Abstract
Understanding and analyzing the changes in vegetation cover is very important in several aspects including climatic changes, water budget, ecological balance and specially to undertake necessary conservation measures. The concept of neural network has gained much significance in the analysis of vegetation dynamics using remote sensing satellite data. In the current study an attempt has been made to predict the vegetation dynamics using MODIS NDVI time series data sets and long short term memory network, an advanced technique adapted from the artificial neural network. The dataset of 861 NDVI images from January 2000 to June 2016 is used for making the time series. The data is segregated into three sets which comprises of training set (70%), validation set (20%), and testing set (10%). To check the reliability of the experiment we have finalised two different regions after extensive research for investigation. These include different terrains in the Great Nicobar Islands, one region along the coast where vegetation has severe ecological damage due to 2004 Indian Ocean tsunami and the other, an interior region which remained imperturbable during the tsunami. Long short term memory network, an advanced neural network is trained with these NDVI values for both the regions separately to predict the future vegetation dynamics. To measure the accuracy of the LSTM network, root mean square error is calculated. The resulting plots from both the experiments indicate that the long short-term memory neural network follows the series in addition to coinciding with the required time series. Also, an unanticipated change in the trend of the NDVI series were well adapted by the network and was able to predict the future NDVI values with good accuracy maintaining RMSE less than 0.03 without providing any supplementary data. By adopting the prescribed method in the paper, anticipation of vegetation changes can be done accurately much ahead of time and take proactive measures accordingly to safeguard and improve the vegetation in any area.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Analysing the vegetation dynamics is important in the context of their sustainability. Vegetation structure changes rapidly due to various reasons influenced by both anthropogenic and natural factors, potentially resulting in the degradation and destruction. Vegetation phenology is impacted by lifecycle patterns, season and climatic conditions resulting in changes of their spectral reflectance patterns (Rußwurm and Korner 2017). Spatio-temporal mapping and monitoring of vegetation helps in assessing the health of vegetation and imply better management practices to safeguard them. The availability of the spatio-temporal satellite data aids in visualizing changing vegetation dynamics in due course of time. Further, development of vegetation indices using remote sensing satellite data provides better insights about the growth patterns, seasonal changes and health conditions of the vegetation (Wang et al. 2016).
Normalised Difference Vegetation Index (NDVI) is one of the most widely acknowledged indices for vegetation related studies (Xue and Su 2017). NDVI is a numerical indicator of the greenness of vegetation derived using the visible and near infrared (NIR) bands (Jeevalakshmi et al. 2016; Eslamian and Eslamian 2017). Typically, the NDVI values are normalized between + 1 and − 1, and a higher NDVI value indicates greener and denser vegetation (Peckham and Jordan 2007). The negative NDVI value denotes other non-vegetated classes like water, urban areas, barren lands, snow, etc (Anonymous 2000). The generalized annual NDVI profile for vegetation increases with the plant growth and reaches a peak or plateau (Jose et al. 2002). Later the profile falls off eventually with plant death or leaf senescence. Thus, NDVI series provides a means to describe plant phenology (Viovy et al. 1992; Wang et al. 2016). However, the phenological changes in concurrence with time appear to be very minimal or obscure, as observed in the vegetation of tropical rainforest due to subtle climatic variations (Valtonen et al. 2013).
The analysis of time series is widely used approach in many studies as they work with data of high temporal resolution and low spatial resolution (Petitjean et al. 2014; Khorasani et al. 2016). The availability of time series NDVI data enhances information about the vegetation conditions as well as in assessing and monitoring its changes (Ivits et al 2013). Interim changes are better inferred by time series analyses which cannot be accounted using low temporal satellite data. Time series includes any temporary changes and thus provides better analysis than any other datasets, as the analysis is based on observations of particular time frame. Major seasonal changes and inter-annual patterns can be well defined by analyzing the time series data of vegetation and surface water bodies (Haas et al. 2009; Tulbure and Broich 2013; Cordeiro et al. 2016; Rembold et al. 2015). NDVI time series analysis offers more accurate and efficient results in detecting the change in vegetation cover (Lyu and Mou 2016; Agone and Bhamare 2012), land use and land cover (LULC) classification (Gómez et al. 2016; Anderson 1976), estimation and prediction of vegetation, mapping forest disturbance (Kennedy et al. 2010), etc. Time series analysis also enables to estimate and model biomass (Gómez et al. 2014), analyze forest degradation (Shimabukuro et al. 2014) and assess forest carbon sinks (Gómez et al. 2012).
Various types of remotely sensed imagery and processing methods have been introduced and used to predict NDVI time series. Autoregressive integrated moving average (ARIMA) models are used for forecasting NDVI time series (Stepchenko 2016; Manobavan et al. 2002). These models use the adjoining data to predict the next values in the time series, but as these models are parametric and assumption of the data to be linear and stationary, makes them inappropriate for precise prediction of time series. Markov chain model simply constructs a probability mass function incrementally across the possible next states (Stepchenko and Chizhov 2015a, b). It is memory-less as it considers only the present state of the process to predict future. It is a simple and effective method for prediction, but uses a fixed window (Kriminger and Latchman 2011).
Neural networks have become popular in the analysis of remote sensing data with the increase in availability of satellite data, as they are non-parametric unlike most of the statistical methods (Foody 2006; Mas and Flores 2008). Many studies have shown that neural networks work well as they are non-linear models and perform well with noisy data as well (Atkinson and Tatnall 1997). Some of the applications of neural networks in remote sensing are land cover mapping (Zhou and Yang 2008), forest change detection (Gopal and Woodcock 1996), and predicting vegetation changes (Kang et al. 2016). Different neural networks from the Multiple Layer Perceptron (MLP) (Atkinson and Tatnall 1997), Artificial Neural Network (ANN) (Silva et al. 2014; Miller et al. 1995; Wagh et al. 2016) to advanced neural networks like Back-propagation (BP) neural networks (Zhang and Chang 2015) and Convolution Neural Networks (Maggiori et al. 2016) are used for classification of remote sensing satellite imagery (Atkinson and Tatnall 1997).
ANNs can be used for forecasting NDVI index (Kang et al. 2016; Nay et al. 2016), but as ANNs have no memory to store the information of the past data in the time series, results are less efficient. Recurrent neural network (RNN), which has memory can also be used for predicting the time series (Stepchenko and Chizhov 2015a, b; Stepchenko 2016), by training the RNN with some past NDVI values but vanishing gradient problem of RNNs makes them less suitable (Bengio et al. 1994). Further, they require a substantial amount of preceding values as input to the network for prediction of the succeeding values and thus involve lot of computational overhead compared to Long Short-Term Memory (LSTM) network. The LSTM is a variant of RNN, which has an internal memory to store the information received till time t, for a long time in the model (Skymind 2016; Budama 2015). This property of LSTM makes them very much preferable for predicting the time series. LSTM is a deep learning neural network which is highly preferred in predicting time series due to its long-term memory (Gamboa 2017).
Monitoring vegetation changes is inevitable in the context of current climatic change conditions and rapid human interventions. Vegetation, especially island ecosystems are vulnerable to both human induced disturbances and natural forces like tsunami and volcanic eruptions. Seasonal changes are common in vegetation. However, it is also effected by climate change and may lead to drastic change due to catastrophes (Verbesselt et al. 2010). Change detection methods help in assessing the change in vegetation (de Beurs and Henebry 2005) and the methods available so far are able to identify changes using low temporal resolution or decadal gap remote sensing data (Coppin et al. 2004). They are able to track the extent of areal changes but are incapable of identifying the minor seasonal or any gradual changes.
Alternatively, time series data with higher temporal resolution depict better changes in vegetation (seasonal, gradual or sudden abrupt changes) and is widely adopted in assessing minor/shorter phenomenal changes (Jana et al. 2016). Further the use of index derived from time series data (such as NDVI) is beneficial in identifying the changes compared to typical satellite data analysis, where only areal extent is calculated and compared. Time series data can be analysed using varied ANN methods and the best one proposed in recent past is the use of LSTM. Though LSTM gained specific significance in different themes (like LULC, soil moisture studies, predictions/forecasting an event), its application in vegetation studies is still in nascent stage. The research of Rußwurm and Korner (2017) can be cited as one example of LSTM study adopted for crop identification using Sentinel data. However, no specific studies have been carried out with reference to use of LSTM in forest vegetation and the current study is first of its kind to be cited as an example of LSTM study in prediction of forest dynamics as well as for the study area selected.
In the view of above context, the main objective of current research is to predict vegetation dynamics using MODIS (Moderate Resolution Imaging Spectroradiometer) NDVI time series data. Time series is a sequence of correlated scalars or vectors which vary with time. In the present study, the sequence consists of the scalar NDVI values. Time series can be predicted, if one knows the past values in the series up to time t, and then estimating the value at time t + s, s = 1, 2... The study considered s = 1. LSTM network is trained to predict the NDVI value at t + 1.
Study area
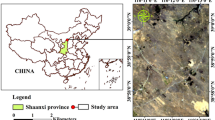
The Nicobar Islands are a group of 28 islands covering an area of 1841 km2, located between 6°45′–9°15′N latitude and 92°42′–93°56′E and are separated from Andaman group of islands by a ten-degree channel (Prasad et al. 2009). The Great Nicobar Island (GNI), the current area of interest is the southernmost island of the Nicobar Islands with an area of 1045 km2 (Fig. 1) extended between 6°45′–7°15′N latitude and 93°38′–93°55′E (Sridhar et al. 2006; Mehal et al. 2009). The GNI has two National Parks (Galathea Bay and Campbell Bay) and was declared as a biosphere reserve (85% of island) in 1989 owing its high biological diversity and richness (Mehal et al. 2009; Sridhar et al. 2006). The climate is typically moist and humid with warm temperatures relatively throughout the year having dry season from October to March followed by rainy season from April to September (Mehal et al. 2009). The seasonal changes are observed to be minimal in these islands as they receive rainfall from both southwest and northeast monsoons representing wet and evergreen conditions with few months of dry conditions (Velmurugan et al. 2015). However, observations of Velmurugan et al. (2015) from the climate data of 30 years, reported climate of Nicobar as hot and humid with temperature ranging between 23 and 30.2 °C with an average relative humidity of 79%, recording maximum rainfall during May to December and dry periods from January to March months. Overall, the climate is unpredictable without clear demarcation of wet and dry seasons affecting the vegetation condition in view of current changing climatic conditions.
The vegetation types of GNI include evergreen, mixed evergreen, moist deciduous, lowland swamp, littoral and mangrove forest (Anonymous 2003). The interior site selected for study shows dominant Andaman evergreen forest, while the coastal site is characterised by lowland swamp forest, Syzigium swamp, mixed mangroves in addition to evergreen forest (Porwal et al. 2012), The GNI was severely affected by the tsunami occurred in 2004 in Indian Ocean and the vegetation along the coastline is damaged, especially mangroves (Prasad et al. 2012). The study of Sridhar et al. (2006) observed a loss of 531.70 ha of mangrove vegetation on south-west and south-eastern side of GNI due to tsunami. While Porwal et al. (2012) reported submergence of 34% of mixed evergreen forest located near the coast. Now the vegetation seems to restore gradually under natural conditions (Prabhakaran and Paramasivam 2015). Thus, the selected sites for present experiment found to best suitable areas to trace the variation in the NDVI patterns capturing seasonal changes along with deforested and regenerating vegetation conditions.
Material and method
Satellite data and processing
The satellite data required for the study is obtained from the data service platform developed by University of Natural Resources and Life Sciences (BOKU), Vienna. The dataset consists of a total of 861 MODIS NDVI images (Vuolo et al. 2012), extracted from MODIS Terra (MOD13Q1) satellite with a spatial resolution of 250 m from January 2000 to June 2016 at an interval of 7 days. The NDVI time series obtained from the satellite imagery is often contaminated by the clouds, haze and other environmental disturbances and thus leads to noise in the data or loss of some NDVI values in the time series (Chen et al. 2004; Cai and Liu 2015). A smoothing technique or noise reduction filter is required to eliminate perturbations and obtain a smooth and continuous time series data (Geng et al. 2014). Whittaker smoothing technique was used to filter the NDVI images and to fill out any gaps in the data (Eilers 2003). The average of the NDVI values is taken to obtain a time series for further analysis. The obtained NDVI values are normalized between 0 and 1.
Method: LSTM
For predicting the vegetation dynamics using NDVI, LSTM network, an advanced technique adapted from ANN was considered. The concept of ANN is inspired by the functionality of neurons in a human brain (Gershenson 2003; Pratap and Shelja 2013). ANN is made up of neurons and links connecting the nodes. Nodes process the information and output the information received by them to other neurons through the links connecting the nodes. All the links are associated with some weights (Agatonovic-Kustrin and Beresford 2000). ANNs learn all the functional relationships of the data by varying the weights associated with the links. This property of ANNs makes them well suited for situations where a lot of data is available without sufficient knowledge about the data. Also, as they learn the hidden dependencies in the data based on the past values, they are very useful in predicting the time series and work fairly even with the noisy data (Obitko 1999; Gamboa 2017).
LSTM network is a modified structure of RNN. RNN is a feed forward network with a feedback loop and internal memory (Lyu and Mou 2016). They remember the information in a sequential order analogous to human beings. (Lipton et al. 2015; Gamboa 2017) They store this sequence in their internal memory for a long time of span which helps in taking the decision for the next step accurately compared to the traditional feed forward network (Skymind 2016). However, the recurrent structure in LSTM is a gated structure unlike RNN (Ordóñez and Roggen 2016). LSTMs have a long-term memory compared to RNN, making them suitable for time series prediction (Wu et al. 2017). Further, the vanishing gradient descent problem in a standard RNN is resolved in LSTM networks by introducing the concept of the memory cell (Wildml 2015; Colah 2015). Memory cell consists of gates which take care of what information should be stored, read or written to the network. They preserve the error that is back propagated for a long time. They can learn from a large number of time steps, over 1000 and so are very suitable for time series prediction.
A memory cell (Fig. 2) is composed of four main components—forget gate, input gate, output gate and a candidate hidden state (Zhou et al. 2016). These gates are composed of a sigmoid layer. They let the information pass through them if the sigmoid layer’s output a non-zero value. If the output is zero, no information is passed through the gate; else if it is one, all the information is passed through the network (Colah 2015).
A standard LSTM network works as follows—The first step is to decide which information is to keep in the network (Xu et al. 2015; Skymind 2016). This is decided by forget gate layer, ft. It takes into account the previous hidden state and the current input and outputs a value between 0 and 1. If the output is zero, then the information is completely forgotten likewise if it’s one, all the information is retained by the network (Jia et al. 2017) (Eqs. 1–7 adopted from Colah 2015; Kang et al. 2016)
Then the network needs to decide what new information should be stored in the cell. First the input gate layer, it decides which values are to be updated and then the new information gt is added to the cell state (Xu et al. 2015).
where ft = forget gate state; it = hidden state at time t; σ = sigmoid function, Wf = weight matrix for previous hidden to current hidden states, Wi = weight matrix for previous hidden to current hidden states ht−1 = hidden state at time t-1, Vf = weight matrix for input to hidden state links, Vi = weight matrix for input to hidden state links, Vg = weight matrix for input to hidden state links, xt = input at time t, bf = bias of the forget gate layer. bi = bias of the input gate layer, bg = bias of the input gate layer, gt = new candidate value.
Then finally, it decides what the network should output. This is done in two steps. First, the output gate layer, ot decides what values to output by the network and then the updated cell state in the previous steps is passed through tanh layer to normalize the output to be between − 1 and 1. This is how the cell states are updated in LSTM.
where ot = output gate output at time t, σ = sigmoid function, Wo = weight matrix for previous hidden to current hidden states, ht−1 = hidden state at time t-1, Vo = weight matrix for input to hidden state links, xt = input at time t, bo = bias of the input gate layer.
Experimental procedure
The focus of this study is to predict the future NDVI value of the time series (i.e. next 7th day), using the past NDVI values taking into consideration the seasonal changes and any abrupt changes in the NDVI series. Also, the research emphasizes the accuracy, adaptability and efficiency of LSTM networks in predicting the time series by reinforcing the internal dependencies and variances in the data.
The dataset under consideration is categorized into training, validation, and testing datasets. The training set is used to make the network learn the data by propagating the error back to the network and updating the weights. The validation set is used to verify the model if the error of the network is below a threshold then training can be terminated. Testing set helps in analysing the performance of the neural network on new data. Studies have shown that dividing the data in 7:2:1 ratio gives a better assessment of the network (Stepchenko 2016). The dataset of 861 images is divided into 601 training set images, 172 images for validation and 86 for testing.
The two experiments have been formulated to test two different regions in the GNI (Fig. 1) In the first case the average NDVI of an interior part of GNI is taken into account which is not affected by the tsunami and in the second only a specific portion of the island along the southern coast is considered which is affected widely by the 2004 Indian Ocean tsunami (Prasad et al. 2012). The first experiment is done to check if the network is able to learn the seasonal changes in vegetation and the latter to verify if it is able to learn the abrupt changes in the vegetation along with the seasonal changes. The input to the network is the current NDVI value and the output is the next NDVI value, which is the NDVI of the next 7th day. Unlike in RNN, here one need not give many past values as input to the network since the LSTM stores all the information received till the previous moment in its internal memory. All the NDVI values are normalized between 0 and 1 and given as input to the network. The network is stopped when it starts to over-fit the data. Over-fitting can be understood when the validation error rises continuously for some epochs in a row.
To measure the accuracy of the prediction by the LSTM network, RMSE is used. By changing the number of epochs to train the network, the RMSE in predicting the next NDVI value in the training, testing and validation phases are calculated using the Eq. (7).
where n = number of values, y = actual values, y’ = predicted values.
Results and observations
The current research is an experimental study to monitor the vegetation dynamics of GNI, typically dominated by evergreen forest in the interior of island and littoral and mangrove towards the coast. The interior forest is subjected to human interactions, while the coastal forest is impacted by natural calamities (Prasad et al. 2012). However, the area along the coast selected for experiment is severely eroded by tsunami of 2004 and proves to be better site for representing abrupt changes in the vegetation. Two experiments are done in two different terrains of GNI. The first experiment shows that the LSTM network has self-trained the seasonal changes well and the second experiment shows that the network along with seasonal changes is also able to learn any sudden changes in the NDVI values (here, due to the tsunami) well and predicted the time series well with an acceptable accuracy.
Experiment 1: seasonal changes
After experimenting with a different number of hidden nodes, the optimal number of hidden nodes is found to be 10. The RMSE for training, validation, and testing for a different number of epochs is shown in Table 1. Based on the RMSE (Table 1), 300 is decided to be the optimal number of epochs for testing. The obtained RMSE shows that the LSTM network is good at predicting the NDVI time series by learning the seasonal changes. The training, validation and testing time series seem to follow the same trend as the actual time series and as the predicted time series seems to be below the actual time series, this concludes that there is no over fitting of the data from the network. Original NDVI time series (blue) versus predicted time series in the training phase, validation phase and testing phase of the LSTM network for 300 epochs are shown in Fig. 3.
Experiment 2: both seasonal and sudden changes
Similar to the previous experiment by varying the number of hidden nodes and epochs, the optimal values are found to be 10 and 400 respectively. Figure 4 shows how the vegetation is affected by the tsunami in December 2004 and also how it is recovering after the disaster. The RMSE for training, validation, and testing for a different number of epochs is shown in Table 2. Based on the RMSE shown in Table 2, 400 is decided to be the optimal number of epochs.
NDVI images highlighting a region along the coast line of the Great Nicobar island affected by tsunami in 2004. a NDVI image of Great Nicobar before tsunami (December 2004). b NDVI image of Great Nicobar just after tsunami (January 2005). c NDVI image showing vegetation recovery in Great Nicobar vegetation (June 2016)
The obtained RMSE show that the LSTM network is satisfying at predicting the NDVI time series by learning both the seasonal changes and any abrupt changes in the NDVI that may be caused by any natural calamity or deforestation by itself without any additional information fed to the network. Clearly there is no over fitting of the data from the network in this experiment too. Original NDVI time series (blue) versus predicted time series in the training phase, validating phase and testing phase of the LSTM network for 400 epochs are shown in Fig. 5.
All the plots in all the three phases of both the experiments show that the LSTM neural network is able to follow the series well and almost coinciding with the required time series. All the unforeseen changes in the trend of the NDVI series are well learned by the network without requirement of any additional information. By implementing the proposed method to predict vegetation dynamics using NDVI time series, there is a way to forecast the vegetation in the unfolding years and take necessary measures accordingly to protect and improve the vegetation in any area. Results evidently show that the network is effective in predicting the future NDVI values by learning all the seasonal and annual changes in vegetation, changes due to deforestation or natural calamities, etc. with an appreciable accuracy. Thus, it is recommended to use LSTM network for reliable and accurate prediction of vegetation dynamics. When the LSTM network is trained with large data, the accuracy is much higher. This makes LSTM very unique and efficient method to use it for prediction of NDVI dynamics than other methods.
Conclusion
The predicted time series shows that LSTM has far better performance than many other traditional neural networks. The long-term internal memory benefits the network in training well for the given data. The gated memory cell structure is a great advantage for LSTM in predicting the time series. Unlike other neural networks, a large number of past values need not be fed to the network as LSTM can store all the information about the previous values in its internal memory. LSTM has varied applications in the areas like water quality assessment, land use and land cover change detection, monitoring land surface temperature, etc. LSTM model can efficiently predict the time series not just for the next time interval, but up to next 50 intervals which will aid the efforts to take care of vegetation well in advance to avoid any disturbance leading to loss of vegetation. The study is an experiment to analyse the use of LSTM for vegetation dynamics prediction. Since the current study typically shows evergreen nature as well as influenced by both the southwest and northeast monsoons, the phenological or seasonal changes are minimum. Further better interpretation can be made by experimenting LSTM in an area predominated by dry deciduous forest as a future investigation. Additionally, integrating climatic factors may yield a high reliable prediction of vegetation dynamics.
References
Agatonovic-Kustrin S, Beresford R (2000) Basic concepts of artificialneural network (ANN) modeling and its application in pharmaceutical research. J Pharm Biomed Anal 22(5):717–727
Agone V, Bhamare SM (2012) Change detection of vegetation cover using remote sensing and GIS. J Res Dev 2(4)
Anderson JR (1976) A land use and land cover classification system for use with remote sensor data. US Government Printing Office, Washington, D.C. ).
Anonymous (2000) Understanding the Normalized Difference VegetationIndex (NDVI). Accessed 03 June 2017
Anonymous (2003) Biodiversity characterization at landscape level in Andaman and Nicobar islands using remote sensing and geographic information system. Indian Institute of Remote Sensing, Dehra Dun, p 304
Atkinson PM, Tatnall ARL (1997) Introduction neural networks in remote sensing. Int J Remote Sens 18(4):699–709
Bengio Y, Simard P, Frasconi P (1994) Learning long-term dependencies with gradient descent is difficult IEEE Trans Neural Netw, 5(2):pp 157–166
Budama N (2015) A deep drive into recurrent neural nets. http://nikhilbuduma.com/2015/01/11/a-deep-dive-into-recurrent-neural-networks/. Accessed 19 Jun 2017
Cai S, Liu D (2015) Detecting change dates from dense satellite time series using a sub-annual change detection algorithm. Remote Sens 7:8705–8727
Chen J, Jonsson P, Tamura M et al (2004) A simple method for reconstructing a high-quality NDVI time-series data set based on the Savitzky–Golay filter. Remote Sens Environ 91:332–344
Colah (2015) Understanding LSTM Networks. http://colah.github.io/posts/2015-08-Understanding-LSTMs/. Accessed 19 Apr 2017
Coppin P, Jonckheere I, Nackaerts K, Muys B, Lambin E (2004) Review ArticleDigital change detection methods in ecosystem monitoring: a review. Int J Remote Sens 25(9):1565–1596
Cordeiro C, Cristina S, Goela PC, Danchenko S, Icely J, Lavender S, Newton A (2016) Time series analysis of remote sensing data. http://www.joclad2016.uevora.pt/index.php?/event/content/download/811/4417/file/. Accessed 19 Jan 2017
De Beurs KM, Henebry GM (2005) A statistical framework for the analysis of long image time series. Int J Remote Sens 26(8):1551–1573
Eilers PH (2003) A perfect smoother. Anal Chem 75(14):3631–3636
Eslamian S, Eslamian FA (2017) Handbook of drought and water scarcity: principles of drought and water scarcity. CRC Press, Florida, p 674
Foody GM (2006) Pattern recognition and classification of remotely sensed images by artificial neural networks. In: Ecological informatics. Springer, Berlin, pp 459–477
Gamboa JCB (2017) Deep Learning for Time-Series Analysis. arXiv preprint arXiv:1701.01887
Geng L, Ma M, Wang X, Yu W, Jia S, Wang H (2014) Comparison of eight techniques for reconstructing multi-satellite sensor time-series NDVI data sets in the Heihe river Basin, China. Remote Sens 6:2024–2049
Gershenson C (2003) Artificial neural networks for beginners. arXiv preprint cs/0308031
Gómez C, Wulder MA, White JC, Montes F, Delgado JA (2012) Characterizing 25 years of change in the area, distribution, and carbon stock of Mediterranean pines in Central Spain. Int J Remote Sens 33(17):5546–5573
Gómez C, White JC, Wulder MA, Alejandro P (2014) Historical forest biomass dynamics modelled with Landsat spectral trajectories. ISPRS J Photogramm Remote Sens 93:14–28
Gómez C, White JC, Wulder MA (2016) Optical remotely sensed time series data for land cover classification: a review. ISPRS J Photogramm Remote Sens 116:55–72
Gopal S, Woodcock C (1996) Remote sensing of forest change using artificial neural networks. IEEE Trans Geosci Remote Sens 34(2):398–404
Haas EM, Bartholomé E, Combal B (2009) Time series analysis of optical remote sensing data for the mapping of temporary surface water bodies in sub-Saharan western Africa. J Hydrol 370(1):52–63
Ivits E, Cherlet M, Sommer S, Mehl W (2013) Addressing the complexity in non-linear evolution of vegetation phenological change with time-series of remote sensing images. Ecol Ind 26:49–60
Jana A, Maiti S, Biswas A (2016) Seasonal change monitoring and mapping of coastal vegetation types along Midnapur-Balasore Coast, Bay of Bengal using multi-temporal landsat data Model. Earth Syst Environ 2:7. https://doi.org/10.1007/s40808-015-0062-x
Jeevalakshmi D, Reddy SN, Manikiam B (2016) Land cover classification based on NDVI using LANDSAT8 time series: a case study Tirupati region. In: Communication and Signal Processing (ICCSP), 2016 International Conference on IEEE, pp 1332–1335
Jia Y, Wu Z, Xu Y, Ke D, Su K (2017) Long Short-Term Memory Projection recurrent neural networkarchitectures for Piano’s continuous note recognition. J Robot
Jose E, Ryutaro T, Renchin T (2002) The polynomial least squares operation (PoLeS): a method for reducing noise in NDVI time series data. In: Proceedings of the 23rd Asian conference on remote sensing, section of data processing, algorithm and modelling, Kathmandu, Nepal
Kang L, Di L, Deng M, Yu E, Xu Y (2016) Forecasting vegetation index based on vegetation-meteorological factor interactions with artificial neural network. In: Agro-Geoinformatics (Agro-Geoinformatics), 2016 Fifth International Conference on IEEE, pp 1–6
Kennedy RE, Yang Z, Cohen WB (2010) Detecting trends in forest disturbance and recovery using yearly Landsat time series:1. LandTrendr—Temporal segmentation algorithms. Remote Sens Environ 114(12):2897–2910
Khorasani M, Ehteshami M, Ghadimi H et al (2016) Simulation and analysis of temporal changes of groundwater depth using time series modeling Model. Earth Syst Environ 2:90. https://doi.org/10.1007/s40808-016-0164-0
Kriminger E, Latchman H (2011) Markov chain model of homeplug CSMA MAC for determining optimal fixed contention window size. In: Power Line Communications and Its Applications (ISPLC), 2011 IEEE International Symposium on IEEE, pp 399–404
Lipton ZC, Berkowitz J, Elkan C (2015) A critical review of recurrent neural networks for sequence learning. arXiv preprint arXiv:1506.00019
Lyu H, Lu H, Mou L (2016) Learning a transferable change rule from a recurrent neural network for land cover change detection. Remote Sens 8(6):506
Maggiori E, Tarabalka Y, Charpiat G, Alliez P (2016). Fully convolutional neural networks for remote sensing image classification. In:Geoscience and Remote Sensing Symposium (IGARSS), 2016 IEEE International (pp 5071–5074). IEEE
Manobavan M, Lucas NS, Boyd DS, Petford N (2002) Forecasting the interannual trends in terrestrial vegetation dynamics using time series modelling techniques. ForestSAT Symposium Heriot Watt University
Mas JF, Flores JJ (2008) The application of artificial neural networks to the analysis of remotely sensed data. Int J Remote Sens 29(3):617–663
Mehal Z, Van Ryn L, Jess K, Hamilton (2009) http://www.psu.edu/dept/nkbiology/India/Great_Nicobar.pdf. Accessed on 22 Jan 2018
Miller DM, Kaminsky EJ, Rana S (1995) Neural network classification of remote-sensing data. Comput Geosci 21(3):377–386
Nay JJ, Burchfield E, Gilligan J (2016) A machine learning approach to forecasting remotely sensed vegetation health. arXiv preprint arXiv:1602.06335
Obitko M (1999) Prediction using neural networks. http://www.obitko.com/ tutorials/neural-network-prediction/introduction.html. Accessed 20 May 2017
Ordóñez FJ, Roggen D (2016) Deep convolutional and LSTM recurrent neural networks for multimodal wearable activity recognition. Sensors 16(1):115
Peckham RJ, Gyozo J (2007) Digital terrain modelling. Springer, Berlin
Petitjean F, Inglada J, Gancarski P (2014) Assessing the quality of temporal high-resolution classifications with low-resolution satellite image time series. Int J Remote Sens 35(7):2693–2712
Porwal MC, Padalia H, Roy PS (2012) Impact of tsunami on the forest and biodiversity richness in Nicobar Islands (Andaman and Nicobar Islands), India. Biodivers Conserv 21(5):1267–1287
Prabakaran N, Paramasivam B (2015) Littoral forest composition and influence of soil characteristics on vegetation succession in the Tsunami impacted coastal habitats of Nicobar Islands, India. J Isl Ecol 1:1–17
Prasad PRC, Nagabhatla N, Reddy CS, Gupta S, Rajan KS, Raza SH, Dutt CBS (2009) Assessing forest canopy closure in a geospatial medium to address management concerns for tropical islands—Southeast Asia. Environ Monit Assess 160:541–553
Prasad PRC, Mamtha Lakshmi P, Rajan KS, Bhole V, Dutt CBS (2012) Tsunami and tropical Island ecosystems—a meta-analysis of the studies in Andaman and Nicobar Islands. Biodivers Conserv 21(2):309–322
Pratap K, Shelja (2013) Artificial Neural Network (ANN) inspired from biological nervous system. Int J Appl Innovation Eng Manag 2
Rembold F, Meroni M, Urbano F, Royer A, Atzberger C, Lemoine G, Haesen D (2015) Remote sensing time series analysis for crop monitoring with the spirits software:New functionalities and use examples. Front Environ Sci 3:46
Rußwurm M, Körner M (2017) Multi-temporal land cover classification with long short-term memory neural networks.International archives of the photogrammetry, remote sensing and spatial information sciences, p 42
Shimabukuro YE, Beuchle R, Grecchi RC, Achard F (2014) Assessment of forest degradation in Brazilian Amazon due to selective logging and fires using time series of fraction images derived from Landsat ETM + images. Remote Sens Lett 5(9):773–782
Silva PR, Acerbi Júnior FW, Carvalho LMTD., Scolforo JRS (2014) Use of artificial neural networks and geographic objects for classifying remote sensing imagery. Cerne 20(2):267–276
Singh (2011) Geography, Tata McGraw Hill Series. TataMcGraw-Hill Education, USA
Skymind (2016) A beginners guide to recurrent networks and LSTMs. http://deeplearning4j.org/lstm.html. Accessed 25 Oct 2016
Solutions HG (2013) Vegetation analysis: using vegetation indices in ENVI. http://www.harrisgeospatial.com/Learn/WhitepapersDetail/TabId/802/ArtMID/2627/ArticleID/13742/Vegetation-Analysis-Using-Vegetation-Indices-in-ENVI.aspx. Accessed 25 Oct 2016
Sridhar R, Thangaradjou T, Kannan L, Ramachandran A, Jayakumar S (2006) Rapid assessment on the impact of tsunami on mangrove vegetation of the Great Nicobar Island. J Indian Soc Remote Sens 34(1):89–93
Stepchenko A (2016) Ndvi index forecasting using a layer recurrent neural net-work coupled with stepwise regression and the pca. The 5th International Virtual Scientific Conference on Informatics and Management Sciences, pp 130–135
Stepchenko A, Chizhov J (2015a) Applying markov chains for NDVI time series forecasting of latvian regions. Inf Technol Manag Sci 18(1):57–61
Stepchenko A, Chizhov J (2015b) NDVI Short-Term Forecasting Using Recurrent Neural Networks. In Environment. Technology.Resources. Proceedings of the International Scientific and Practical Conference 3:180–185
Tulbure MG, Broich M (2013) Spatiotemporal dynamic of surface water bodies using Landsat time-series data from 1999 to 2011. ISPRS J Photogramm Remote Sens 79:44–52
Valtonen A, Molleman F, Chapman CA, Carey JR, Ayres MP, Roininen H (2013) Tropical phenology: Bi-annual rhythms and interannual variation in an Afrotropical butterfly assemblage. Ecosphere 4:36
Velmurugan A, Dam Roy S, Krishnan P, Swarnam TP, Jaisankar I, Singh AK, Biswas TK (2015) Climate change and Nicobar islands: impacts and adaptation strategies. J Andaman Sci Assoc 20(1):7–18
Verbesselt J, Hyndman R, Newnham G, Culvenor D (2010) Detecting trend and seasonal changes in satellite image time series. Remote Sens Environ 114:106–115
Viovy N, Arino O, Belward AS (1992) The Best Index Slope Extraction (BISE): a method for reducing noise in NDVI time-series. Int J Remote Sens 13(8):1585–1590
Vuolo F, Mattiuzzi M, Klisch A, Atzberger C (2012) Data service platform for MODIS Vegetation Indices time series processing at BOKU Vienna: current status and future perspectives. In: SPIE Remote Sensing. International Society for Optics and Photonics, 85380A-85380A
Wagh VM, Panaskar DB, Muley AA et al (2016) Prediction of groundwater suitability for irrigation using artificial neural network model: a case study of Nanded tehsil, Maharashtra, India Model. Earth Syst Environ 2:196. https://doi.org/10.1007/s40808-016-0250-3
Wang S, Yang B, Yang Q, Lu L, Wang X, Peng Y (2016) Temporal trends and spatial variability of vegetation phenology over the northern hemisphere during 1982–2012. PLoS One 11(6):e0157134. https://doi.org/10.1371/journal.pone.0157134
Wildml (2015) Recurrent neural networks tutorial, Part 3 Backpropagation Through Time And Vanishing Gradients. http://www.wildml.com/. Accessed 19 Oct 2016
Wu C-Y, Ahmed A, Beutel A, Smola AJ, Jing H (2017) Recurrent recommender networks. In: Proceedings of the tenth ACM international conference on web search and data mining, pp 495–503
Xu Y, Lili M, Ge L, Yunchuan C, Hao P, Zhi J (2015) Classifying relations via long short term memory networks along shortest dependency paths. In: Proceedings of the 2015 conference on empirical methods in natural language processing, pp 1785–1794
Xue J, Su B (2017) Significant remote sensing vegetation indices: a review of developments and applications. J Sens, 2017
Zhang SL, Chang TC (2015) A study of image classification of remote sensing based on back-propagation neural network with extended delta bar delta. Math Problems Eng
Zhou L, Yang X (2008) Use of neural networks for land cover classification from remotely sensed imagery.the international archivesof the photogrammetry, remote sensing and spatial information sciences, p 37
Zhou GB, Wu J, Zhang CL, Zhou ZH (2016) Minimal gated unit for recurrent neural networks. Int J Autom Comput 13(3):226–234
Acknowledgements
We are thankful to University of Natural Resources and Life Sciences (BOKU), Vienna for providing us with the MODIS NDVI dataset for free. Also, grateful to anonymous reviewers for their constructive suggestions in revising the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Reddy, D.S., Prasad, P.R.C. Prediction of vegetation dynamics using NDVI time series data and LSTM. Model. Earth Syst. Environ. 4, 409–419 (2018). https://doi.org/10.1007/s40808-018-0431-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40808-018-0431-3