subtropical countries (D’Hont et al. 2012). They are the sixth-ranked food crop produced worldwide following maize, rice, wheat, potatoes, and cassava, with 139 million tons produced in 2012 (Kumar et al. 2015). Bananas are cultivated in nearly 120 countries (Rustagi et al. 2015) and are the major staple food and income for millions of people (Chai et al. 2015). Bananas are propagated by suckers, division of the rhizome (corm), or by micropropagation (Israeli et al. 1995). The use of suckers and rhizomes as planting stock, however, is responsible for the spread of many pests and pathogens, especially viruses (Kumar et al. 2015).
Banana bunchy top virus (BBTV), banana streak viruses (BSVs), and cucumber mosaic virus (CMV) are frequently reported infecting bananas globally (Lockhart 2000; Thiribhuvanamala and Doraisamy 2001; Helliot et al. 2002; Kumar et al. 2009; Adegbola et al. 2013; Amrita et al. 2014; Boloy et al. 2014; Javer-Higginson et al. 2014; Wang et al. 2014). BBTV, the causal agent of banana bunchy top disease, belongs to the type species Banana bunchy top virus, genus Babuvirus, in the family Nanoviridae (King et al. 2012). Its spherical virions are ~18–19 nm in diameter (Thomas and Dietzgen 1991) and the genome is composed of at least six individual single-stranded DNAs, each about 1 kb in size (Xie and Hu 1995) and packaged in separate particles. BSVs cause banana streak disease and include virus isolates from nine species: Banana streak MY virus, Banana streak OL virus, Banana streak UA virus, Banana streak UI virus, Banana streak UL virus, Banana streak UM virus, Banana streak VN virus, Banana streak GF virus and Banana streak IM virus. These species are in the genus Badnavirus, family Caulimoviridae (Lockhart 1986). Their non-enveloped bacilliform virions measure 30 nm in width by 150 nm in length (Geering et al. 2000) and contain a circular, double-stranded DNA genome of 7.2–7.8 kb (Kumar et al. 2015). The sequences of banana streak virus isolates from Hawaii were closely related to those of banana streak OL virus (BSV-OL), and some undetermined BSV isolates from India, China, and Colombia. Thus, the sequences of the isolates in this species were used to develop the detection assays in this study. CMV causes a mosaic-like chlorosis and heart rot of bananas. In China, CMV can infect 40–90% of the banana plants in fields (Hu et al. 1995). It belongs to the type species, Cucumber mosaic virus, genus Cucumovirus in the family Bromoviridae. CMV has a segmented, tripartite, linear, single-stranded, positive-sense RNA genome composed of RNA1 (3.4 kb), RNA2 (3.1 kb), and RNA3 (2.2 kb) (Palukaitis et al. 1992; Hu et al. 1995).
Although resistant cultivars are the most convenient and effective way to control many plant diseases, banana plants with a high resistance to these viruses are not currently available (Kumar et al. 2015). Effective control is possible only through early identification and elimination of virus-infected plants followed by propagation and planting of virus-free material (Hu et al. 1995). To aid in the rapid, convenient, sensitive, and reliable identification of virus infections, we have developed a method for multiplex detection of these three viruses in banana.
Loop-mediated isothermal amplification (LAMP) is a recently developed molecular tool that uses a DNA polymerase with DNA displacement activity to generate amplification products at a single temperature (Notomi et al. 2000). Compared to conventional PCR, LAMP is simple, rapid, specific, and cost-effective (Tomita et al. 2008). It has been used successfully to detect viruses from several plant species (Fukuta et al. 2003). A modification of the LAMP reaction known as reverse transcription LAMP (RT-LAMP) relies on the synthesis of cDNAs from RNA templates, followed by LAMP technology to detect RNA viruses in plants using a one-step reaction in a single tube (Parida et al. 2004). It has been used to detect CMV (Fukuta et al. 2005), plum pox virus (Varga and James 2006), several viruses of rice (Le et al. 2010), and cymbidium mosaic virus from orchids (Lee et al. 2011). This study describes our development of a RT-LAMP multiplex assay to simultaneously detect different DNA and RNA viruses in banana. The assay can be used for the rapid and sensitive indexing of BBTV, BSV-OL, and CMV in banana plants that are used for high-throughput, virus-free propagation.
Materials and methods
Plant materials
To develop single and multiplex RT-LAMP assays, we collected plant material from various Musa genotypes growing at the Waimanalo Research Station, College of Tropical Agriculture and Human Resources (CTAHR), University of Hawaii at Manoa, located in Honolulu County, HI, USA. The material included twelve samples from plants with typical symptoms of banana bunchy top disease: ‘Mysore’ (AAB Mysore), ‘Giant Plantain’ (AAB Plantain), ‘Kifutu’ (AAB Pome), ‘Foconah’ (AAB Pome), ‘Pome’ (AAB Pome), ‘Goldfinger’ (AAAB hybrid), and several others; four from plants that had symptoms resembling banana streak disease: ‘Mysore’, ‘Giant Plantain’, ‘Pome’, and ‘Goldfinger’; and four from plants that had typical symptoms of cucumber mosaic disease. Virus-free tissue-culture samples of ‘Dwarf Brazilian’ (AAB, Pome subgroup) were used as controls.
To screen samples from the field using the newly developed multiplex assay, we collected nineteen banana samples from different locations on the island of Oahu. These samples included plants with symptoms of BBTV, BSVs, or CMV infections, plus asymptomatic plants. We also collected two samples of honohono grass (Commelina diffusa N.L. Burm.) with symptoms of CMV. Honohono grass is a known weedy reservoir of CMV, which can be transmitted to banana (Ferreira 1992).
Total DNA and RNA exaction
Total DNA and RNA were isolated from banana samples respectively using the DNeasy Plant Mini kit (Qiagen) and RNeasy Plant Mini kit (Qiagen), according to manufacturer’s instructions. Isolated RNAs were then treated with RQ1 RNase-free DNase I (Promega) prior to conducting RT-LAMP and RT-PCR assays. The DNase I digestion reactions were as follows: ~100 ng RNA, 1.0 μL 10× reaction buffer, 1.0 μL RQ1 RNase-free DNase I and nanopure H2O to a final volume of 10 μL (final concentration of RNA ~10 ng/μL). Samples were then incubated at 37 °C for 30 min and the reaction stopped by adding 1.0 μL of RQ1 DNase I stop solution (Promega), and then incubated for 10 min at 65 °C.
PCR and RT-PCR
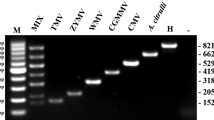
First-strand cDNA synthesis was initiated with 2 μL of DNase I-treated RNA and 1.0 μL of random primers (50 ng/μL) (Promega), heated at 70 °C for 5 min and then quenched on ice. Next, 5 μL M-MLV 5× reaction buffer (Promega), 5 μL dNTPs (2.5 μM each), 0.5 μL recombinant RNasin ribonuclease inhibitor (40 U/μL), and 1.0 μL M-MLV reverse transcriptase (200 U/μL) were added and the reaction brought to a final volume of 25 μL with nanopure H2O. This assembled reaction was added to the annealed primer/template and incubated for 60 min at 37 °C. The PCR reaction conditions were: 10 μL 2× GoTaq Green master mix (Promega), 0.5 μL each forward/reverse primer, 1.0 μL DNA or cDNA and 8 μL nanopure H2O. The BBTV primers were CPXI.PRI (5′- GCT AGG TAT CCG AAG AAA TCC -3′) and BBTV3C.EXP (5′- ATA AAG CTT TCA AAC ATG ATA TGT-3′), with a predicted product size of 513 bp (Wanitchakorn et al. 2000). The BSOLV primers were P1 (5′- AAG CTT CGG GTA CAC AAA ATA TCA TC -3′) and P2 (5′- GGA TCC GGT TTT CTT AAC TTC TTC -3′), with a predicted product size of 945 bp, based on the conserved sequence of the coat protein of an isolate from China previously tested positive for BSV-OL (Tan et al. 2010). The CMV primers were P1 (5′- TAT GAT AAG AAG CTT GTT TCG CG -3′) and P2 (5′- GCC GTA AGC TGG ATG GAC AA -3′), with a predicted product size of 480 bp (Tan et al. 2010). The PCR program was as follows: 95 °C for 5 min; 35 cycles at 95 °C for 30 s; annealing at 49 °C (BBTV), or 51 °C (BSV-OL), or 56 °C (CMV) for 30 s; 72 °C for 1 min; and a final extension at 72 °C for 10 min.
LAMP and RT-LAMP
Conserved regions of the BBTV, BSV-OL, and CMV genome sequences were identified with Vector NTI (Invitrogen) by alignment of: 42 BBTV coat protein sequences; 70 BSV RNase H sequences that included BSV-OL isolates and undetermined BSV isolates from various genotypes of banana from India, China and Colombia; and 44 CMV coat protein sequences available from NCBI. These conserved regions were used to design LAMP primers using Primer Explorer 4.0 (Eiken Chemical Co., http://primerexplorer.jp/e/) (Table 1). Each 25-μL LAMP reaction contained 2.5 μL 10× ThermoPol reaction buffer (Mg2+-free, New England Biolabs), MgSO4 (10 mM), dNTPs (1.2 mM each), primers LAMP-F3/B3 (0.2 μM each), LAMP-FIP/BIP (1.6 μM each), and LAMP-LF/LB (0.8 μM each), betaine (1 M) (Affymetrix), Bacillus stearothermophilus DNA polymerase (8 U) (New England Biolabs), EvaGreen (450 nM; Biotium), DNA or RNA templates (1.0 μL) and nanopure H2O to a final volume of 25 μL. For RT-LAMP, M-MLV reverse transcriptase (40 U) (Promega) was added to the reaction mix. Negative controls contained 1.0 μL of nanopure H2O as a template. LAMP and RT-LAMP were performed on an iQ5 Multicolor Real-Time PCR Detection System (Bio-Rad) under reaction conditions of 65 °C for 60 min (one minute/cycle) with fluorescence readings taken after each minute, followed by final inactivation at 85 °C for 2 min.
Specificity of RT-LAMP
To determine the specificity of the RT-LAMP reactions, we used plant samples that were each infected with isolates of one of the following viruses: BBTV, BSV-OL, CMV from banana, tomato spotted wilt virus (TSWV) from tomato and basil, maize mosaic virus (MMV) and maize chlorotic mottle virus (MCMV) from maize, or pineapple mealybug wilt-associated virus-1 (PMWaV-1) from pineapple. Each set infected with one of the viruses included two plant samples and was considered a double repetition. Virus-free banana samples and non-template controls were used as negative controls. The procedure was the same as used in the LAMP and RT-LAMP assays described above.
Sensitivity of RT-LAMP
RQ1 DNase I-treated RNA samples were serially diluted to assess the sensitivity of the RT-LAMP assay. Because the RNA concentrations in the samples were diluted by the DNase I treatment, the RNA concentrations were adjusted to reflect this dilution (~10 ng/μL). Tenfold serial dilutions of the RNA templates in nanopure H2O (100–10−9) were used to assess the sensitivity of RT-LAMP assay for each virus.
Multiplex RT-LAMP
We used primer sets specific for each of the three viruses in the single-virus assay and combined them in a multiplex assay. Each of the three primer sets used in the multiplex assay contained either the same concentrations and volumes (i.e., 3 × 0.2 μM LAMP-F3/B3; 3 × 1.6 μM LAMP-FIP/BIP; 3 × 0.8 μM LAMP-LF/LB) as were used in the individual specific RT-LAMP assays, or 0.5-fold concentrations and volumes (i.e., 3 × 0.1 μM LAMP-F3/B3; 3 × 0.8 μM LAMP-FIP/BIP; 3 × 0.4 μM LAMP-LF/LB) as were used in the individual specific RT-LAMP assays. H2O-only templates and RNAs from uninfected plant tissues were used as negative controls. The samples collected from the field (see Plant Materials section above) were tested using the developed multiplex assay.
Results
Molecular identification of virus-infected samples using RT-LAMP
We found that DNA-based PCR and LAMP assays consistently produced BSV amplicons from virus-free, asymptomatic banana samples (data not shown), suggesting that endogenous BSV sequences were being amplified in these assays. To avoid this potential problem, we developed RNA-based RT-LAMP assays that incorporated a DNase I pretreatment. Because BBTV and BSV-OL replication includes RNA intermediates, RNA templates can be used to develop a multiplex detection assay for these viruses. All RNA templates from the samples that generated PCR amplicons in BBTV assays (samples B1-B12) and RT-PCR amplicons in CMV assays (samples C1-C4) produced exponential amplification curves within 60 min in RT-LAMP reactions (Fig. 1). No RNAs from healthy controls and H2O-only templates produce amplicons within 60 min. However, only four of five PCR-positive BSV-OL samples, S1, S3, S4 and B8, showed positive amplification using RT-LAMP (Fig. 2a). When RT-LAMP assays were longer than 60 min, non-specific amplification products were produced by the primers, as indicated for the H2O-only templates. The virus could not be detected from sample S2 within 60 min. Comparison of the DNase I-treated to the non-treated RNA templates from healthy controls (Fig. 2b), confirmed that DNase I treatment effectively minimized the false positives in the BSV-OL RT-LAMP assay.
RT-LAMP amplification using BBTV and CMV primers. a Twelve PCR-positive BBTV samples (B1–B12) with exponential amplification standard curves; b Four RT-PCR-positive CMV samples (C1–C4) with exponential amplification standard curves. All produced within 60 min. The healthy control and nanopure H2O-only samples showed only baseline fluorescence (RFU = 0), indicating negative amplification. Green curves = BBTV PCR-positive banana samples, blue curves = CMV RT-PCR-positive banana samples, orange curves = healthy controls, and gray curves = nanopure H2O-only templates
RT-LAMP amplification with BSV-OL primers. a Four PCR-positive BSV-OL samples (S1, S3, S4, B8) with exponential amplification standard curves generated within 60 min, except for S2; b Healthy control RNA with DNase I treatment was negative, while healthy control RNA without DNase I treatment produced positive reactions. Red curves = BSV-OL PCR-positive samples, orange curves = healthy control, and gray curves = nanopure H2O-only templates
Comparison of RT-LAMP with RT-PCR for detecting DNA viruses
Four PCR-positive BBTV samples (B1-B4) and two PCR-positive BSV-OL samples (S1-S2), were assayed using RT-PCR and results were compared to those from RT-LAMP assays using the same RNA samples as templates. The four BBTV-infected samples and one of the BSV-OL-infected samples (S1) produced detectable amplicons when tested using RT-PCR with non-DNase I-treated RNA samples (Fig. 3a). Viruses in the DNase I-treated samples that were difficult to detect with RT-PCR (Fig. 3b) were easily detected by the RT-LAMP assay (Figs. 1a and 2a).
RT-PCR detection of BBTV and BSV-OL. a Non-DNase I-treated RNA templates of S1–S2 (BSV-OL PCR-positive) and B1–B4 (BBTV PCR-positive) tested with RT-PCR; b DNase I-treated RNA templates tested with RT-PCR. Upper panel = BBTV, lower panel = BSV-OL. H = DNase I-treated healthy control, H* = DNase I-untreated healthy control, C = cDNA synthesized with nanopure H2O instead of RNA, N = nanopure H2O as template
Specificity and sensitivity of RT-LAMP for each virus
RNA templates of two BBTV-infected samples produced exponentially increasing fluorescence in RT-LAMP assays specific for BBTV, while samples infected with BSV-OL, CMV, and the other tested viruses, TSWV, MMV, MCMV and PMWaV-1, produced no amplified products in BBTV-specific assays (Table 2). RT-LAMP assays designed specifically for BSV-OL or CMV produced results similar to the results from the BBTV-specific assays; only samples infected with BSV-OL or CMV were detected when using BSV-OL- or CMV-specific primers (Table 2). These results showed that each of the three assays was specific for its target virus.
Tenfold serial dilutions of the RNA templates in H2O, beginning with an RNA concentration of ~10 ng/μL, were used to determine the sensitivity of each assay. The BBTV (Fig. 4a) and CMV (Fig. 4c) RT-LAMP assays produced amplicons within 60 min with 102-fold template dilutions (~0.1 ng/μL). The RT-LAMP assay for BSV-OL (Fig. 4b), produced amplicons within 60 min with tenfold dilutions of the RNA templates (~1.0 ng/μL).
Sensitivity determination of RT-LAMP for BBTV, BSV-OL, and CMV. a BBTV RT-LAMP assay; b BSV-OL RT-LAMP assay; c CMV RT-LAMP assay. Initial RNA template concentrations were 10 ng/μL. Tenfold serial dilutions (100–109) were made in nanopure H2O. Green curves = BBTV-positive banana samples, red curves = BSV-OL-positive banana samples, blue curves = CMV-positive banana samples, and gray curves = nanopure H2O-only templates
Development of a multiplex detection assay for BBTV, BSV-OL, and CMV
We based development of the assay to simultaneously detect all three viruses on the individual RNA-based RT-LAMP assays. We then added equal volumes of each primer set and tested the multiplex assay. However, H2O-only templates and RNA templates from healthy controls also produced false-positive, non-specific amplicons within 60 min (Fig. 5a). When all of the primer concentrations were reduced one half (0.5-fold), only RNA templates from virus-infected samples produced amplicons within 60 min (Fig. 5b). The healthy and H2O-only controls were both negative within this time. This confirmed that the lower concentrations of primers used in the multiplex RT-LAMP assay was sufficient to reliably detect the viruses within 60 min, with no false positives from the healthy or H2O-only templates.
Evaluation of primer concentrations and reaction times in the development of the multiplex assay. a Multiplex assay using equal volumes of each primer-set used in the individual specific RT-LAMP assays; b Multiplex assay using 0.5-fold lower volumes of each primer-set used in the individual specific RT-LAMP assays. Green curves = BBTV-positive banana samples, red curves = BSV-OL-positive banana samples, blue curves = CMV-positive banana samples, orange curves = healthy control, and gray curves = nanopure H2O-only templates
Field screening using the multiplex detection assay
The virus status of nineteen symptomatic and non-symptomatic banana samples and two symptomatic honohono grass samples collected from the field was validated by PCR, RT-PCR and LAMP prior to being assayed by multiplex RT-LAMP. Of the 19 banana samples, five had typical symptoms of banana bunchy top disease, five had symptoms of banana streak disease, and two showed symptoms of CMV, as did both of the honohono grass samples. When assayed by multiplex RT-LAMP, each symptomatic sample tested positive for at least one of the three viruses, while all asymptomatic samples tested free of viruses (Fig. 6). These results demonstrated that the multiplex assay could efficiently and reliably detect viruses from infected banana and other plants. This screening was repeated four times with similar results, indicating that the multiplex assay was robust and repeatable.
A multiplex assay of banana samples from different locations on Oahu, Hawaii, USA. Banana samples included plants symptomatic for BBTV, BSV-OL, and CMV, plus asymptomatic plants and virus-free plants. Four replicates of each sample were analyzed in four individual assays, and this figure represents a typical assay. Green curves = banana samples with BBTV symptoms, red curves = samples with BSV-OL symptoms, blue curves = banana samples with CMV symptoms, yellow curves = honohono grass samples with CMV symptoms, purple curves = asymptomatic banana samples, orange curves = healthy control, and gray curves = nanopure H2O-only templates
Discussion
Integrated BSV sequences within banana genomes, referred to as endogenous BSV sequences (eBSVs), are widespread in banana species and cultivars (Duroy et al. 2016). The genomes of Musa acuminata and M. balbisiana both contain these endogenous sequences (Gayral et al. 2010; D’Hont et al. 2012), but eBSVs in bananas that have the ability to form virions have been documented only in M. balbisiana (Gayral et al. 2008; Chabannes et al. 2013; Kumar et al. 2015). The accurate diagnosis of plant-pathogenic BSV infections is therefore a major challenge, primarily due to the presence of these endogenous sequences. Immunocapture PCR (IC-PCR) (Le et al. 2006) and rolling circle amplification (RCA) assays (James et al. 2011) are presently considered the most reliable methods for detecting active BSV infections in banana. However, RCA is expensive, time consuming, difficult to perform reliably (Sharma et al. 2014), and is not applicable for detecting BBTV or CMV infections. IC-PCR assays rely on virus-specific serological methods, but considerable serological variability exists among known BSV strains. In this study, we used RNA templates isolated from banana instead of DNA templates to minimize the potential of eBSVs amplification.
Using total RNA instead of DNA as a template removed most of the ambiguity created by eBSVs (Liu et al. 2012), yet still allowed amplification of BBTV and episomal BSV-OL. The contamination of RNA by trace amounts of DNA in RT-LAMP (Fig. 2b) and RT-PCR (Fig. 3b) reactions can be problematic unless nucleic acid preparations are pre-treated with DNase I (Kumar et al. 2015). However, DNase I treatment diluted the RNA templates and made the viral sequences difficult to detect with RT-PCR (Fig. 3b). In our study, the RT-LAMP assay was still capable of detecting RNA templates at the lower concentration (Fig. 1a and 2a), indicating the RT-LAMP assay was more sensitive than regular RT-PCR.
The BSV-OL-RT-LAMP assay developed in this study used primers based on 70 BSV RNase H gene sequences that included BSV-OL isolates and undetermined BSV isolates from various genotypes of banana from India, China, and Colombia. More highly variable RNase H gene sequences among other BSV species, such as banana streak GF virus and banana streak Uganda virus, were excluded from the sequence alignment. The relatively large genomic variation among BSVs makes it difficult to design RT-LAMP assay capable of detecting all BSV species.
Banana plants occasionally are infected by several viruses in the field (Brioso et al. 2000; Carnelossi et al. 2014). Most multiplex assays developed to detect virus infections in banana are based on PCR (Tan et al. 2010; Prema et al. 2012). These assays are able to detect two or three banana viruses simultaneously from a single source. A multiplex IC-PCR has also been developed for the simultaneous detection of three viruses from banana and plantain using crude sap extracts (Sharman et al. 2000). However, the procedures of PCR amplification followed by visualization of amplicons by gel electrophoresis make these methods time-consuming. The RT-LAMP assay developed in this study used relatively simple procedures, required less time to conduct, and still provided adequate detection sensitivity (Yasuhara-Bell et al. 2013). Reliable results, however, depend on the quality of the RNA extracts.
LAMP was used previously to detect BBTV (Peng et al. 2012a) and BSV (Peng et al. 2012b) from banana, and RT-LAMP has been used to detect CMV from banana (Peng et al. 2012c). These BSV-LAMP assays did not account for the large variations among BSV species or the false positives caused by eBSVs. Neither did they specify which BSV species could be detected by this assay or how to avoid eBSVs. There are no reports of RT-LAMP assays that have been developed to detect BBTV or BSVs from banana, or of LAMP/RT-LAMP assays developed to simultaneously detect BBTV, BSVs, and CMV from banana. Our multiplex assay minimized false positives due to eBSVs by incorporating a DNaseI pretreatment step. It also simplified the process by using RNA templates to detect both DNA and RNA viruses, and reduced detection times to less than 60 min (Fig. 5b).
This RT-LAMP multiplex assay can significantly reduce the time needed to index plants potentially infected by single or multiple viruses, and fulfills the needs of industry to detect an array of plant-pathogenic viruses in plant propagation material or during quarantine inspections. If samples are positive in this multiplex assay, they may be further evaluated as needed using individual assays for the three viruses.
References
Adegbola RO, Ayodeji O, Awosusi OO, Atiri GI, Kumar PL (2013) First report of banana bunchy top virus in banana and plantain (Musa spp.) in Nigeria. Plant Disease 97:290
Amrita B, Somnath R, Behere GT, Roy SS, Dutta SK, Ngachan SV (2014) Identification and characterization of a distinct banana bunchy top virus isolate of Pacific-Indian Oceans group from North-East India. Virus Research 183:41–49
Boloy FN, Nkosi BI, Losimba JK, Bungamuzi CL, Siwako HM, Balowe FW, Lohaka JW, Djailo BD, Lepoint P, Sivirihauma C, Blomme G (2014) Assessing incidence, development and distribution of banana bunchy top disease across the main plantain and banana growing regions of the Democratic Republic of Congo. African Journal of Agricultural Research 9:2611–2623
Brioso PST, Cordeiro ZJM, Rezende JAM, Kitajima EW, Pimentel JP, Figueiredo AR (2000) Mixed infection by cucumber mosaic (CMV) and banana streak (BSV) viruses in banana in Brazil. Summa Phytopathologica 26:254–257
Carnelossi PR, Bijora T, Facco CU, Silva JM, Picoli MHS, Souto ER, Oliveira FT (2014) Episomal detection of Banana streak OL virus in single and mixed infection with Cucumber mosaic virus in banana 'Nanicão Jangada'. Tropical Plant Pathology 39:342–346
Chabannes M, Baurens FC, Duroy PO, Bocs S, Vernerey MS, Rodiergoud M, Barbe V, Gayral P, Iskracaruana ML (2013) Three infectious viral species lying in wait in the banana genome. Journal of Virology 87:8624–8637
Chai J, Feng R, Shi H, Ren M, Zhang Y, Wang J (2015) Bioinformatic identification and expression analysis of banana microRNAs and their targets. PLoS One 10:e0123083
D'Hont A, Denoeud F, Aury JM, Baurens FC, Carreel F, Garsmeur O, Noel B, Bocs S, Droc G, Rouard M, Da Silva C, Jabbari K, Cardi C, Poulain J, Souquet M, Labadie K, Jourda C, Lengellé J, Rodier-Goud M, Alberti A, Bernard M, Correa M, Ayyampalayam S, Mckain MR, Leebens-Mack J, Burgess D, Freeling M, Mbéguié-A-Mbéguié D, Chabannes M, Wicker T, Panaud O, Barbosa J, Hribova E, Heslop-Harrison P, Habas R, Rivallan R, Francois P, Poiron C, Kilian A, Burthia D, Jenny C, Bakry F, Brown S, Guignon V, Kema G, Dita M, Waalwijk C, Joseph S, Dievart A, Jaillon O, Leclercq J, Argout X, Lyons E, Almeida A, Jeridi M, Dolezel J, Roux N, Risterucci AM, Weissenbach J, Ruiz M, Glaszmann JC, Quétier F, Yahiaoui N, Wincker P (2012) The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 488:213–219
Duroy PO, Perrier X, Laboureau N, Jacquemoud-Collet JP, Iskra-Caruana ML (2016) How endogenous plant pararetroviruses shed light on Musa evolution. Annals of Botany 117:625–641
Ferreira SA (1992) Cucumber mosaic virus. Available at: http://www.extento.hawaii.edu/Kbase/Crop/Type/cucvir.htm. Accessed on December 12, 2017
Fukuta S, Kato S, Yoshida K, Mizukami Y, Ishida A, Ueda J, Kanbe M, Ishimoto Y (2003) Detection of tomato yellow leaf curl virus by loop-mediated isothermal amplification reaction. Journal of Virological Methods 112:35–40
Fukuta S, Nimi Y, Oishi K, Yoshimura Y, Anai N, Hotta M, Fukaya M, Kato T, Oya T, Kambe M (2005) Development of reverse transcription loop-mediated isothermal amplification (RT-LAMP) method for detection of two viruses and chrysanthemum stunt viroid. Annual Report of The Kansai Plant Protection Society 47:31–36
Gayral P, Noa-Carrazana JC, Lescot M, Lheureux F, Lockhart BE, Matsumoto T, Piffanelli P, Iskra-Caruana ML (2008) A single banana streak virus integration event in the banana genome as the origin of infectious endogenous pararetrovirus. Journal of Virology 82:6697–6710
Gayral P, Blondin L, Guidolin O, Carreel F, Hippolyte I, Perrier X, Iskra-Caruana ML (2010) Evolution of endogenous sequences of banana streak virus: what can we learn from banana (Musa sp.) evolution? Journal of Virology 84:7346–7359
Geering ADW, McMichael LA, Dietzgen RG, Thomas JE (2000) Genetic diversity among banana streak virus isolates from Australia. Phytopathology 90:921–927
Helliot B, Panis B, Poumay Y, Swennen R, Lepoivre P, Frison E (2002) Cryopreservation for the elimination of cucumber mosaic and banana streak viruses from banana (Musa spp.). Plant Cell Reports 20:1117–1122
Hu JS, Li HP, Barry K, Wang M (1995) Comparison of dot blot, ELISA, and RT-PCR assays for detection of two cucumber mosaic virus isolates infecting banana in Hawaii. Plant Disease 79:902–906
Israeli Y, Lahav E, Reuveni O (1995) In vitro culture of bananas. Fruits 43:219–223
James P, Geijskes RJ, Dale JL, Harding RM (2011) Development of a novel rolling-circle amplification technique to detect banana streak virus that also discriminates between integrated and episomal virus sequences. Plant Disease 95:57–62
Javer-Higginson E, Acina-Mambole I, González JE, Font C, González G, Echemendía AL, Muller E, Teycheney PY (2014) Occurrence, prevalence and molecular diversity of banana streak viruses in Cuba. European Journal of Plant Pathology 138:157–166
King AMQ, Adams MJ, Lefkowitz EJ, Carstens EB (2012) Virus taxonomy: ninth report of the international committee on taxonomy of viruses. Elsevier Academic Press, New York
Kumar PL, Ayodele M, Oben TT, Mahungu NM, Beed F, Coyne D, Londa L, Mutunda MP, Kiala D, Maruthi MN (2009) First report of banana bunchy top virus in banana and plantain (Musa spp.) in Angola. Plant Pathology 58:402
Kumar PL, Selvarajan R, Iskra-Caruana ML, Chabannes M, Hanna R (2015) Biology, etiology, and control of virus diseases of banana and plantain. Advances in Virus Research 91:229–269
Le PG, Iskra-Caruana ML, Acina I, Teycheney PY (2006) Improved detection of episomal banana streak viruses by multiplex immunocapture PCR. Journal of Virological Methods 137:7–13
Le DT, Netsu O, Uehara-Ichiki T, Shimizu T, Choi IR, Omura T, Sasaya T (2010) Molecular detection of nine rice viruses by a reverse-transcription loop-mediated isothermal amplification assay. Journal of Virological Methods 170:90–93
Lee MS, Yang MJ, Hseu YC, Lai GH, Chang WT, Hsu YH, Lin MK (2011) One-step reverse transcription loop-mediated isothermal amplification assay for rapid detection of Cymbidium mosaic virus. Journal of Virological Methods 173:43–48
Liu FX, Feng LX, Chen X, Han YC, Li WD, Xu W, Cai B, Lin MG (2012) Simultaneous detection of four banana viruses by multiplex PCR. Journal of Phytopathology 160:622–627
Lockhart BEL (1986) Purification and serology of a bacilliform virus associated with banana streak disease. Phytopathology 76:995–999
Lockhart BEL (2000) Virus diseases of Musa in Africa: epidemiology, detection and control. Acta Horticulturae 540:355–359
Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T (2000) Loop-mediated isothermal amplification of DNA. Nucleic Acids Research 28:e63
Palukaitis P, Roossinck MJ, Dietzgen RG, Francki RI (1992) Cucumber mosaic virus. Advances in Virus Research 41:281–348
Parida M, Posadas G, Inoue S, Hasebe F, Morita K (2004) Real-time reverse transcription loop-mediated isothermal amplification for rapid detection of West Nile virus. Journal of Clinical Microbiology 42:257–263
Peng J, Fan ZF, Huang JS (2012a) Rapid detection of banana streak virus by loop-mediated isothermal amplification assay in South China. Journal of Phytopathology 160:248–250
Peng J, Shi MJ, Xia ZH, Huang JS, Fan ZF (2012b) Detection of cucumber mosaic virus isolates from banana by one-step reverse transcription loop-mediated isothermal amplification. Archives of Virology 157:2213–2217
Peng J, Zhang JF, Xia ZH, Li YQ, Huang JS, Fan ZF (2012c) Rapid and sensitive detection of banana bunchy top virus by loop-mediated isothermal amplification. Journal of Virological Methods 185:254–258
Prema GU, Rangaswamy KT, Pruthvi TPM (2012) Multiplex polymerase chain reaction technique for simultaneous detection of the banana streak virus (BSV) and cucumber mosaic virus (CMV) in banana from Karnataka. Current Biotica 6:141–151
Rustagi A, Jain S, Kumar D, Shekhar S, Jain M, Bhat V, Sarin NB (2015) High efficiency transformation of banana [Musa acuminata L. cv. Matti (AA)] for enhanced tolerance to salt and drought stress through overexpression of a peanut Salinity-Induced Pathogenesis-Related Class 10 protein. Molecular Biotechnology 57:27–35
Sharma SK, Kumar PV, Baranwal VK (2014) Immunodiagnosis of episomal banana streak MY virus using polyclonal antibodies to an expressed putative coat protein. Journal of Virological Methods 207:86–94
Sharman M, Thomas JE, Dietzgen RG (2000) Development of a multiplex immunocapture PCR with colourimetric detection for viruses of banana. Journal of Virological Methods 89:75–88
Tan XY, Ruan XL, Wu LT, Zhai GH, Li HP (2010) Development of a multiplex PCR protocol for the detection of three viruses in banana. Acta Horticulturae Sinica 37:829–834
Thiribhuvanamala G, Doraisamy S (2001) Survey for important virus diseases of banana. Madras Agricultural Journal 88:148–149
Thomas JE, Dietzgen RG (1991) Purification, characterization and serological detection of virus-like particles associated with banana bunchy top disease in Australia. Journal of General Virology 72:217–224
Tomita N, Mori Y, Kanda H, Notomi T (2008) Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nature Protocols 3:877–882
Varga A, James D (2006) Use of reverse transcription loop mediated isothermal amplification for the detection of Plum pox virus. Journal of Virological Methods 138:184–190
Wang D, Ma W, Yin X, Gu G (2014) Investigation and detection of banana virus disease from Hainan. Journal of Southern Agriculture 45:209–213
Wanitchakorn R, Harding RM, Dale JL (2000) Sequence variability in the coat protein gene of two groups of banana bunchy top isolates. Archives of Virology 145:593–602
Xie WS, Hu JS (1995) Molecular cloning, sequence analysis, and detection of banana bunchy top virus in Hawaii. Phytopathology 85:339–347
Yasuhara-Bell J, Kubota R, Jenkins DM, Alvarez AM (2013) Loop-mediated amplification of the Clavibacter michiganensis subsp. michiganensis micA gene is highly specific. Phytopathology 103:1220–1226
Acknowledgements
This work was supported in part by grants from the National Natural Science Foundation of China (31300118), Natural Science Foundation of Guangdong Province, China (2015A030312002), the Innovation Team Program of Modern Agricultural Science and Technology of Guangdong Province (2016LM2147, 2016LM2149), the National Institute of Food and Agriculture, U.S. Department of Agriculture, Hatch HAW09025-H under Accession no. 1001478, and from the U.S. Department of Agriculture, Agricultural Research Service under award number 58-5320-4-012.
Author information
Authors and Affiliations
Corresponding author
Additional information
Section Editor: Juliana Freitas-Astua
Rights and permissions
About this article
Cite this article
Zhang, J., Borth, W., Lin, B. et al. Multiplex detection of three banana viruses by reverse transcription loop-mediated isothermal amplification (RT-LAMP). Trop. plant pathol. 43, 543–551 (2018). https://doi.org/10.1007/s40858-018-0257-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40858-018-0257-6