Abstract
Since Asian soybean rust (ASR) isolates in South America are highly virulent, diverse, and distantly related to Japanese ones, limited numbers of resistance resources are available in soybean breeding in that region. Pyramiding of available ASR resistance genes (Rpp) in a single soybean genotype may provide wider spectrum and higher level of ASR resistance to soybean. However, the desired combinations of genes conferring adequate resistance to highly virulent or distantly related ASR isolates have not yet been studied. In this study, seven pyramided lines carrying multiple Rpp genes have been developed and evaluated for their resistance against one ASR isolate from Japan and two from Brazil. Significantly higher resistance was observed in the pyramided lines, No6-12-B (Rpp4 + Rpp5), Oy49-4 (Rpp2 + Rpp3 + Rpp4), and No6-12-1 (Rpp2 + Rpp4 + Rpp5) compared to the original resistance sources, PI 230970 (Rpp2), Hyuuga (Rpp3), PI 459025 (Rpp4), and Kinoshita (Rpp5) carrying single Rpp genes. Although infection of the resistance sources with the highly virulent Brazilian ASR isolates resulted in susceptible phenotypes with moderate to abundant sporulation, highly resistant phenotypes with almost no sporulation were observed in the three Rpp-pyramided lines. Therefore, pyramided lines carrying these Rpp gene combinations are useful in soybean breeding for conferring broad spectrum, high resistance to ASR isolates that are virulent to the varieties carrying single resistance genes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Asian soybean rust (ASR), caused by the biotrophic basidiomycete Phakopsora pachyrhizi Syd. & P. Syd., occurs in major soybean-growing regions of all tropical and sub-tropical areas. Severe yield losses are especially common in South America when environmental conditions are conducive for ASR development (Yorinori 2008). Hence, ASR is considered as one of the most serious economic threats for soybean growers in that region (Goellner et al. 2010). Several management tactics have been employed to control ASR and to minimize the impact of this disease. Chemical treatment with fungicides has been perceived as the first line of defense against ASR (Levy 2005). However, limited number of appropriate fungicides, specific application requirements, increased production costs, environmental pollution and development of fungicide resistant strains are the main concerns of using fungicides (Schneider et al. 2008). Hence, an environmentally-friendly, cost-effective and long-term management of the disease can be achieved through utilization of host genetic resistance to ASR (Ribeiro et al. 2007).
Seven dominant genes (Rpp1 to Rpp6 and Rpp1b) controlling race-specific resistance to ASR have so far been identified and mapped at different loci (Hyten et al. 2007, 2009; Garcia et al. 2008; Silva et al. 2008; Chakraborty et al. 2009; Li et al. 2012; Hossain et al. 2015). Although these major Rpp genes are now available for breeding, they rarely offer durable resistance to the highly variable ASR pathogen (Oliveira et al. 2005). These genes provide effective resistance to some P. pachyrhizi races, but were ineffective when challenged with other races. This has limited the use of single genes for resistance in soybean especially in South America, where ASR populations are highly virulent and diverse (Yamanaka et al. 2010; Akamatsu et al. 2013). These commonly encountered problems associated with the ineffectiveness of the specific resistance genes and difficulties in the identification of durable resistance against ASR has led to the continuous search for new resistance genes. Development of ASR resistant cultivars has been an important aspect of breeding programs in soybean in the present days and would be augmented by the identification of gene conditioning the ASR resistance in a wide range of soybean varieties. Although single resistance genes can be overcome by specific races of P. pachyrhizi, broad spectrum resistance may be created by pyramiding multiple resistance genes into modern cultivars.
Gene pyramiding has been successfully applied in combining multiple disease resistance genes in several previous experimental studies. Pyramiding of soybean mosaic virus (SMV) resistance genes (Rsv) by marker-assisted selection brought durable and wide spectrum of resistance to several strains of SMV (Maroof et al. 2008). In rice, pyramided lines showed not only a wider spectrum but also a higher level of bacterial blight resistance compared with lines with only a single gene (Huang et al. 1997). A higher level of resistance in pyramided wheat lines was also observed against cereal cyst nematode (Barloy et al. 2007). In soybean, pyramiding of Rpp2, Rpp4, and Rpp5 in a single genotype was shown to provide higher resistance to ASR (Lemos et al. 2011). Higher resistance of this pyramided line to Brazilian ASR isolates was also confirmed in subsequent experiments (Yamanaka et al. 2013b). Maphosa et al. (2012) reported a low level of ASR severity and sporulation through Rpp2, Rpp3 and Rpp4 pairwise gene pyramiding. Thus, Rpp gene pyramiding is expected to bring broad-spectrum and higher resistance to the ASR pathogen in soybean. Nevertheless, studies to pyramid desired gene combinations for conferring adequate resistance to ASR are still lacking. In the present work, we report the development of several Rpp-pyramided lines and their evaluation using three ASR isolates.
Materials and methods
Plant materials
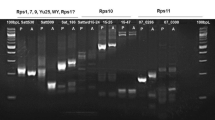
The soybean genotypes used in this study include five ASR-resistant plant introductions and varieties carrying single Rpp gene; PI 200492 (Rpp1), PI 230970 (Rpp2), Hyuuga (Rpp3), PI 459025 (Rpp4) and Kinoshita (Rpp5) (Fig. 1, Table 1). These sources were crossed to obtain Rpp-pyramided lines and therefore were named as ‘resistance source’ in this paper. The soybean variety BRS 184 was used as susceptible control. Four Rpp-pyramided lines carrying two Rpp genes [Mo84-6 (Rpp1 + Rpp2), An76-1 (Rpp2 + Rpp4), No12-1-A (Rpp2 + Rpp5), and No6-12-B (Rpp4 + Rpp5)], and three Rpp-pyramided lines carrying three Rpp genes [Mo42-1 (Rpp1 + Rpp2 + Rpp4), Oy49-4 (Rpp2 + Rpp3 + Rpp4), and No6-12-1 (Rpp2 + Rpp4 + Rpp5)], were obtained from crosses between resistance sources or between resistance sources and Rpp-pyramided line through marker-assisted selection (MAS) to F2 or F3 progenies (Fig. 1). Among the seven Rpp-pyramided lines, An76-1 and No6-12-1 have been obtained from previous studies (Lemos et al. 2011; Yamanaka et al. 2011, 2013b), while the other five were newly developed from three kinds of F2 populations: 1) No population consisting of 140 plants derived from a cross ‘An76-1’ × ‘Kinoshita’, 2) Oy population of 94 plants from ‘An76-1’ × ‘Hyuuga,’ and 3) Mo population of 93 plants from ‘PI 200492’ × ‘An76-1’. All seven Rpp-pyramided lines used in this study were derived from a single F3 plant. For the evaluation of resistance to ASR isolates, F4 plants from No12-1-A (Rpp2 + Rpp5), No6-12-B (Rpp4 + Rpp5), Oy49-4 (Rpp2 + Rpp3 + Rpp4), Mo42-1 (Rpp1 + Rpp2 + Rpp4), and Mo84-6 (Rpp1 + Rpp2), F5 plants from An76-1 (Rpp2 + Rpp4) and F6 plants from No6-12-1 (Rpp2 + Rpp4 + Rpp5) were used (Fig. 1). Three plants each from resistance sources, Rpp-pyramided lines, and the susceptible control were grown in an ASR-free growth chamber as described by Yamanaka et al. (2010) and used for evaluation of ASR resistance.
Pedigree of Rpp-pyramided lines used in this study. The resistant Rpp alleles that each genotype carries in homozygous state are indicated in parentheses. For each cross, the ovule parent is shown above the pollen parent. P and SSD mean parental variety and single-seed decent, respectively. Pedigree of An76-1 and No6-12-1 has been reported in previous studies (Lemos et al. 2011; Yamanaka et al. 2011, 2013b)
MAS of Rpp-pyramided lines
DNA was extracted from the parental genotypes (Fig. 1) as well as from individual F2 and F3 plants. Marker-assisted selection (MAS) using simple sequence repeat (SSR) markers linked to five Rpp loci (Table 1) was applied to obtain F2 or F3 plants carrying two or three Rpp genes as homozygous resistant. All SSR markers used in this study were co-dominant for parents. In each Rpp locus, at least two SSR markers which are polymorphic between parents and sandwiching Rpp locus were used to identify the presence of Rpp genes. If recombination was observed between the markers in the F2 or F3 plants, they were excluded from screening because of difficulties to determine the genotype with Rpp gene. PCR and subsequent electrophoresis were performed following the procedures described by Yamanaka et al. (2013a).
Two F2 plants, ‘No6-12’ and ‘No12-1’, were selected from the ‘No population’ used in the previous study (Lemos et al. 2011) and their progenies were screened to obtain Rpp-pyramided lines (Table 1, Fig. 1). No6-12 carried Rpp2 as heterozygous and Rpp4 and Rpp5 as homozygous resistant (Rpp2rpp2Rpp4Rpp4Rpp5Rpp5). Twelve F3 plants produced from this plant were screened and three of them were found to show Kinoshita genotype in the three Rpp2 (rpp2rpp2)-linked SSR markers, Satt380, Satt620, and Sat366. Two SSR markers for each of the Rpp4 and Rpp5 were checked to confirm whether the F3 plants carry these genes as homozygous resistant (Rpp4Rpp4Rpp5Rpp5). One of these three F3 plants, ‘No6-12-B’ (rpp2rpp2Rpp4Rpp4Rpp5Rpp5) was cultivated to obtain F4 plants and evaluated for ASR resistance. On the other hand, two of nine F3 plants from No12-1 (Rpp2Rpp2Rpp4rpp4Rpp5Rpp5) were identified as homozygous resistant for Rpp2 and Rpp5, and homozygous susceptible for Rpp4 (Rpp2Rpp2rpp4rpp4Rpp5Rpp5) based on screening with two Rpp4-linked SSR markers, Satt288 and AF162283. A total of five SSR markers for Rpp2 and Rpp5 (Table 1) were also used to validate whether these plants carry these two genes as resistant homozygous. One of these two F3 plants, ‘No12-1-A’ (Rpp2Rpp2rpp4rpp4Rpp5Rpp5) was cultivated to obtain F4 plants and used for evaluating ASR resistance.
The ‘Mo population’ developed in this study was screened with SSR markers linked to Rpp1, Rpp2, and Rpp4 to obtain Rpp-pyramided lines carrying Rpp1 and Rpp2, and Rpp1, Rpp2 and Rpp4, respectively (Table 1). Two F2 plants, ‘Mo42’ and ‘Mo84’, were identified to have Rpp1 and Rpp2 as homozygous resistant and Rpp4 as heterozygous (Rpp1Rpp1Rpp2Rpp2Rpp4rpp4). Twelve F3 plants from each of ‘Mo42’ and ‘Mo84’ were screened again by SSR markers linked to Rpp4. Three F3 plants from Mo42 and one from Mo84 were identified to have Rpp4 as homozygous resistant (Rpp1Rpp1Rpp2Rpp2Rpp4Rpp4). Only one plant, ‘Mo84-6’, of the 24 F3 progenies tested, carried the Rpp4 as homozygous susceptible (Rpp1Rpp1Rpp2Rpp2rpp4rpp4). One plant from each of the four different F3 genotypes was cultivated further to obtain F4 plants and used for evaluating their ASR resistance.
The ‘Oy population’ developed in this study was also screened with SSR markers for obtaining Rpp-pyramided lines carrying Rpp2, Rpp3 and Rpp4 (Table 1). A single F2 plant, ‘Oy49’, was identified to have Rpp2 and Rpp4 as homozygous resistant and Rpp3 as heterozygous (Rpp2Rpp2Rpp3rpp3Rpp4Rpp4). Twenty-two F3 plants were screened again with SSR markers linked to Rpp3. Three of seven F3 plants from Oy49 were identified to carry Rpp3 as homozygous resistant (Rpp2Rpp2Rpp3Rpp3Rpp4Rpp4). One of them, ‘Oy49-4’, was used to produce F4 plants, followed by evaluation for ASR resistance.
Pathogen inoculation and resistance evaluation
One Japanese ASR isolate (T1-2) and two Brazilian ASR isolates (BRP-2.5 and BRP-2.6) used in the previous studies (Yamanaka et al. 2013b; Hossain et al. 2015) were used for inoculation. Single-lesion isolate T1-2 was obtained from ASR population T1 originally collected from a soybean field in Tsukuba, Japan on September 2007 (Yamaoka et al. 2014). Single-lesion isolates BRP-2.5 and BRP-2.6 were obtained from ASR population BRP-2 originally collected at a greenhouse in Embrapa Soja, Brazil on August 2008 (Yamanaka et al. 2010). When plants reached the V3 to V4 growth stage (approximately 3 weeks after sowing), a total of nine leaflets were excised from three plants (three leaflets × three plants) in each genotype, respectively. Then three detached leaflets derived from three independent plants were inoculated with one of three ASR isolates using the detached-leaf method as described by Yamanaka et al. (2013b). The final spore concentration used for each inoculation was 6.5 × 104 urediniospores/mL. Two weeks after inoculation, each single lesion was scored for the numbers of uredinia and sporulation (0: no spores to 3: abundant spores; Yamanaka et al. 2013a). Then, the numbers of uredinia per lesion (NoU) and sporulation level (SL) were obtained based on 30 lesions for each plant (replication). Finally, average NoU and SL of three replications were compared among genotypes in each isolate inoculation. Significant differences (p < 0.05) in NoU and SL among genotypes was confirmed by ANOVA or Kruskal-Wallis test. Significance levels in NoU and SL between genotypes were determined by Tukey HSD test (p < 0.05). These tests were conducted in R software v. 3.0.1 (R Development Core Team 2013).
Results
Resistance to ASR isolate T1-2
A level of susceptibility similar to the control variety BRS 184 was observed in PI 230970 (Rpp2) and An76-1 (Rpp2 + Rpp4) with regards to NoU and in PI 230970 (Rpp2) with regards to SL during infection by ASR isolate T1-2 (Fig. 2). The other resistance sources and Rpp-pyramided lines showed a lower level of NoU and SL than BRS 184 (Fig. 2), although they differed widely with a range of 0.0 ≦ NoU ≦ 1.2 and 0.0 ≦ SL ≦ 1.6. No uredinia and sporulation were observed in three Rpp-pyramided lines: No6-12-B (Rpp4 + Rpp5), Oy49-4 (Rpp2 + Rpp3 + Rpp4) and No6-12-1 (Rpp2 + Rpp4 + Rpp5). Some of their resistance source varieties, i.e., Hyuuga (Rpp3) and Kinoshita (Rpp5), also had very low levels of NoU and SL, and no significant differences between these three Rpp-pyramided lines and their resistance source varieties were observed (Fig. 2).
Mean values of number of uredinia per lesion (NoU) and sporulation level (SL) with standard deviations against one Japanese (T1-2) and two Brazilian ASR isolates (BRP-2.5 and BRP-2.6) in Rpp-pyramided lines and their resistance sources. The asterisks in the Rpp-pyramided lines mean that the resistance was significantly higher than in their respective resistance sources. ‘S’ means that the resistance level was the same as or lower than BRS 184
Resistance to ASR isolate BRP-2.5
All resistance sources except PI 459025 (Rpp4) showed the same or higher level of NoU and SL compared to the susceptible control variety BRS 184 after infection by ASR isolate BRP-2.5 (Fig. 2). PI 459025 (Rpp4) had significantly lower levels of NoU and SL than BRS 184 and was considered as resistant. On the other hand, six and five out of seven Rpp-pyramided lines showed significantly lower levels of NoU and SL, respectively, than BRS 184. Only one pyramided line, Mo84-6 (Rpp1 + Rpp2), and two pyramided lines, Mo84-6 (Rpp1 + Rpp2) and No12-1-A (Rpp2 + Rpp5), did not have not significantly lower SL and NoU, respectively, than BRS 184. All five Rpp-pyramided lines that had significantly lower SL than BRS184 carried the Rpp4 gene. Neither uredinial formation nor sporulation was observed in two Rpp-pyramided lines, No6-12-B (Rpp4 + Rpp5) and No6-12-1 (Rpp2 + Rpp4 + Rpp5), as observed for T1-2 infection. However, a fewer uredinia and little sporulation were observed in Oy49-4 (Rpp2 + Rpp3 + Rpp4).
Resistance to ASR isolate BRP-2.6
The NoU in all five resistance source varieties (Rpp1-5) did not appear to differ significantly with that in the susceptible cultivar BRS 184 after infection by BRP-2.6 (Fig. 2). Only PI 459025 (Rpp4) showed a significant difference in SL over BRS 184 and the other four resistance source varieties, producing moderate sporulation with a SL value of 2.0. However, the three Rpp-pyramided lines, No6-12-B (Rpp4 + Rpp5), Oy49-4 (Rpp2+ Rpp3+ Rpp4) and No6-12-1 (Rpp2 + Rpp4 + Rpp5), were highly resistant and had significantly lower NoU and SL than BRS 184. Additionally, the NoU and SL in these pyramided lines were also significantly lower than those of their resistance source varieties, PI 230970 (Rpp2), Hyuuga (Rpp3), PI 459025 (Rpp4) and Kinoshita (Rpp5). The other four Rpp-pyramided lines, Mo84-6 (Rpp1 + Rpp2), An76-1 (Rpp2 + Rpp4), No12-1-A (Rpp2 + Rpp5) and Mo42-1 (Rpp1 + Rpp2+ Rpp4), did not show significantly lower NoU and/or SL than BRS 184. Thus, no significant enhancement in ASR resistance was observed in these four Rpp-pyramided lines in comparison with their resistance source varieties, PI 200492 (Rpp1), PI 230970 (Rpp2), PI 459025 (Rpp4) and Kinoshita (Rpp5).
Discussion
The use of ASR resistance genes (Rpp) should be an important management component for control of the disease, but the seven unique Rpp genes that were identified so far rarely confer sufficient resistance to highly virulent and diverse South American ASR populations and isolates (Yamanaka et al. 2010, 2011; Akamatsu et al. 2013). Therefore, searching for new breeding resources and strategies is required to provide durable and high impact resistance to ASR in soybean production. Previous studies have indicated the usefulness of pyramiding different Rpp genes in one genotype, as it improved the efficiency of plant breeding leading to the development of broad spectrum and high level of resistance capabilities against ASR (Lemos et al. 2011; Maphosa et al. 2012; Yamanaka et al. 2013b). However, it has not yet been investigated which combinations of Rpp genes confer higher resistance to multiple ASR races and whether pyramiding Rpp genes brings high resistance to the ASR races that are virulent to individual Rpp genes.
In this study, the resistance to three ASR isolates was compared among four combinations of double Rpp genes and three combinations of triple Rpp genes. These combinations were Rpp1 + Rpp2, Rpp2 + Rpp4, Rpp2 + Rpp5, Rpp4 + Rpp5, Rpp1 + Rpp2 + Rpp4, Rpp2 + Rpp 3+ Rpp4, and Rpp2 + Rpp4 + Rpp5. Among them, the Rpp4 + Rpp5, Rpp2 + Rpp3 + Rpp4, and Rpp2 + Rpp4 + Rpp5 combinations showed the highest potential of resistance to all tested ASR isolates (Fig. 2). These pyramided lines were more ASR resistant than their source varieties (BRP-2.6 in Fig. 2), however, the relative resistance in the Rpp2 + Rpp4 and Rpp2 + Rpp5 combinations was not as high as that in the Rpp4 + Rpp5 and Rpp2 + Rpp4 + Rpp5 combinations. Thus, pyramiding Rpp4 + Rpp5 expressed additional synergistic effects. Regarding the combination of Rpp2 + Rpp3 + Rpp4, the digenic or trigenic effects of these genes were not realized, since the genotypes with Rpp2 + Rpp3 and Rpp3 + Rpp4 were not included in the present study. However, Maphosa et al. (2012) compared the effects of pairwise gene combination using three genes, Rpp2, Rpp3 and Rpp4, against an ASR population in Kabanyolo, Uganda. They concluded that the Rpp2 + Rpp3 genotype had significantly lower severity, lesions per square centimeter and frequency of sporulating lesion than those of the parents (Rpp2, Rpp3 or Rpp4) and of the Rpp2 + Rpp4 genotype. This supports presence of improved ASR resistance in the Rpp2 + Rpp3 combination. Thus, high ASR resistance observed in Oy49-4 (Rpp2 + Rpp3 + Rpp4) could be derived from the possible digenic interaction between Rpp2 and Rpp3 as implied by Maphosa et al. (2012).
The combinations of Rpp1 + Rpp2, Rpp2 + Rpp4, Rpp2 + Rpp5 and Rpp1 + Rpp2 + Rpp4 were not effective to the highly virulent Brazilian ASR isolates used in this study. Their Rpp2 resistance source, PI 230970, was also susceptible to all three tested ASR isolates. However, since other Rpp2-carrying varieties such as Iyodaizu B and Hougyoku were known to show different reactions to Brazilian ASR isolates (Yamanaka et al. 2015), they may be effective to the highly virulent Brazilian isolates used in this study. In addition, digenic interactions in genotypes with Rpp2 + Rpp4 and Rpp2 + Rpp5 were observed to play significant roles in reducing NoU and SL during infection by Brazilian ASR populations in a previous study (Lemos et al. 2011). Therefore, the gene combinations Rpp1 + Rpp2, Rpp2 + Rpp4, Rpp2 + Rpp5 and Rpp1 + Rpp2 + Rpp4 could be effective against different ASR isolates from those used in this study.
In this study we did not include Rpp1-b and Rpp6 for Rpp-pyramiding. The Rpp1-b gene from PI 594767A, PI 587905 and PI 587880A is closely located to Rpp1 in chromosome 18 and has shown resistance to 89–96 % of South American ASR population samples (Hossain et al. 2015). Similarly, soybean variety PI 567102B carrying Rpp6 is currently included in differential varieties (Yamanaka et al. 2013a) and found to show high resistance against Paraguayan ASR populations (Miles et al. 2008). These resistant varieties could be useful and should be included as resistance sources for future Rpp-pyramiding breeding programs. In addition, several varieties have been identified to share same Rpp genes, e.g., four and five varieties were reported to share Rpp5 (Garcia et al. 2008) and Rpp3 (Brogin 2005; Monteros et al. 2007; Hyten et al. 2009; Ray et al. 2011; Hossain et al. 2015), respectively. The allelic differences of these single genes in different varieties (or tightly linked independent genes) may influence the range of ASR pathogens to which they show resistance and also change the degree of resistance derived from genetic interactions due to Rpp-pyramiding.
Since all Rpp-pyramided lines except for An76-1 are derived from crosses with An76-1, half of the genome (except for Rpp loci) among all seven Rpp-pyramided lines are theoretically the same. Minor effects of genetic background on resistance phenotypes have been observed in Rpp-pyramided lines (Yamanaka et al. 2013b). Differences on the phenotypes among the lines might be influenced by such effects as well as environmental factors. In order to estimate the pyramiding effect more exactly, Rpp-pyramided line series having the same genetic background should be developed by the use of isogenic lines or linebreeding.
In conclusion, our study successfully observed that pyramided lines carrying two or three Rpp genes showed higher resistance to three ASR isolates with different origins and pathogenicity, although the ASR isolates were virulent to varieties carrying the single Rpp genes used for the pyramiding. Pyramiding with the combinations Rpp4 + Rpp5, Rpp2 + Rpp3 + Rpp4 and Rpp2 + Rpp4 + Rpp5 was most useful for enhancing the resistance to ASR isolates in a less race-specific manner. These Rpp- pyramided lines have practical breeding value by providing broad-spectrum and high level of resistance against ASR. However, validation tests under field conditions with natural ASR infection are necessary to demonstrate the pyramiding effect observed in this study. In addition, further experiments should be performed using additional Rpp combinations, genetic backgrounds of pyramided lines, and ASR isolates to identify the most effective combination of Rpp genes for ASR resistance.
References
Akamatsu H, Yamanaka N, Yamaoka Y, Soares RM, Morel W, Ivancovich AJG, Bogado AN, Kato M, Yorinori JT, Suenaga K (2013) Pathogenic diversity of soybean rust in Argentina, Brazil, and Paraguay. J Gen Plant Pathol 79:28–40
Barloy D, Lemoine J, Abelard P, Tanguy AM, Rivoal R, Jahier J (2007) Marker-assisted pyramiding of two cereal cyst nematode resistance genes from Aegilops variabilis in wheat. Mol Breed 20:31–40
Brogin RL (2005) Mapeamento de genes de resistência à ferrugem e de QTLs envolvidos na resistência à septoriose em soja. M.Sc. Dissertation, Universidade de São Paulo. São Paulo, Brazil.
Chakraborty N, Curley J, Frederick R, Hyten D, Nelson R, Hartman G, Diers B (2009) Mapping and confirmation of a new allele at Rpp1 from soybean PI 594538A conferring RB lesion-type resistance to soybean rust. Crop Sci 49:783–790
R Development Core Team (2013) R: a language and environment for statistical computing. Available at: http://www.R-project.org. Accessed 16 May 2013.
Garcia A, Calvo ES, Kiihl RAS, Harada A, Hiromoto DM, Vieira LG (2008) Molecular mapping of soybean rust (Phakopsora pachyrhizi) resistance genes: Discovery of a novel locus and alleles. Theor Appl Genet 117:545–553
Goellner K, Loehrer M, Langenbach C, Conrath U, Koch E, Schaffrath U (2010) Phakopsora pachyrhizi, the causal agent of Asian soybean rust. Mol Plant Pathol 11:169–177
Hossain MM, Akamatsu H, Morishita M, Mori T, Yamaoka Y, Suenaga K, Soares RM, Bogado AN, Ivancovich AJG, Yamanaka N (2015) Molecular mapping of Asian soybean rust resistance in soybean landraces PI 594767A, PI 587905 and PI 416764. Plant Pathol 64:147–156
Huang N, Angeles ER, Domingo J, Magpantay G, Singh S, Zhang G, Kumaravadivel N, Bennett J, Khush GS (1997) Pyramiding of bacterial blight resistance genes in rice: marker-assisted selection using RFLP and PCR. Theor Appl Genet 95:313–520
Hyten D, Hartman G, Nelson R, Frederick RD, Concibido VC, Narvel JM, Cregan PB (2007) Map location of the Rpp1 locus that confers resistance to soybean rust in soybean. Crop Sci 47:837–840
Hyten D, Smith J, Frederick R, Tucker M, Song Q, Cregan P (2009) Bulked segregant analysis using the GoldenGate assay to locate the Rpp3 locus that confers resistance to soybean rust in soybean. Crop Sci 49:265–271
Lemos NG, Braccini AL, Abdelnoor RV, Oliveira MCN, Suenaga K, Yamanaka N (2011) Characterization of genes Rpp2, Rpp4, and Rpp5 for resistance to soybean rust. Euphytica 182:53–64
Levy C (2005) Epidemiology and chemical control of soybean rust in southern Africa. Plant Dis 89:669–674
Li S, Smith J, Ray J, Frederick R (2012) Identification of a new soybean rust resistance gene in PI 567102B. Theor Appl Genet 125:133–142
Maphosa M, Talwana H, Tukamuhabwa P (2012) Enhancing soybean rust resistance through Rpp2, Rpp3 and Rpp4 pair wise gene pyramiding. Afr J Agric Res 7:4271–4277
Maroof S, Jeong SC, Gunduz I, Tucker DM, Buss GR, Tolin SA (2008) Pyramiding of soybean mosaic virus resistance genes by marker-assisted selection. Crop Sci 48:517–526
Miles MR, Morel W, Ray JD, Smith JR, Frederick RD, Hartman GL (2008) Adult plant evaluation of soybean accessions for resistance to Phakopsora pachyrhizi in the field and greenhouse in Paraguay. Plant Dis 92:96–105
Monteros M, Missaoui A, Phillips D, Walker D, Boerma H (2007) Mapping and confirmation of the ‘Hyuuga’ red-brown lesion resistance gene for Asian soybean rust. Crop Sci 47:829–836
Oliveira ACB, Godoy CV, Martins MC (2005) Avaliação da tolerância de cultivares de soja à ferrugem asiática no Oeste da Bahia. Fitopatol Bras 30:658–662
Ray JD, Smith JR, Morel W, Bogado N, Walker DR (2011) Genetic resistance to soybean rust in PI 567099A in at or near the Rpp3 locus. J Crop Improv 25:219–231
Ribeiro AS, Moreira JUV, Pierozzi PHB, Rachid BF, De Toledo JFF, Arias CAA, Soares RM, Godoy CV (2007) Genetic control of Asian rust in soybean. Euphytica 157:15–25
Schneider R, Sikora E, Padgett B, Sciumbato G (2008) Managing late-season soybean diseases and soybean rust: a southern perspective. In: Dorrance AE, Draper MA, Hershman DE (eds) Using of foliar fungicides to manage soybean rust. Ohio State University, Columbus, pp 72–75
Silva DCG, Yamanaka N, Brogin RL, Arias CAA, Nepomuceno AL, Di Mauro AO, Pereira SS, Nogueira LM, Passianotto ALL, Abdelnoor RV (2008) Molecular mapping of two loci that confer resistance to Asian rust in soybean. Theor Appl Genet 117:57–63
Yamanaka N, Yamaoka Y, Kato M, Lemos NG, Passianotto ALL, Santos JVM, Benitez ER, Abdelnoor RV, Soares RM, Suenaga K (2010) Development of classification criteria for resistance to soybean rust and differences in virulence among Japanese and Brazilian rust populations. Trop Plant Pathol 35:153–162
Yamanaka N, Lemos NG, Akamatsu H, Yamaoka Y, Silva DCG, Passianotto ALL, Abdelnoor RV, Soares RM, Suenaga K (2011) Soybean breeding materials useful for resistance to soybean rust in Brazil. Jpn Agric Res Q 45:385–395
Yamanaka N, Akamatsu H, Yamaoka Y (2013a) Laboratory manual for studies on soybean rust resistance. Available at: http://www.jircas.affrc.go.jp/english/manual/soybean_rust/soybean_rust.html. Accessed 1 Dec 2014.
Yamanaka N, Lemos NG, Uno M, Akamatsu H, Yamaoka Y, Abdelnoor RV, Braccini AL, Suenaga K (2013b) Resistance to Asian soybean rust in soybean lines with the pyramided three Rpp genes. Crop Breed Appl Biotechnol 13:75–82
Yamanaka N, Hossain MM, Yamaoka Y (2015) Molecular mapping of Asian soybean rust resistance in Chinese and Japanese soybean lines, Xiao Jing Huang, Himeshirazu, and Iyodaizu B. Euphytica. doi:10.1007/s10681-015-1377-4
Yamaoka Y, Yamanaka N, Akamatsu H, Suenaga K (2014) Pathogenic races of soybean rust Phakopsora pachyrhizi collected in Tsukuba and vicinity in Ibaraki, Japan. J Gen Plant Pathol 80:184–188
Yorinori JT (2008) Soybean germplasms with resistance and tolerance to Asian rust and screening methods. In: Kudo H, Suenaga K, Soares RM, Toledo A (eds) JIRCAS Working Report No.58: Facing the Challenge of Soybean Rust in South America. Tsukuba, Japan. JIRCAS. pp 70–87
Acknowledgments
We are grateful to EMBRAPA in Brazil and the National Institute of Crop Science (NICS) in Japan for providing seeds of soybean varieties. This study was financially supported and conducted by the JIRCAS research project “Development of Breeding Technologies toward Improved Production and Stable Supply of Upland Crops.” NGL and MMH were financially supported by the JIRCAS Visiting Research Fellowship Program.
Author information
Authors and Affiliations
Corresponding author
Additional information
Section Editor: Elaine Souza
Rights and permissions
About this article
Cite this article
Yamanaka, N., Morishita, M., Mori, T. et al. Multiple Rpp-gene pyramiding confers resistance to Asian soybean rust isolates that are virulent on each of the pyramided genes. Trop. plant pathol. 40, 283–290 (2015). https://doi.org/10.1007/s40858-015-0038-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40858-015-0038-4