Abstract
Asian soybean rust (ASR) is caused by the fungal pathogen Phakopsora pachyrhizi Sydow & Sydow. It was first identified in Brazil in 2001 and quickly infected soybean areas in several countries in South America. Primary efforts to combat this disease must involve the development of resistant cultivars. Four distinct genes that confer resistance against ASR have been reported: Rpp1, Rpp2, Rpp3, and Rpp4. However, no cultivar carrying any of those resistance loci has been released. The main objective of this study was to genetically map Rpp2 and Rpp4 resistance genes. Two F2:3 populations, derived from the crosses between the resistant lines PI 230970 (Rpp2), PI 459025 (Rpp4) and the susceptible cultivar BRS 184, were used in this study. The mapping populations and parental lines were inoculated with a field isolate of P. pachyrhizi and evaluated for lesion type as resistant (RB lesions) or susceptible (TAN lesions). The mapping populations were screened with SSR markers, using the bulk segregant analysis (BSA) to expedite the identification of linked markers. Both resistance genes showed an expected segregation ratio for a dominant trait. This study allowed mapping Rpp2 and Rpp4 loci on the linkage groups J and G, respectively. The associated markers will be of great value on marker assisted selection for this trait.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Brazil is the second largest soybean producer in the world and still has a great potential to increase production in the future given the existence of areas not cropped and the increasing world demand for soybean products. Among the problems threatening soybean production and expansion in Brazil, the fungal disease Asian soybean rust (ASR), caused by Phakopsora pachyrhizi, is responsible for severe economic losses. Until 2000, significant yield reductions caused by P. pachyrhizi had been reported only in the Eastern Hemisphere, where yield losses ranged from 5 to 95% (Hartman et al. 1991, 1997). After the first identification of the disease in Paraguay, and in Paraná, Brazil, in 2001, ASR rapidly infected soybean areas all over Brazil, Paraguay, Bolivia, and some regions of Argentina, causing yield losses of up to 75% (Yorinori et al. 2005) and making this disease as the main threat affecting Latin American soybean production. With the recent detection of P. pachyrhizi in continental USA (Schneider et al. 2005), ASR is now present in most of the soybean growing areas of the world.
The only method used to control ASR is fungicide applications, which substantially increase production costs and are a continuous source of environmental contamination. The use of resistant cultivars is seemed to be the most promising method to control this disease. Four distinct major genes, that confer resistance against ASR, have been reported to date: Rpp1, Rpp2, Rpp3, and Rpp4 (Bromfield and Hartwig 1980; McLean and Byth 1980; Hartwig and Bromfield 1983; Hartwig 1986). Several other resistance sources have been also identified abroad (Miles et al. 2006) and in Brazil (C.A.A. Arias, personal communication). However, no commercial cultivar carrying any of these resistance genes has been released.
The usual reactions associated with this disease are the presence of reddish-brown lesions (RB lesions) on resistant reactions and the presence of tan color lesions (TAN) on the susceptible ones. In some cases, the resistance is also associated with an immune response (no visible symptoms), as is the case of Rpp1 in the presence of certain isolates (Miles et al. 2006). To date, all known ASR resistance loci evaluated have been overcome by at least one isolate throughout the world (Miles et al. 2006; Yamaoka et al. 2002). However, Rpp2 and Rpp4 loci remain effective against the Brazilian isolates, whereas Rpp1 and Rpp3 were defeated in 2003, just 2 years after ASR detection in Brazil (José Tadashi Yorinori, personal communication). Rpp2 and Rpp4 loci were identified on the lines PI 230970 and PI 459025, respectively, and behave as a single dominant allele (Bromfield and Hartwig 1980; Hartwig 1986).
Monogenic rust resistance has been a volatile trait on other species. In wheat, the race-specific rust leaf resistance has a short-lived nature and it has been frequently associated to other types of resistance in order to have a more stable resistance (Kolmer 1996). Lately, efforts have been made by several breeding programs to introduce ASR resistance loci into elite soybean cultivars, and the stacking of different loci could result in more stable ASR resistance, capable of overcoming the historical failure of many monogenic resistance mechanisms in plants, especially those genes leading to a hypersensitive response (Niks and Rubiales 2002). The main difficult of this process is to select the plants carrying more than one locus by conventional methods, and techniques that could assist the introgression of multiple loci would be very useful in this manner.
DNA markers have great potential for assisting selection of resistant genotypes in breeding programs and have been used in several crops, including soybean (Concibido et al. 2004), and could facilitate stacking of different resistance loci in a single cultivar, which could provide a more sustainable resistance to P. pachyrhizi in the future. Microsatellites or simple sequence repeats (SSRs) are the preferred markers presently used to map host resistance to soybean diseases because of their high polymorphism, co-dominance, reproducibility, and good distribution throughout the soybean genome. Furthermore, over a thousand well-distributed SSR markers are already mapped in the soybean genetic map (Song et al. 2004). The present study describes the mapping of Rpp2 and Rpp4 loci on the soybean genetic map using SSR markers.
Materials and methods
Plant materials
The mapping population for Rpp2 consisted of 130 F2:3 families derived from a cross between the ‘BRS 184’, a Brazilian cultivar susceptible to soybean rust, and PI 230970, the genotype carrying the resistant dominant allele Rpp2. For Rpp4 locus, the mapping population consisted of 80 F2:3, derived from the cross between the same susceptible line as used for Rpp4 mapping, ‘BRS 184’, and PI 459025, the genotype carrying the resistant dominant allele Rpp4. Each F2:3 family consisted of 15 plants. The parental lines used on the crosses were obtained from Embrapa-Soybean Germplasm Collection, located at Londrina, PR, Brazil. The two F2:3 mapping populations and the parental lines for each of Rpp2 and Rpp4, were cultivated in greenhouse for evaluating their resistance to ASR, and the F2 populations were cultivated for leaf sampling. Leaf tissues were stored at −80°C for DNA extraction.
ASR inoculation and evaluation
Inoculum consisted of a fungal population collected in commercial field in the state of Mato Grosso, and maintained on cultivar BRSMS-Bacuri in a greenhouse, for artificial inoculation. Urediniospores were collected by tapping infected leaves over a plastic tray and diluted in distilled water with 0.05% of polyoxyethylenesorbitan monolaurate (Tween 20) (v/v) to a final concentration of 80,000 spores/mL. This suspension of spores was sprayed onto the plants at V5 developmental stage. Twelve days after inoculation, the parental lines and the F2:3 families were evaluated by classifying the plants showing reddish-brown lesions (RB) and low sporulation as resistant, whereas plants showing tan lesions (TAN) and high sporulation were classified as susceptible. The 15 plants per each family were independently evaluated by two people, and any discrepancy on the result was resolved by a third person evaluation. Families showing no segregation at F3 generation were classified as resistant and susceptible homozygous, whereas families showing segregation were classified as resistant heterozygous. The inoculum production and F2:3 evaluations were performed in a greenhouse with inside temperature ranging from 19 to 26°C.
The rust isolate used in this study is being maintained in a susceptible cultivar (BRMS-Bacuri) in the greenhouse. In addition, a sample of the isolate urediniospores is stored frozen at −80°C and can be obtained upon request. The ITS (Internal Transcribed Spacer) regions were cloned from P. pachyrhizi urediniospores by PCR with primers ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) and ITS5 (5′-GGAAGTAAAAGTCGTAACAAGG-3′), as described by Frederick et al. (2002). The sequencing reaction was performed in the genetic analyzer ABI PRISM 3100, with the Big Dye Terminator Cycle Sequencing Kit (Applied Biosystems, Inc., Foster City, CA), according to manufacturer’s instructions.
DNA extraction and mapping strategy
DNA of each F2 plant was extracted using a modified CTAB protocol (Keim et al. 1988). Approximately 0.1 g of leaf tissue was ground with liquid nitrogen and mixed with CTAB extraction buffer (1% CTAB, 1.4 M NaCl, 0.1 M Tris–HCl pH 8.0, 0.5 M EDTA, 0.1% mercaptoethanol). This mixture was heated, centrifuged, and the supernatant was extracted twice with chloroform–isoamyl alcohol (24:1 v/v). The DNA was precipitated with isopropanol and treated with RNase A. DNA concentration and integrity were estimated by spectrophotometer analysis and gel electrophoresis, respectively.
The soybean SSR markers used in this study were developed by the Beltsville Agricultural Research Center—BARC/ASR (Cregan et al. 1999a; Song et al. 2004). Primer sequence information for all SSR markers is available at Soybase Web Site (http://soybase.agron.iastate.edu/resources/ssr.php; Soybase 2006). To guarantee a sufficient coverage of the soybean genome, at least one polymorphic SSR marker in each 20 cM interval was used, for each one of the 20 linkage groups. In order to quickly find SSR markers associated to the target loci, markers known for their linkage to loci for resistance to soybean diseases were preferentially tested, particularly on linkage groups A2, F, G and J. Similar strategies have been also used to map other resistance loci in soybean (Mian et al. 1999; Gordon et al. 2006). The bulked segregant analysis (BSA; Michelmore et al. 1991) strategy was used to expedite the search for linked markers to ASR resistance loci. Four bulks consisting of equal amounts of DNA from six plants were made: two bulks for the resistant phenotype and two for the susceptible. The bulks were constructed based on the F2:3 ASR evaluations, allowing the selection of only homozygous plants for each differing phenotype. DNA samples from the two parental lines and from the four bulks, for each cross, were used for SSR analysis. In order to confirm putative linkage, markers found polymorphic among the contrasting bulks were used to amplify the full F2 populations.
For SSR analysis, 30 ng of DNA was used as template in a 10 μl reaction containing buffer (100 mM Tris–HCl, 500 mM KCl), 1.5 mM MgCl2, 32.5 μM of each dNTP, 0.2 μM of each primer, and 1 U of Taq DNA polymerase. The cycling consisted of 5 min at 94°C; 35 cycles of 1 min at 94°C, 1 min at 50°C, 1 min at 72°C; followed by 7 min at 72°C. The amplified fragments were separated by electrophoresis in 10% polyacrylamide gels or 3% agarose-synergel, stained with ethidium bromide, and visualized under UV light.
Data analysis
All SSR markers were scored as co-dominant markers. The ASR phenotype was evaluated as a qualitative trait and also scored as co-dominant trait, according to F2:3 progeny evaluation. Segregation ratio for SSR markers and observed ASR phenotype were tested for goodness of fit to expected ratios for segregation of a single gene using Chi-square test. A P-value greater than 0.05 in the Chi-square test reflects that the segregation fits the expected ratio. Linkage analysis and map construction were performed with the program GQMOL 9.1 (Cruz and Schuster 2006) using Kosambi function. The linkage criteria were a LOD score >3.0 and a maximum distance of 37.2 cM.
Results
Asian soybean rust evaluation
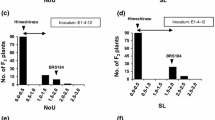
Asian soybean rust reaction was assessed following inoculation with an isolate of P. pachyrhizi, maintained on the cultivar BRSMS-Bacuri. This cultivar is known for its high susceptibility to the isolate of soybean rust used in this study (J.T. Yorinori, personal communication). This cultivar was also resistant to the first isolate identified in Brazil, in 2001. However, its resistance was broken by the field isolate that also overcame Rpp1 and Rpp3 resistance loci. The use of this cultivar to maintain the inoculum insured the use of the current isolate present in most Brazilian soybean areas. Parental lines PI 230970 (Rpp2) and PI 459025 (Rpp4) showed typical resistant reaction while ‘BRS 184’ showed the expected susceptible reaction to ASR. Identical susceptible reaction was found on PI 200492 (Rpp1) and ‘Ankur’ (Rpp3). For Rpp2 and Rpp4 mapping populations, a progeny of 15 plants was evaluated per F3 family, allowing the ASR phenotype to be scored as a qualitative trait. Both genes satisfactorily fit the expected ratio for single dominant gene segregation (Table 1). Twenty-five out of 130 F2:3 lines, evaluated for the Rpp2 mapping population, were considered homozygous resistant, 77 were heterozygous resistant and 28 were homozygous susceptible, respectively. For Rpp4 mapping population, 29 were homozygous resistant, 32 were heterozygous resistant, and 19 were homozygous susceptible in 80 F2:3 lines evaluated.
SSR screening and linkage analysis
In the Rpp2 mapping population, a total of 177 SSR markers were screened and 77 were found polymorphic between the parental lines PI 230970 and ‘BRS 184’ while for Rpp4 mapping population, 65 out of 175 SSR markers were polymorphic between the parental lines (PI 459025 and BRS 184). The four DNA bulks for each population were initially screened with the polymorphic markers. Two markers, Satt431 and Satt406, on linkage group J, were found polymorphic between the pairs of bulks for the Rpp2 mapping. For Rpp4 mapping, three SSR markers were polymorphic between the resistant and susceptible bulks; however, only two of them, Satt199 and Satt288, were actually linked to Rpp4 on linkage group G. Once their linkage was confirmed by testing the full mapping populations, other SSR markers for linkage groups G and J were chosen on basis of the available integrated genetic map of soybean (Song et al. 2004). All SSR markers mapped in this work satisfactorily fit the expected ratio for co-dominant inheritance (1:2:1) on the Rpp2 and Rpp4 mapping populations (Table 1). The program GQMOL 9.1 was used to perform the linkage analysis and mapped Rpp2 at 7.3 cM from Sat_255 and at 5.6 cM from Satt620, on the linkage group J, while Rpp4 was mapped at 1.9 cM from Satt288 and at 12.8 cM from AF162283, on the linkage group G (Fig. 1).
Linkage maps of the Rpp2 (a) and Rpp4 (b) genomic regions. On the left side is displayed the genetic map generated by this study and on the right side is an inset of the consensus linkage groups J (a) and G (b), displaying the SSR markers mapped on the region with the cumulative distances in centimorgan. On the right side of the inset are displayed the QTLs to resistance loci mapped on these regions (BSR—brown stem rot; Mi—M. incognita; SCN—soybean cyst nematode; Sclero—Sclerotinia stem rot). These insets were clipped from the Soybase webpage (Williams physical map feature; Soybase 2006). Asterisk the marker Satt620 was not present on the Williams physical map, but its position was inferred based on the composite genetic map (Song et al. 2004), available at http://www.ars.usda.gov/SP2UserFiles/person/1190/soymap2.mht
Discussion
Asian soybean rust resistance is an important trait in most soybean breeding programs worldwide. However, no resistant cultivars have been released to date. From the four previously identified resistance loci, two were mapped in this study, Rpp2 and Rpp4. This study revealed that Rpp2 and Rpp4 loci are located on linkage groups J and G, respectively. These resistance loci have been known for over 20 years, but only with the recent surge of ASR in South America, they have been focus of more intense molecular studies. Previous efforts, in Thailand, have also located a resistance locus in the linkage group G, in a close association to Satt012 and Satt472 (Nuntapunt et al. 2004). However, that resistance source was generated by mutagenesis, and whether it is allelic to Rpp4 remains to be discovered. Several other resistance loci are also under investigation. Recently, a resistance loci found on the Brazilian cultivar ‘FT-2’ has been mapped on the linkage group C2 (Brogin et al. 2004). Coincidently, a resistance loci found on the Japanese cultivar ‘Hyuuga’ was also mapped on the same genomic location, between SSR markers Satt460 and Satt307 (Monteros et al. 2007). If these two loci are allelic or are independent resistance genes is still unknown. However, while ‘Hyuuga’ remains resistant to the ASR isolate used in this study (data not shown), the resistance of ‘FT-2’ has been already defeated. Recently, the Rpp1 locus was mapped on the bottom part of linkage group G (Hyten et al. 2007), but, differently from other resistance loci, Rpp1 confers an immune response to the India-73-1 ASR isolate, used on the mapping experiment.
Since there is no study on ASR races in Brazil yet and, the fact Rpp2 and Rpp4 confer resistance to most isolates characterized abroad (Yamaoka et al. 2002), this study was performed with a field isolate of the fungus, maintained on the cultivar BRSMS-Bacuri in the greenhouse. Based on ITS sequence, this isolate is probably related to the MUT Zimbabwe isolate (GenBank accession no. AF333499), showing an identity of 99.8% (data not shown). Although the ITS sequences from all 13 isolates available on GenBank are very similar (about 98% of identity), the MUT Zimbabwe isolate contains a four nucleotide deletion compared to all other isolates (Frederick et al. 2002). The isolate used in this study (GenBank accession no. EU523736) contains the same four nucleotide deletion, making it almost identical to MUT Zimbabwe (data not shown). A more detailed investigation of the genetic diversity of South American isolates is currently under way and will be reported elsewhere. No differentiation on the disease severity was found among the two resistance sources used in this study. Both resistance loci fit the expected patterns showing typical resistant reactions (RB lesions). Rpp2 and Rpp4 also behaved as dominant traits, confirming previous genetic studies on these two loci (Hartwig 1986; Hartwig and Bromfield 1983).
Both resistance loci, Rpp2 and Rpp4, are located in regions considered hot spots for resistance genes, meaning that the strategy of using markers previously known to be linked to other resistance loci was successful. Among the markers chosen to make the initial screening, based on information of mapped resistance loci, Satt431, linked to Rbs1 locus (Bachman et al. 2001) on linkage group J, and Satt199, linked to a QTL for Sclerotinia stem rot (Arahana et al. 2001) and also for Southern root-knot nematode (Li et al. 2001), on linkage group G, were found polymorphic between the resistant and susceptible bulks.
Actually, the chromosomal region in the vicinity of Rpp4 contains a cluster of resistance genes. Significant quantitative trait loci (QTLs) for resistance to different diseases have been reported in this region. Within 20 cM interval harboring the Rpp4 locus, QTLs for resistance to the Southern root-knot nematode (M. incognita, Tamulonis et al. 1997; Li et al. 2001), Sclerotinia stem rot (Arahana et al. 2001), and soybean cyst nematode (Yue et al. 2001; Wang et al. 2001) can be found (Fig. 1b). In addition, the resistance genes rhg1 (SCN; Concibido et al. 1994; Cregan et al. 1999b); Rps4, Rps5, and Rps6 (Phytophthora root rot; Demirbas et al. 2001); Rfs (Sudden Death Syndrome; Chang et al. 1997); and the recently mapped Rpp1 (Hyten et al. 2007) are also located on this linkage group.
Linkage group J is also known to contain resistance clusters, with most of the resistance loci localized at the lower part of the group. The resistance loci Rps2 (conditions resistance to Phytophthora root rot), Rmd (powdery mildew) and Rj2 (that controls nodulation by Bradyrhizobium japonicum) were all located within 3.8 cM interval on the bottom of this linkage group (Polzin et al. 1994). The Rpp2 locus was mapped around 25 cM from this resistance cluster. In addition, the resistance loci Rbs1 and Rbs3 (Brown stem rot, Bachman et al. 2001; Patzoldt et al. 2005) and Rcs3 (Frogeye leaf spot, Mian et al. 1999) are mapped near this region, and also a recently identified QTL for soybean cyst nematode resistance (Guo et al. 2005). Kanazin et al. (1996) and Graham et al. (2002) also demonstrated cluster of R-genes in soybean. Clustering of resistance genes is not an uncommon situation and has been reported in several plant species (Michelmore and Meyers 1998).
The linkage maps generated by this study showed that the order of the molecular markers was consistent with the soybean consensus linkage map (Song et al. 2004) with few inversions and small differences on map distance (Fig. 1). Differences on mapping position are expected since the linkage analyses were performed on different mapping populations. These maps may be useful in selecting adequate markers to be used on marker assisted selection in breeding programs that aim resistance to this pathogen. Although some of the markers mapped on these populations are not closely linked to the resistance loci, other markers located on the same region may be useful in different germplasms. The Rpp4 locus was mapped between Satt288 and AF162283. Other two markers, Sat_143 and Satt612, located on either side of Satt288 (Fig. 1b), were not polymorphic on the population used in this study. Similar situation occurred in the Rpp2 region. Several markers, mapped in a 2 cM interval, were located 7.3 cM upstream the Rpp2 locus (Fig. 1a). Three additional markers on this region, Satt215, Sat_361, and Satt621 were not polymorphic on this population, and could, perhaps, reveal a closer association to Rpp2. No additional BARC_SSR markers are available in the interval between Sat_093/Sat_255/Sct_001 and Satt620. However, in order to have a saturated map of this region, additional mapping efforts involving different populations and other kind of molecular markers are needed. Currently, new SSR markers based on BAC-End sequencing are being developed by the Southern Illinois University (Shultz et al. 2006) and a soybean transcript map has been released (Choi et al. 2007). These may be useful to increase the marker density on these regions. The SSR markers linked to Rpp2 and Rpp4 described here will be of great utility in assisting the introgression of multiple loci on a single cultivar and will be of great value as a start point toward the cloning of these resistance genes.
References
Arahana VS, Graef GL, Specht JE, Steadman JR, Eskridge KM (2001) Identification of QTLs for resistance to Sclerotinia sclerotiorum in soybean. Crop Sci 41:180–188
Bachman MS, Tamulonis JP, Nickell CD, Bent AF (2001) Molecular markers linked to brown stem rot resistance genes, Rbs(1) and Rbs(2), in soybean. Crop Sci 41:527–535
Brogin RL, Arias CAA, Vello NA, Toledo JFF, Pipolo AE, Catelli LL, Marin SRR (2004) Molecular mapping of a gene conferring resistance to soybean rust. In: VII world soybean research conference, Abstracts of contributed papers and posters, Foz do Iguassu, p 318
Bromfield KR, Hartwig EE (1980) Resistance to soybean rust and mode of inheritance. Crop Sci 20:254–255
Chang SJC, Doubler TW, Kilo VY, Abuthredeih J, Prabhu R, Freire V, Suttner R, Klein J, Schmidt ME, Gibson PT, Lightfoot DA (1997) Association of loci underlying field resistance to soybean sudden death syndrome (SDS) and cyst nematode (SCN) Race 3. Crop Sci 37:965–971
Choi IY, Hyten DL, Matukumalli LK, Song Q, Chaky JM, Quigley CV, Chase K, Lark KG, Reiter RS, Yoon MS, Hwang EY, Yi SI, Young ND, Shoemaker RC, van Tassell CP, Specht JE, Cregan PB (2007) A soybean transcript map: gene distribution, haplotype and single-nucleotide polymorphism analysis. Genetics 176:685–696
Concibido VC, Diers BW, Arelli PR (2004) A decade of QTL mapping for cyst nematode resistance in soybean. Crop Sci 44:1121–1131
Concibido VC, Denny RL, Boutin SR, Hautea R, Orf JH, Young ND (1994) DNA marker analysis of loci underlying resistance to soybean cyst-nematode (Heterodera glycines ichinohe). Crop Sci 34:240–246
Cregan PB, Jarvik T, Bush AL, Shoemaker RC, Lark KG, Kahler AL, Kaya N, Vantoai TT, Lohnes DG, Chung L, Specht JE (1999a) An integrated genetic linkage map of the soybean genome. Crop Sci 39:1464–1490
Cregan PB, Mudge J, Fickus EW, Marek LF, Danesh D, Denny R, Shoemaker RC, Matthews BF, Jarvik T, Young ND (1999b) Targeted isolation of simple sequence repeat markers through the use of bacterial artificial chromosomes. Theor Appl Genet 98:919–928
Cruz CD, Schuster I (2006) GQMOL: aplicativo computacional para análise de dados moleculares e de suas associações com caracteres quantitativos, Versão 9.1. http://www.ufv.br/dbg/gqmol/gqmol.htm. Cited 2 Apr 2007
Demirbas A, Rector BG, Lohnes DG, Fioritto RJ, Graef GL, Cregan PB, Shoemaker RC, Specht JE (2001) Simple sequence repeat markers linked to the soybean rps genes for phytophthora resistance. Crop Sci 41:1220–1227
Frederick RD, Snyder CL, Peterson GL, Bonde MR (2002) Polymerase chain reaction assays for the detection and discrimination of the soybean rust pathogens Phakopsora pachyrhizi and P. meibomiae. Phytopathology 92:217–227
Gordon SG, Martin SKS, Dorrance AE (2006) Rps8 maps to a resistance gene rich region on soybean molecular linkage group F. Crop Sci 46:168–173
Graham MA, Marek LF, Shoemaker RC (2002) Organization, expression and evolution of a disease resistance gene cluster in soybean. Genetics 162:1961–1977
Guo B, Sleper DA, Arelli PR, Shannon JG, Nguyen HT (2005) Identification of Qtls associated with resistance to soybean cyst nematode races 2, 3 and 5 in soybean PI 90763. Theor Appl Genet 111:965–971
Hartman GL, Wang TC, Tschanz AT (1991) Soybean rust development and the quantitative relationship between rust severity and soybean yield. Plant Dis 75:596–600
Hartman GL, Wang TC, Shanmugasundaram S (1997) Soybean rust research: progress and future prospects. In: World soybean research conference V, proceedings, Kasetsart, pp 180–186
Hartwig EE (1986) Identification of a 4th major gene conferring resistance to soybean rust. Crop Sci 26:1135–1136
Hartwig EE, Bromfield KR (1983) Relationships among 3 genes conferring specific resistance to rust in soybeans. Crop Sci 23:237–239
Hyten DL, Hartman GL, Nelson RL, Frederick RD, Concibido VC, Narvel JM, Cregan PB (2007) Map location of the Rpp1 locus that confers resistance to soybean rust in soybean. Crop Sci 47:837–838
Kanazin V, Marek LF, Shoemaker RC (1996) Resistance gene analogs are conserved and clustered in soybean. Proc Nat Acad Sci USA 93:11746–11750
Keim P, Olson TC, Shoemaker RC (1988) A rapid protocol for isolating DNA. Soybean Genet Newslett 15:150–152
Kolmer JA (1996) Genetics of resistance to wheat leaf rust. Annu Rev Phytopathol 34:435–455
Li Z, Jakkula L, Hussey RS, Tamulonis JP, Boerma HR (2001) SSR mapping and confirmation of the qtl from PI 96354 conditioning soybean resistance to southern root-knot nematode. Theor Appl Genet 103:1167–1173
Mclean RJ, Byth DE (1980) Inheritance of resistance to rust (Phakopsora pachyrhizi) in soybeans. Aust J Agric Res 31:951–956
Mian MAR, Wang TY, Phillips DV, Alvernaz J, Boerman HR (1999) Molecular mapping of the Rcs3 gene for resistance to frogeye leaf spot in soybean. Crop Sci 39:1687–1691
Michelmore R, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis—a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 88:9828–9832
Michelmore RW, Meyers BC (1998) Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res 8:1113–1130
Miles MR, Frederick RD, Hartman GL (2006) Evaluation of soybean germplasm for resistance to Phakopsora pachyrhizi. Plant Health Progr Online. doi:10.1094/PHP-2006-0104-01-RS
Monteros MJ, Missaoui AM, Phillips DV, Walker DR, Boerma HR (2007) Mapping and confirmation of the ‘Hyuuga’ red-brown lesion resistance gene for Asian soybean rust. Crop Sci 47:829–834
Niks RE, Rubiales D (2002) Potentially durable resistance mechanisms in plants to specialized fungal pathogens. Euphytica 124:201–216
Nuntapunt M, Srisombun S, Chunwongse J (2004) Soybean breeding for rust resistance in Thailand and extent of rust resistant cultivars used. In: VII world soybean research conference, proceedings, Foz do Iguassu, pp 423–430
Patzoldt ME, Grau CR, Stephens PA, Kurtzweil NC, Carlson SR, Diers BW (2005) Localization of a quantitative trait locus providing brown stem rot resistance in the soybean cultivar Bell. Crop Sci 45:1241–1248
Polzin KM, Lohnes DG, Nickell CD, Shoemaker RC (1994) Integration of Rps2, Rmd, and Rj2 into linkage group-J of the soybean molecular map. J Hered 85:300–303
Schneider RW, Hollier CA, Whitam HK, Palm ME, Mckemy JM, Hernandez JR, Levy L, Devries-Paterson R (2005) First report of soybean rust caused by Phakopsora pachyrhizi in the continental United States. Plant Dis 89:774
Shultz JL, Kurunam D, Shopinski K, Iqbal MJ, Kazi S, Zobrist K, Bashir R, Yaegashi S, Lavu N, Afzal AJ, Yesudas CR, Kassem MA, Wu CC, Zhang HB, Town CD, Meksem K, Lightfoot DA (2006) The soybean genome database (SoyGd): a browser for display of duplicated, polyploid, regions and sequence tagged sites on the integrated physical and genetic maps of Glycine max. Nucleic Acids Res 34:758–765
Song QJ, Marek LF, Shoemaker RC, Lark KG, Concibido VC, Delannay X, Specht JE, Cregan PB (2004) A new integrated genetic linkage map of the soybean. Theor Appl Genet 109:122–128
Soybase (2006) Soybase and the soybean breeder’s toolbox. Ames, IA. http://soybase.org/. Cited 05 March 2007
Tamulonis JP, Luzzi BM, Hussey RS, Parrott WA, Boerma HR (1997) DNA markers associated with resistance to javanese root-knot nematode in soybean. Crop Sci 37:783–788
Wang D, Arelli PR, Shoemaker RC, Diers BW (2001) Loci underlying resistance to race 3 of soybean cyst nematode in Glycine soja plant introduction 468916. Theor Appl Genet 103:561–566
Yamaoka Y, Fujiwara Y, Kakishima M, Katsuya K, Yamada K, Hagiwara H (2002) Pathogenic races of Phakopsora pachyrhizi on soybean and wild host plants collected in Japan. J Gen Plant Pathol 68:52–56
Yorinori JT, Paiva WM, Frederick RD, Costamilan LM, Bertagnoli PF, Hartman GL, Godoy CV, Nunes JJ (2005) Epidemics of soybean rust (Phakopsora pachyrhizi) in Brazil and Paraguay from 2001 to 2003. Plant Dis 89:675–677
Yue P, Arelli PR, Sleper DA (2001) Molecular characterization of resistance to Heterodera glycines in soybean PI 438489b. Theor Appl Genet 102:921–928
Acknowledgments
This study was partly supported by Japan International Research Center for Agricultural Sciences (JIRCAS) international project (Identification of Stable Resistance to Soybean Rust for South America), National Council for Scientific and Technological Development (CNPq) and Studies and Projects Financing Entity (FINEP). Approved for publication by the editorial board of Embrapa Soybean as manuscript 12/2007.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by D. A. Lightfoot.
Rights and permissions
About this article
Cite this article
Silva, D.C.G., Yamanaka, N., Brogin, R.L. et al. Molecular mapping of two loci that confer resistance to Asian rust in soybean. Theor Appl Genet 117, 57–63 (2008). https://doi.org/10.1007/s00122-008-0752-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-008-0752-0