Abstract
This paper proposes a multispectral magnetic resonance imaging (MRI) image fusion scheme for improved visualization of anatomical and pathological information of meningioma (MG) brain tumors that combines the contourlet transform and fuzzy statistics. The proposed fusion technique mainly targets the tumor and its surrounding hyperintense (edema) region, which leads to improved brain imaging informatics for radiologists. The developed methodology mainly consists of the contourlet transform for multiscale and directional decomposition, fuzzy entropy for fusing approximation coefficients, and region-based fuzzy energy for fusing detailed coefficients of two input images with the same orientations. Two fusion rules are established here in order to fuse corresponding lower- and higher-frequency subbands of images. The proposed methodology is applied to five various combinations (such as T1-weighted and T2-weighted, T1 post-contrast and T2-weighted etc.) generated from four modalities of MRI images (T1-weighted, T1 post-contrast, T2-weighted, and fluid-attenuated inversion recovery (FLAIR)). A total of 150 MRI images (30 images from each of five combinations) are considered from 20 cases of MG brain tumors. A quantitative evaluation of the proposed method is performed in terms of three performance measures. The performance is compared with that of existing medical image fusion techniques tested on the same dataset. Experimental results show the superiority of the proposed methodology in terms of both qualitative and quantitative measures, which also indicates that fused images contain enriched diagnostic information that can aid the detection of tumors and edema. A fusion of post-contrast T1-weighted MRI images with FLAIR and T2-weighted MRI images provided clinically relevant information.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Brain tumors are solid masses that result from uncontrolled cell division, which leads to abnormal growth of brain cells. They are categorized as benign (non-cancerous) or malignant (cancerous). Their invasiveness and aggressiveness obstruct the normal functionalities of the brain, making them life-threatening. In addition, fluid accumulation surrounding the tumor region, called edema, leads to dysfunction of healthy tissues by creating pressure. Edema plays an important role in diagnosis, because radiologists can correlate it with the size and growth of a tumor. Among various brain imaging techniques, magnetic resonance imaging (MRI) is regarded as the most effective modality for evaluating brain tumors due to its impressive soft tissue contrast. Moreover, it is non-invasive and radiation-free. Conventional MRI scans (e.g., T1-weighted (T1), T2- weighted (T2), T1 post-contrast (T1C), and fluid-attenuated inversion recovery (FLAIR)) provide differential intensity profiles of tumors and edema [1]. For T1C, a paramagnetic substance, gadolinium contrast agent (which shortens relaxation times of neighboring protons), is injected. It crosses the blood–brain barrier and enhances the visualization of the active vascular region of a tumor. Experimentally, it is well understood that a tumor becomes hypointense in T1 whereas the same tumor gets enhanced in T1C. Interestingly, edema is almost invisible in both scans. In the FLAIR imaging protocol, the effects of the cerebrospinal fluid (CSF) are suppressed and hyperintense (finely heterogeneous) edema with an isointense tumor can be seen. In some situations, hyperintense edema is found in T2, but the visibility of isointense/hypointense (with gray matter) lesion or active region suffers due to an overlap of intensities. Therefore, the identification of an exact margin of a solid tumor region from T2 seems to be an impossible task. This kind of situation is very common in MG brain tumors, which are defined as extra-axial brain tumors that originate from arachnoidal cells. About 88–94 % of MGs are benign and a very small percentage are atypical and malignant [2]. This kind of tumor is more common in women than men. An MG is a highly vascularised lesion, and perilesional edema is very common. Sometimes, calcification appears within the lesion, but MRI has limitations in visualizing this. In practice, radiologists usually examine all types of MRI scans for anatomical and pathological evaluation of MG tumors and edema sequentially. The evaluation is not only subjective but also very time-consuming to obtain accurate diagnosis. Under such circumstances, it would be more effective to evaluate the abnormality (MG) if both the enhancing lesion and edema can be represented in a single image. In view of this, our research aims at integrating an enhancing vascular region and its surrounding edema of an MG brain tumor into a single image by keeping their original location intact. As an enhancing lesion is hyperintense and has a well-defined boundary, it can be identified from the surrounding region as well as edema in a single image. In order to achieve this, various combinations of different scans are taken into account by deriving an effective image fusion methodology.
Volumes of white matter, gray matter, and CSF have been evaluated through the fusion of different brain MRI images using a context-based fusion operator [3]. Generally, image fusion can be done at four levels, namely the signal level, pixel level, feature level, and decision level [4, 5]. Pixel-level image fusion is simple and easy to implement, but it introduces loss of information and blurring of edges due to its simplicity [6, 7] which degrades the quality of fusion. In feature-level fusion, features are extracted from different types of brain imaging data and fused using a data-driven analysis technique to obtain a more clinically significant image [8]. The problems of pixel-level fusion can be overcome by implementing multiscale decomposition along with pixel-level image fusion. Multiscale decomposition can be achieved using Fourier, Laplacian pyramid, or wavelet transforms. A one-dimensional (1D) wavelet is very efficient in capturing the edge or contour information of a 1D signal. Particularly, this leads to fast transformation and a convenient tree data structure. Hence, 1D wavelets are popular in many signal processing and communication applications. When a 1D wavelet is extended to a two-dimensional (2D) wavelet for three-band MRI image fusion by using the tensor product, it is efficient in isolating discontinuities at edge points, but lacks directionality and efficiency [9]. Therefore, several new transforms have been developed to solve the problem of 2D wavelet transform. The problem of directionality can be solved using a dual-tree complex wavelet transform (CWT) [10]. The dual-tree discrete wavelet transform [11] has been developed for image coding. As the coefficients of CWT are complex, a large amount of data is required to record the imaginary part of the coefficients. The contourlet transform [12] has better directionality and efficiency, and can preserve the geometrical structure of the image. Other approaches, such as bandlets [13], wedgelets [14], and quadtree coding [15], can be employed for representing geometrical regularities. The major drawback of these approaches is the requirement of an edge detection stage, which is very noise sensitive. Being a fixed transformation, the contourlet transform does not require edge detection, which is responsible for its popularity in a wide range of medical image fusion applications [16, 17]. The dual-tree complex contourlet transform is employed for multi-modal medical image fusion by using principle component analysis (PCA) for generating fusion rules [18]. Fuzzy logic is a well-appreciated method for estimating the transform coefficients of the contourlet transform [19] and the non-subsampled contourlet transform [20] for developing the fusion rules in medical image fusion. A fusion algorithm based on a fuzzy inference system [21] has been proposed for multi-modal medical image fusion. Li et al. proposed a novel approach of ensembling a dictionary learning technique with group sparsity and graph regularization for combining hard and soft tissues through the fusion of T2 MRI images with computed tomography (CT) images [22]. Due to the lack of anatomical information in single-photon emission computed tomography (SPECT) images, researchers have developed various fusion schemes for fusing them with MRI images to obtain an informative image [23, 24]. Shen et al. developed a fusion rule based on cross-scale for multispectral volumetric MRI image fusion and showed the applicability of their method for CT-T1, SPECT-T2, and positron emission tomography (PET)-T2 fusion [25].
This paper proposes an efficient technique for multispectral MRI image fusion to improve the visualization of MG brain tumors that combines an enhancing lesion and surrounding edema based on the contourlet transform driven by fuzzy logic. In the proposed method, two multispectral MRI images are decomposed into one low-frequency and many high-frequency subbands using the contourlet transform. For fusing the corresponding low-frequency subbands of both input images, fuzzy entropy is applied to each coefficient for obtaining its degree of participation in the fusion. This paper proposes a fusion rule based on a fuzzy entropy function for low-frequency subband fusion. A fusion rule based on a fuzzy energy function is proposed for high-frequency subbands. Experimental results show the efficacy and superiority of the proposed methodology in comparison to existing fusion methods.
2 Materials and Methods
2.1 MRI Image Database
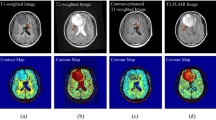
Brain MRI images were produced using a 1.5-T MRI scanner in Digital Imaging and Communications in Medicine (DICOM) format with a resolution of 512 × 512. The field of view (FOV), slice thickness, flip angle, and magnification of four types of MRI image (T1, T2, T1C, and FLAIR) were kept at 240 × 180 mm2, 5 mm, 90°, and 1.2 respectively. During the scan, the repetition time (TR) and echo time (TE) for T1 were maintained in the ranges of 250–500 and 10–30 ms, respectively. Similarly, for the T2 scan, TR and TE were fixed in the ranges of 2000–5000 and 100–200 ms, respectively. In the post-contrast T1 scan, generally 10 ml of gadolinium (Gd)-dimeglumine was injected. The usual prescribed dose for this contrast medium is 0.1 mmol/kg of body weight. 1 ml of contrast medium corresponds to 469.01 mg of Gd-dimeglumine. The inversion recovery (IR) time for FLAIR was kept at 750 ms for all cases. For each MRI protocol, 19 slices were acquired for each patient in a particular plane. 20 cases of human MG brain tumors were considered in this study. During image processing, the T1 MRI images were kept as the reference images and all other modalities (T2, T1C, and FLAIR) were co-registered with T1 using a rigid registration technique for each case. This task was accomplished with three-dimensional (3D) slicer software [26].
2.2 Proposed Methodology
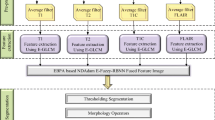
The main goal of this study is to fuse complementary information by removing redundant information from the source registered images. The contourlet transform is a multiscale and directional decomposition framework in the transform domain, which provides a sparse representation of an image. Moreover, it preserves the smooth contours of an image, which avoids the difficulties encountered with the wavelet and curvelet transforms [27]. The contourlet transform is also referred to as a pyramidal directional filter bank (PDFB), which is a combination of a Laplacian pyramid (LP) [28] and a directional filter bank (DFB) [29]. PDFB thus has a double filter bank structure. The multiscale decomposition is obtained from LP by generating the down-sampled low-pass version of the source image. The difference between the source and low pass filtered image represents the band-pass image. This resulting image is fed into the DFB to obtain the directional subbands (i.e., high-frequency components). DBF comprises two-channel quincunx filter banks with fan filters, which do not modulate the image. At each level l, DFB decomposes the image into 2l directional subbands. The contourlet transform returns the contourlet coefficients in the form of cell vector \(\left\{ {a_{L} ,d_{l,k} } \right\}\) of length n + 1, where n is the total number of decomposition levels. The first element of the cell represents low-frequency (approximation) coefficients; the remaining cell corresponds to each pyramidal level, representing a cell vector that contains band-pass directional subbands (detailed coefficients) from the DFB at that particular level. If l is the level of decomposition, the directional subbands at this level must satisfy k = 2l. The key difference between the contourlet transform and other transforms is that the contourlet transform allows different numbers of directions at each scale.
In the context of multiscale and directional decomposition, a fusion rule has an important impact on the quality of the fused image. The high-frequency components contain discontinuity lines, such as smooth curves and edges, which are key for visualizing geometric shapes. Here, high-frequency components play a pivotal role in enhancing the visual quality of MG tumor regions along with edema in a single image. Therefore, the development of separate fusion rules for the two coefficients is more efficient approach in fusion. According to this, coefficients with similar characteristics are fused with the same fusion rule. In this regard, separate fusion rules are developed for the two coefficients to achieve the desired goal. Figure 1 shows the step-by-step operations of the proposed methodology.
2.2.1 Input Images
Consider two input MRI images, A and B, that are decomposed into low- and high-frequency subbands as \(\{ a_{L}^{A} ,d_{l,k}^{A} \}\) and \(\{ a_{L}^{B} ,d_{l,k}^{B} \}\), respectively, using the contourlet transform. The coefficients of the fused image are denoted as \(\{ a_{L}^{F} ,d_{l,k}^{F} \}\), where L represents level of decomposition, k indicates the number of decomposition direction, and 1 ≤ l < L.
2.2.2 Approximation Coefficient Fusion Rule
The conventional way of averaging the corresponding approximation coefficients \(a_{L}^{A}\) and \(a_{L}^{B}\) of two inputs leads to major information loss, which in turn degrades the quality of the fused image. Therefore, the proposed method estimates the contribution of each approximation coefficient towards fusion. This estimation process is done by employing a fuzzy entropy function. To achieve this, these coefficients are considered as a fuzzy set and the membership of each is obtained from the Gaussian membership function [30]. Fuzzy entropy is then derived from the membership value of each approximation coefficient as:
This entropy function \(h(\mu_{A} (x))\) is monotonically increasing, but once μ reaches 0.5, it is at its maximum (i.e., 1); it then starts decreasing in the same manner. The integral entropy function [31] (H(A)) of a fuzzy image set (A), which is defined over a universe of discourse, say U, is:
where λ (> 0) and n are a positive constant and the total number of elements in the set.\(p(x_{i} )\) and \(\mu_{A} (x_{i} )\) denote the probability distribution and fuzzy membership functions of the i th element of x in set A, respectively. Finally, the integral entropy of all approximation coefficients is obtained from the sum of the product of individual entropies, the probability distribution of each coefficient, and a fixed value. A similar process is repeated for obtaining the integral fuzzy entropy (H(B)) of the second input image. The proposed design rule for obtaining the fused approximation coefficient from the input coefficients of two MRI images is defined as:
Multiplication of fuzzy entropy with the approximation coefficient enhances the effect of the coefficient having more information and reduces the effect of the coefficient having less information.
2.2.3 Detailed Coefficient Fusion Rule
Detailed coefficients contain directional subbands at different decomposition levels. The fuzzy information energy of each coefficient is computed as the initial stage of fusion. In this context, detailed coefficients are considered as the elements of a fuzzy set and again the Gaussian membership function is employed to generate the membership of each element. The information energy [32] of a fuzzy set A for an element x is derived from its membership and its complement \((\mu_{A}^{c} (x))\) as:
The value of information energy is in the range of 0 to 1. The local information energy, independent of \(x\), as a measure of the total fuzzy energy of a set A, is obtained by integrating the product of individual energy and the corresponding probability distribution within the limit of \([a,b]\) as:
To compute this for an individual coefficient, we use a neighborhood-based processing technique, where a square neighborhood with dimensions of w × w (w is an odd integer) is chosen. In this work, a normalized window with dimensions of 3 × 3 is considered. The local energy (E(v)) at every element (i, j) of the detailed coefficient matrix with dimensions M × N for image A with respect to a square neighborhood is computed as:
The first term \(w(i,j)\) is the weight \((w(i,j) = 1/9)\), the second term denotes the fuzzy information energy of the coefficient, and last term is the probability distribution. After obtaining the local energy of each element of the detailed coefficient matrix in image B, an element-based comparison of energy is carried out to identify the coefficients with high energy. These coefficients will be considered for fusion. The mathematical process of this comparison is:
The proposed design rule for generating the fused detailed coefficients is defined as:
For the fusion of detailed coefficients, we consider coefficients with high information energy for the fused image while discarding coefficients with low information energy. After obtaining the fused approximation and detailed coefficients at all decomposition levels, an inverse contourlet transform is applied to reconstruct the fused image (F).
2.3 Performance Evaluation
Three performance measures, namely universal image quality index (UIQI), entropy, and fusion factor (FF), were employed for the quantitative assessment of the proposed fusion technique. UIQI is employed to measure the structural distortion that appears during the process of fusion [33]. Entropy is evaluated to quantify the amount of rich information present in the fused image [34] and FF signifies how well the input images are fused in terms of mutual information [35]. The ideal value of UIQI is one and higher values are desirable for the other two measures. A statistical analysis was conducted to test the significance of the obtained results. The 95 % confidence interval (CI) for the mean of the difference in the performances of two methods with respect to a performance measure was calculated and the p value was computed for the corresponding CI in order to test the hypothesis (hypothesis: null hypothesis is rejected if the proposed approach outperforms an existing method (p-value <0.05); if both methods perform equally, the test statistic fails to reject the null hypothesis) [36].
3 Results and Discussion
In this experiment, five combinations of axial slices of MRI images (T1C-FLAIR, T1C-T2, T1-FLAIR, T1-T2, and T2-FLAIR) were used. Slice selection plays a key role in this study, because a tumor and edema do not exist in all slices. In this regard, radiologist’s assistance has been taken to consider the input slices. The corresponding slices of two MRI sequences were nominated for fusion. For example, if the radiologist chose the 11th MRI image for a particular case, then T2 and T1C of the 11th slice would be fused. To test the efficiency of the proposed methodology, a performance evaluation was conducted with UIQI, entropy, and FF for 20 cases for each combination and the performance was compared with that of conventional techniques, i.e., the average method (AVG) and PCA [17]. The quantitative assesments of fusion methods (AVG, PCA, and the proposed methodology) for all combinations in terms of UIQI, entropy, and FF for each case are presented in Tables 1, 2, and 3, respectively. The outcomes of fusion obtained using the proposed, AVG, and PCA methods for all five combinations of a particular case are depicted in Fig. 2. Similarly, the results of T1C-T2 fusion obtained using the proposed, AVG, and PCA methods are presented in Fig. 3 for two different cases. From Tables 1, 2, and 3, it can be observed that the proposed methodology has the highest mean and lowest standard deviation (Std) values of all three performance measures for all five combinations as compared to those of the AVG and PCA methods. On the basis of these results, the proposed methodology outperforms the AVG and PCA techniques in fusing multispectral information into a single image. The statistical analysis of the experimental results (Tables 1, 2, and 3) is presented in Table 4, which highlights the test outcomes based on 95 % CI of the mean of the difference. This mean of the difference was computed from the obtained values of performance measures evaluated for the fused images obtained using three fusion methods (proposed, AVG, and PCA). From Table 4, we can be 95 % certain that the proposed methodology is superior to AVG and PCA for all input combinations based on the differences in FF and UIQI measures (because the null hypothesis was rejected for all input combinations). However, for entropy, the test statistic failed to reject the null hypothesis for all combinations except for T1C-FLAIR when comparing the proposed methodology with PCA. From Table 2, there is no significant difference between the performance of the proposed method and the PCA method, even though the proposed method is better than PCA in terms of the mean and standard deviation. Therefore, due to the very little difference (in entropy), the two methods perform almost equally based on the statistical test. The statistical test demonstrates the superiority of the proposed method over AVG on the basis of entropy for all input combinations. Therefore, the statistical tests show the effectiveness and superiority of the proposed approach. From this quantitative and statistical comparison of fusion algorithms, it can be concluded that the proposed methodology is efficient and superior to AVG and PCA for multispectral MRI image fusion for all combinations. The selection of the decomposition level and number of directions for computing the contourlet transform is discussed next.
3.1 Effects of Decomposition Level on Fusion Quality
In this study, the decomposition level used to compute the contourlet transform for all input combinations was selected on the basis of finding the highest entropy in the corresponding level. A fusion experiment using the proposed algorithm was conducted on our dataset at different levels of decomposition. The computed entropy at each decomposition level is represented by the error bar for all combinations, respectively (see Figs. 4a–e). Figures 4a–e show how the entropy of fused image varies with the decomposition level. Initially, entropy increases with the level of decomposition up to a certain value, and then decreases gradually as the level is further increased. Figures 4a and c show that the mean entropy reaches a maximum value with the minimum standard deviation at the 4th level of decomposition for the T1C-FLAIR and T1-FLAIR combinations; the T1C-T2, T1-T2, and T2-FLAIR combinations have the highest mean entropy with the lowest standard deviation at level 6 (see Figs. 4b, d, and e, respectively). This strategy of finding the maximum value entropy is used for selecting the level of decomposition (the level at which entropy is maximum). The processing time increases almost linearly with the level of decomposition, as shown in Fig. 4f. During the experiment, the processing time was independent of MRI image combination.
3.2 Effects of Number of Decomposition Directions on Fusion Quality
In the contourlet transform, it is possible to change the number of directions at every multiscale decomposition level. In this study, the decomposition levels were fixed at 4 and 6, for which we obtained the desired results, as presented in Tables 1, 2, and 3. The number of decomposition directions was selected on the basis of decomposition levels according to basic property of contourlet transform. An experiment was performed using proposed algorithm to examine the changes in the performance measures (entropy and UIQI) with variations in the number of decomposition directions. Initially, there were variations in entropy with increasing number of directions. However, it was observed that when the number of decomposition directions was increased to beyond 128, entropy varied very slowly for all combinations, as shown in Fig. 5a. Figure 5b shows that for all five input combinations, UIQI increases with the number of decomposition directions for 64–128 directions, and then remains constant or slowly decreases with a further increase in the number of directions.
The primary goal of this study was to develop an image fusion technique for clinically enhancing the visualization of MG brain tumors and edema based on the contourlet transform and fuzzy statistics. The clinical contribution of this research is to aid surgical planning, therapeutic intervention, and prognosis of MG brain tumors. From the viewpoint of a radiologist, the T1C-FLAIR and T1C-T2 combinations are clinically significant, as the resulting fused images integrate anatomical and pathological information by combining the enhancing lesion and perilesional edema (see Figs. 2a, b and 3, respectively). But other three combinations are taken into account to test the efficiency and robustness of the proposed algorithm. The contourlet transform was implemented because of its ability to determine the edges in all directions, which a wavelet cannot do because it represents edges only in the vertical, horizontal, and 45° directions. In the context of this research problem, edges play a key role in the visualization of a tumor region from surrounding tissues and hence the contourlet transform is well suited for this application. Fuzzy statistics are adopted for fusing the coefficients of the contourlet transform. Fuzzy logic can be used to deal with uncertainty or ambiguity, making it an efficient approach for estimating the contribution of coefficients towards the fusion. Fuzzy integral entropy and local energy are employed for designing separate fusion rules for low- and high-frequency components. The results of fusion of all five combinations are presented in Fig. 2. The AVG method produces low-contrast images, so we cannot clearly identify edema and the lesion in the fusion of T1C-T2 and T1C-FLAIR combinations. Similarly, the PCA method introduces large artifacts, which degrade the visual quality of the fused image. The proposed technique gives impressive results for all combinations, improving the visualization of tumors and edema in a single image. Figure 3 shows the results of the T1C-T2 combination for two different cases. Each axial slice of T2 indicates homogeneous water density in the vasogenic edema that surrounds the mixed-density isointense lesion, whereas the T1C image shows an enhancing lesion surrounded by brain tissues, as depicted in Fig. 3. The edema and lesion in the fused image obtained using AVG method are more hypointense than input T2 and T1C slices. The PCA method produces a highly distorted image, in which none of the characteristics are clearly visible. The proposed methodology produces a well-defined hyperintense lesion surrounded by well-delineated perilesional edema. Therefore, the fused image obtained using the proposed algorithm presents both the lesion and edema in a single image, which would be really helpful for delineating the whole tumor region of MG. The performance of the proposed algorithm was compared with that of two existing medical image fusion techniques, namely a fuzzy inference system (FIS)-based technique [21], which was developed for multi-modal CT-MRI fusion, and the Neuro-FIS system [37], which was proposed for CT-MRI and SPECT-MRI image fusion. The performance of these two methods was evaluated using our dataset for multispectral MRI image fusion for improved visualization of MG brain tumors. At the beginning of this section, we compared the performance of the proposed methodology with that of a PCA-based method [17]. Al-Azzawi et al. proposed this technique for multi-modal CT-MRI image fusion. The PCA-based technique was also tested using our MG dataset. The results of PCA are included in the comparison with existing medical image fusion algorithms. This comparison (see Table 5) was conducted in terms of the entropy measure of the fused images. The results show the effectiveness and applicability of the proposed technique for multispectral MRI image fusion.
The main focus of the proposed methodology is to bring both the enhancing lesion and perilesional edema of an MG brain tumor into a single image plane so that the pathological and anatomical information can be integrated, improving imaging informatics. This fusion method can be used as an assistive technique for clinicians in surgical and therapeutic interventions. The identification of the boundary of a complete tumor region (enhancing lesion and edema) will aid diagnosis. The efficiency of proposed fusion technique was determined by testing the performance of a segmentation process implemented over the fused images. To accomplish this task, a popular unsupervised clustering algorithm, k-means [38], was employed. The performance validation was done with respect to the ground truth images, which are the standard reference frames made by the radiologist signifying the exact contour of a complete tumor region. Two statistical measures, namely Jaccard and Dice scores [39], were used for the quantitative evaluation of segmentation accuracy. These two performance measures compute the similarity between the outputs of k-means and the ground truths. A higher value (closer to one) of either measure indicates a better quality result. Figure 6 shows the segmentation outcomes of the k-means algorithm applied over the fused images obtained using the proposed method. From the quantitative validation of performance over 30 fused images of the T1C-T2 combination, we obtained a mean Jaccard score of 0.88 with a standard deviation of 0.05 and a mean Dice score of 0.93 with a standard deviation of 0.03. This computational analysis proves that the detection of a complete tumor area is achievable from the fused images (T1C-T2 combination) obtained using the proposed technique. The proposed methodology can thus be applied to MRI image fusion for better visualization of MG brain tumors and edema. In this study, the size and shape of the tumor varied from image to image. The imaging parameters, described in the MRI image database section, were fixed in a specified range during the scanning of the 20 cases of MG brain tumors. The proposed methodology and the existing techniques mentioned in this study were tested on this real dataset. The proposed methodology outperforms some of the existing fusion algorithms, which proves its efficiency.
We tested the proposed method for all five combinations to prove its robustness, efficiency, and flexibility. From the quantitative performance comparison with existing methodologies and the radiological significance of the fused images obtained using the proposed methodology, the proposed algorithm is promising. Segmentation of hypointense or isointense tumors is very difficult because of the overlap of intensities with neighboring tissues, so the presence of an enhancing lesion along with edema in a single image plane can help at the next level machine vision application, i.e., segmentation of the outer boundary of a complete tumor region. The performance of k-means clustering on the fused images obtained using the proposed methodology confirms this statement.
4 Conclusion
This study proposed a multispectral MRI image fusion algorithm for integrating anatomical and pathological features of two MRI scans of a given slice of an MG brain tumor. This methodology improves radiological significance by identifying the entire tumor region that includes an enhancing lesion and edema in the fused image. The major contribution of this research is the design of separate fusion rules for low- and high-frequency subbands. The proposed method outperforms the traditional technique and some existing medical image fusion methods in terms of quantitative analysis and radiological significance. In addition, the segmentation of a complete tumor region in the fused images (fusion of the T1C-T2 combination) obtained using the proposed method was accomplished using the clustering method. The proposed multispectral MRI image fusion, which combines the contourlet transform and fuzzy statistics, is a novel and coherent approach for improving the visualization of MG brain tumors along with the edema region, facilitating surgical and therapeutic interventions.
References
Osborn, A. G. (1994). Diagnostic neuroradiology: A text/atlas. St.Loui: Mosby.
Zee, C. S., Kim, P., Go, J. L., & Conti, P. (2003). Imaging of intracranial and spinal neoplasms. In Z. Petrovich, L. W. Brady, M. L. Apuzzo, & M. Bamberg (Eds.), Combined modality therapy of central nervous system tumors (pp. 109–143). Berlin: Springer.
Barra V., & Boire, J.-Y. (2000). Quantification of brain tissue volumes using MR/MR fusion. Conference Proceedings of The 22nd Annual International Conference of the IEEE Engineering in Medicine and Biology Society, 2, 1451–1454.
Piella, G. (2003). A general framework for multiresolution image fusion: from pixels to regions. Information Fusion, 4(4), 259–280.
Yang, B., Jing, Z., & Zhao, H. (2010). Review of pixel-level image fusion. Journal of Shanghai Jiaotong University (Science), 15(1), 6–12.
Irshad, H., Kamran, M., Siddiqui, A. B., & Hussain, A. (2009). Image fusion using computational intelligence: A survey. Conference Proceedings of Second International Conference on Environmental and Computer Science, 128–132.
Gonzalez, R. C., & Woods, R. E. (2009). Digital image processing. India: Pearson Education.
Calhoun, V. D., & Adali, T. (2009). Feature-based fusion of medical imaging data. IEEE Transactions on Information Technoogy in Biomedicine, 13(5), 711–720.
Zheng, Y., Elmaghraby, A. S., & Frigui, H. (2006). Three-band MRI Image fusion utilizing the wavelet-based method optimized with two quantitative fusion metrics. Conference Proceedings of SPIE 6144, 61440R–61440R–12.
Selesnick, I. W., Baraniuk, R. G., & Kingsbury, N. C. (2005). The dual-tree complex wavelet transform. IEEE Signal Processing Magazine, 22(6), 123–151.
Yang, J., Wang, Y., Xu, W., & Dai, Q. (2008). Image coding using dual-tree discrete wavelet transform. IEEE Transactions on Image Processing, 17(9), 1555–1569.
Do, M. N., & Vetterli, M. (2005). The contourlet transform: an efficient directional multiresolution image representation. IEEE Transactions on Image Processing, 14(12), 2091–2106.
Zhu, W., Li, Q., Liu, S., Xu, K., & Li, T. (2010). Image fusion algorithm based on the second generation bandelet. Conference Proceedings of International Conference on E-Product E-Service and E-Entertainment, 1–3.
Liu, F., Liu, J., & Gao, Y. (2007). Image fusion based on wedgelet and wavelet. Conference Proceedings of International Symposium on Intelligent Signal Processing and Communication Systems, 682–685.
Shusterman, E., & Feder, M. (1994). Image compression via improved quadtree decomposition algorithms. IEEE Transactions on Image Processing, 3(2), 207–215.
Chai, Y., He, Y., & Ying, C. (2008). CT and MRI image fusion based on contourlet using a novel rule. Conference Proceedings of The 2nd International Conference on Bioinformatics and Biomedical Engineering, 2064–2067.
Al-Azzawi, N. A., Sakim, H. A. M., & Wan Abdullah, A. K. (2009). An efficient medical image fusion method using contourlet transform based on PCM. Conference Proceedings of IEEE Symposium on Industrial Electronics & Applications, 1, 11–14.
Al-Azzawi, N., Sakim, H. A. M., Wan Abdullah, A. K. & Ibrahim, H. (2009). Medical image fusion scheme using complex contourlet transform based on PCA. Conference Proceedings of Annual International Conference of the IEEE Engineering in Medicine and Biology Society, 5813–5816.
Chen S., & Wu, Y. (2009). image fusion based on contourlet transform and fuzzy logic. Conference Proceedings of 2nd International Congress on Image and Signal Processing, 1–5.
Ren, X., Zheng, Y., Hu, T. & Zhang, J. (2010). Image fusion based on NSCT and fuzzy logic. Conference Proceedings of International Conference on Multimedia Technoogy, 1–4.
Teng, J., Wang, S., Zhang, J., & Wang, X. (2010). Fusion algorithm of medical images based on fuzzy logic. Conference Proceedings of Seventh International Conference on Fuzzy Systems and Knowledge Discovery, 2, 546–550.
Li, S., Yin, H., & Fang, L. (2012). Group-sparse representation with dictionary learning for medical image denoising and fusion. IEEE Transactions on Biomedical Engineering, 59(12), 3450–3459.
Li, T., & Wang, Y. (2012). Multiscaled combination of MR and SPECT images in neuroimaging: A simplex method based variable-weight fusion. Computer Methods and Programs in Biomedicine, 105(1), 31–39.
Calvini, P., Massone, A. M., Nobili, F. M., & Rodriguez, G. (2006). Fusion of the MR image to SPECT with possible correction for partial volume effects. IEEE Transactions on Nuclear Science, 53(1), 189–197.
Shen, R., Cheng, I., & Basu, A. (2013). Cross-scale coefficient selection for volumetric medical image fusion. IEEE Transactions on Biomedical Engineering, 60(4), 1069–1079.
Pieper, S., Halle, M., & Kikinis, R. (2004). 3D SLICER. Conference Proceedings of IEEE International Symposium on Biomedical Imaging: Nano to Macro, 1, 632–635.
Ma, J., & Plonka, G. (2010). The curvelet transform. IEEE Signal Processing Magazine, 27(2), 118–133.
Burt, P., & Adelson, E. (1983). The Laplacian pyramid as a compact image code. IEEE Transactions on Communications, 31(4), 532–540.
Bamberger, R., & Smith, M. (1992). A filter bank for the directional decomposition of images: theory and design. IEEE Transactions on Signal Processing, 40(4), 882–893.
Chaira, T., & Ray, A. K. (2009). Fuzzy image processing and applications with MATLAB. Florida: CRC Press.
Gang, C. (2009). Discussion on New Integral Entropy and Energy of Fuzzy Sets. Conference Proceedings of Sixth International Conference on Fuzzy Systems and Knowledge Discovery, 6, 181–185.
Wang, W.-J., & Chiu, C.-H. (1999). Entropy and information energy for fuzzy sets. Fuzzy Sets and Systems, 108(3), 333–339.
Wang, Z., & Bovik, A. (2002). A universal image quality index. IEEE Signal Processing Letters, 9(3), 81–84.
Shannon, C. E. (1948). A mathematical theory of communication. The Bell System Technical Journal, 27, 623–656.
Seetha, M., MuraliKrishna, I. V., & Deekshatulu, B. L. (2005). Data fusion performance analysis based on conventional and wavelet transform techniques. Conference Proceedings of IEEE International Geoscience and Remote Sensing Symposium, 4, 2842–2845.
Altman, D. G., & Bland, J. M. (2011). How to obtain the P value from a confidence interval. BMJ, 343, d2304.
Teng, J., Wang, S., Zhang, J., & Wang, X. (2010). Neuro-fuzzy logic based fusion algorithm of medical images. Conference Proceedings of 3rd International Congress on Image and Signal Processing, 4, 1552–1556.
Hartigan, J. A., & Wong, M. A. (1979). Algorithm AS 136: A k-means clustering algorithm. Applied statistics, 28, 100–108.
Thada, V., & Jaglan, D. V. (2013). Comparison of Jaccard, Dice, Cosine similarity coefficient to find best fitness value for web retrieved documents using genetic algorithm. International Journal of Innovations in Engineering and Technology, 2(4), 202–205.
Acknowledgments
The authors would like to acknowledge the EKO CT & MRI Scan Centre at Medical College and Hospitals Campus, Kolkata, for providing brain MRI images. The corresponding author would like to acknowledge DAE-Young Scientist Research Award Scheme (2013/36/38-BRNS/2350, dt.25-11-2013) by the Board of Research in Nuclear Sciences (BRNS), Department of Atomic Energy, for financially supporting this research work.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Koley, S., Galande, A., Kelkar, B. et al. Multispectral MRI Image Fusion for Enhanced Visualization of Meningioma Brain Tumors and Edema Using Contourlet Transform and Fuzzy Statistics. J. Med. Biol. Eng. 36, 470–484 (2016). https://doi.org/10.1007/s40846-016-0149-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40846-016-0149-5