Abstract
Purpose of Review
Successful, durable cancer treatment is limited by drug resistance. Cancer stem cells (CSC) comprise (typically) a rare tumor subpopulation that contributes both intrinsic drug resistance and tumor re-initiation after therapy. Emerging evidence suggests that drug resistance is more complex than this single-cell level might suggest, and is likely governed by dynamics encompassing the entire tumor population. Here, we discuss the complexity of drug resistance by focusing on efforts that interface biology (wet lab) with mathematical modeling and simulation (dry lab) to study and explain the role of CSC and other cancer cells in the context of the entire ecosystem.
Recent Findings
Starting from biological evidence, we review the current state of cancer research from the perspective of the single-cell level, “The cancer cell,” its intrinsic physiopathology and its response to drug exposure. We discuss insufficiencies of this level of observation, in particular, the unaccounted for resistance to targeted therapies, and show why it is necessary to consider the entirety of the cell population, which is the only way to capture the role of biological heterogeneity. Importantly, we review how mathematical models have been implemented to elucidate mechanisms of drug resistance, and efforts made to validate biological experiments. Finally, we present emerging biological models, and therapeutic strategies inspired by mathematics, with the goal of improving the clinical management of cancer.
Summary
Over the past century, we have learned that cancer drug resistance is extraordinarily complex and requires an interdisciplinary scientific effort to unmask. The network of communication between and among cells within the diverse tumor heterogeneity drives acquired and intrinsic mechanisms of resistance. Harnessing biology and math to simulate, study, and explain the mechanisms of resistance, by considering the whole tumor population, is providing new clues to overcome it.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Drug resistance is the underlying cause of failure when treating disease, sharing commonality between viral pathogens, bacterial infection, and cancer. Elucidating drug resistance in cancer is particularly unique; however, since it evolves as an inherent disease, the mechanisms are often as individual as the person himself. The notion that cancer drug resistance is complex is not a new concept. Indeed, the history of our knowledge into resistance is fraught with complication and decades of evolving paradigms (Fig. 1). Researchers spent decades viewing drug resistance as a consequence of single, aberrant drug-effluxing proteins, subverting toxicity at a genetic level [1]. Other attempts have been classically aimed at targeting specific mutations or amplifications that increase the oncogenicity of only small populations of cells. A classic example of this is the ongoing challenge with, and generations of, new drugs for the molecular targeting of BCR-ABL in chronic myelogenous leukemia (CML), which are continually hampered by point mutations and gene amplifications [2]. Different reasons for therapy failure have been proposed, such as redundancy in the diseased intracellular pathways, de novo mutated clones, and generalized escape from (in particular epigenetic) control mechanisms at the whole genome level [3, 4], but another possible cause has also been proposed that needs to consider cancer cells at the population level. Indeed, classic paradigms are fading as researchers favor new, more complex biological models that explain drug resistance.
We present a short timeline of scientific discoveries, which have impacted the course of our understanding of cancer therapy resistance. The major turning points in our history are marked by a switch from genetic mechanisms to epigenetic (non-genetic) mechanisms of resistance, which represent a shift from binary resistance (i.e., on-off switch) to a new field that studies a continuum of phenotypes, respectively
Here, we present two unique, competing, and sometimes synergistic perspectives to the problem of drug resistance. These include (1) a “binary” view, which embraces the notion that resistance evolves at the single-cell level (some cells are totally resistant to a given drug; the others are totally sensitive to it), and as a corollary that a “silver bullet” enabled by molecularly targeting single proteins (so-called druggable targets) may underpin a cure, and (2) a “continuum” view, which highlights evidence that cancer is an extremely plastic, adaptable, and heterogeneous disease as seen both at the single-cell levels (plastic cells) due to variable epigenetic mechanisms, and at the cell population level when one aims to describe the distribution of a continuous phenotype, e.g., of resistance to a given drug, in a cell population (reviewed in Marjanovic et al. and Meacham et al.) [5, 6]. Indeed, this latter perspective suggests that cancer is a function of the whole population, and the dynamics of the cells within the “community” contributes to the development of resistance. When experimental evidence is insufficient, what other tools do we need to employ to study cancer at the population-based level? Below, we will show how biological evidence is being seamlessly integrated with mathematical and computational modeling to explain new phenomena of resistance, and therapeutic strategies to overcome it.
Despite millennia of treating cancer [7], only in the last 150 years have we begun to characterize and address the challenges associated with it. In fact, Stephen Paget was the first to describe that resistance to the surgical removal of cancer is underpinned by early dissemination of tumor cells (reviewed by Fidler and colleagues [8]). Sparked by these observations, we experienced the first chemotherapy revolution in effort to treat cancer as a systemic disease [9].
Genes, Mutations, and a Binary View of Resistance
After decades of experimenting with chemotherapy, motivated by Luria and Delbrück’s original observations in microorganisms [10], researchers began to draw unique correlations between Darwinian natural selection and mutations as an underlying driver of therapy resistance [11, 12]. Indeed, early observations suggested that tumor cell “heterogeneity” could support the expansion of lineages with unique, independent drug resistance phenotypes. Goldie and Coldman used these evidences in parallel with mathematical modeling to support the somatic mutation theory, which postulates that spontaneous mutations in tumor cells will drive acquired drug resistance [13]. These lessons drove the discovery for gene amplification of multi-drug resistance protein 1 (MDR1), which mediates efflux of toxic agents from cancer cells [14], and dihydrofolate reductase (DHFR), which results in metabolic inactivation of therapeutic agents [15]. These findings were penultimate to a new revolution in cancer treatment, paving the way for “targeted” drugs, which attack specific proteins as the drivers of resistance. Indeed, a pivotal discovery in this era is marked by the introduction of Gleevec, an inhibitor of the BCR-Abl gene translocation [16]. Despite early success, researchers and clinicians quickly realized challenges to targeted therapy due to emerging evidence for secondary, acquired mutations [17]. In these original studies, resistance was perceived as an “on-off” switch, in which single mutations and drivers of tumorigenicity could be scuttled to improve outcome.
Epigenetics and a more Fluidic View of Resistance
A surprising finding in the past several decades is that of mutation-less resistance, which can even occur while a patient is being treated [18, 19]. These clinical data have opened the door to new hypotheses that support a greater continuum of phenotypic rather than genetic mechanisms of resistance. A critical discovery in this new era was made by Hirschmann and colleagues who elucidated the cancer stem cell (CSC) hypothesis in drug resistance [20]. The notion that a pervasive, varying phenotypic state could drive resistance led to robust drug screening efforts [21], and the subsequent observation that feedback activation of oncogenes [22], and epigenetic modifications (i.e., DNA methylation) permit inherently resistant phenotypes [23•]. These early evidence into the phenotype-driven model has been challenged; however, most recently by Gupta and colleagues who applied mathematical modeling to demonstrate that cell plasticity and reversibility can create a CSC-like state in stochastic fashion [24]. Even more recently, Huang et al. and Goldman et al. discovered that therapy itself can induce an “adaptive” resistance, a Lamarckian rather than Darwinian mechanism, controlled by cell plasticity [25, 26]. Interestingly, using a strategy that incorporated a mathematical approach to simulate therapy-induced phenotypic shifting, it was determined that adaptive resistance can be driven by a population of non-CSC [25].
Bridging Biology with Mathematics
Starting at the Single-Cell Level
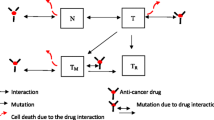
The outdated belief that cancer is a disease of single cells has recently taken center stage at the debate of drug resistance. For example, the cancer stem cell (CSC) theory, originally introduced more than a decade ago [20], takes the view that a minor population of cells within a tumor harbor inherently malignant advantages such as drug efflux, low cell cycling, and high degree of plasticity, and is also considered the subset of cells responsible for both drug resistance and tumor regrowth following unsuccessful therapy [27,28,29]. However, emerging tools such as bioinformatics and computational biology are revealing new clues for the role of CSC in drug resistance. For example, in a retrospective study by Kern and Shibata, it was revealed that drug resistance is essentially a numerical challenge. Using an unbiased approach that systematically analyzes the rate of resistance with the relatively minor population of CSC, the authors determined that it is a mathematical impossibility for CSC, alone, to drive resistance and relapse [30]. Indeed, such findings raise the question of what criteria define the CSC state, and do these criteria need to be expanded? Exhaustive efforts have been made to characterize CSC based on cell surface glycoproteins such as CD44, CD24, and CD133, metabolic proteins such as ALDH1, drug efflux MDR1, and other MDR, and many other groups of proteins that are dependent on tumor indication and stage [31]. Despite these discrepancies, the notion that a CSC-like state confers resistance is universally accepted [32].
Given the impossibility of studying cellular behavior in real time under dynamic conditions, researchers have begun to adopt computational approaches, which can simulate, predict, and model the growth and response dynamics of single cells, given parameters that are only loosely associated. For example, Wichmann and Loeffler [33], as well as Ganguly and Puri [34], developed a multi-compartment model for stem cells, early and late progenitor cells, and mature cells, to study the CSC hypothesis and the effects of chemotherapy [34] in the context of brain tumor. Moreover, Turner et al. [35] used stochastic and continuous models to study the behavior of brain CSC and their response to treatment strategies. Several other mathematical models have also been used to study dynamics of stem cells in cancer initiation and progression, as well as the treatment response and the evolution of drug resistance in chronic myeloid leukemia (CML), see for example Michor et al. [36]. In addition, the dynamics of stem cells has been widely discussed in colorectal cancer modeling, see for example Boman et al. and van Leeuwen et al. [37, 38]. Taken together, these studies provide unique evidence for the behavior of single-cell populations, which classically represent a stem-like phenotype. However, these data do not take into account that stem-like cells harbor inherent mechanisms that enable dynamic, and stochastic phenotypic variation, a property that can enable different states of resistance to environmental pressure.
Inherent Single-Cell Plasticity
The ability of cells to shift their phenotype, differentiate, and de-differentiate in the absence of genetic mutations has become known as “cell plasticity.” Indeed, this property, shared by CSC and endogenous stem cells in healthy tissue, may be observed morphologically or through phenotypic alterations such as dynamic re-wiring of the intracellular signaling networks that govern states of differentiation [39]. Transient and stochastic cell state transitions are often governed by changes in gene expression that lead to different phenotypic states and are linked to different states of DNA methylation, acetylation of histones, and activity of histone methyltransferases, demethylases, acetyltransferases, and deacetylases [40]. In the context of cancer, these epigenetic properties can govern drug resistance. For example, Sharma and colleagues identified a role for the lysine methyltransferase KDM5A in non-small cell lung cancer as a driver of quiescence leading to therapy evasion [23•].
Given the complexity of dynamic cell state transitions, and the difficulty to study them using conventional experimental techniques, numerous studies are now turning towards mathematical models as platforms for discovery. Turner and Kohandel used a computational model to generalize the hierarchical model of CSCs (stem cells, progenitor cells, and mature cells) to include the transition (dedifferentiation?) from non-CSCs to CSCs [41]. Furthermore, Chaffer et al. and Gupta et al. also developed non-hierarchical deterministic models to quantitatively describe phenotype switching between subpopulations of cancer cells, reversibly creating a CSC-like state in stochastic fashion [24, 42]. More specifically, in the context of cancer evolution, plasticity and dedifferentiation have been explored using mathematical models of diffusion approximation [43] and replicator equations [44].
Therapy-Induced Single-Cell Plasticity
More recently, it has been proposed that CSC-like phenotypes can develop under therapy pressure, which can potentially be acquired in deterministic, rather than stochastic, fashion. For example, Goldman and colleagues described, in several therapeutic contexts, that non-Darwinian dynamics can drive a temporary state of stemness, which is driven through a CD44HiCD24Hi phenotypic state [25, 45, 46]. Importantly, these authors show, using computational models that predict phenotypic transitioning rates, that combinations of drugs given in temporal sequence improve the outcome of therapy [25].
While CSC may represent an inherently “advantaged” population, emerging evidence now suggests that they may not function as a single, static population. Indeed, recent studies that rely on a mathematical approach, which seeks to simulate cell phenotype dynamics, have determined that CSC may switch behaviors as a consequence of stochasticity [24]. Such cell transitioning could explain variability in resistance, and adapting new growth potentials to accommodate changes to the microenvironment under drug pressure. This latter view challenges the “binary” hypothesis. Indeed, other evidence using a multiscale model that comprises a set of stochastic differential equations to describe pharmacokinetics, cellular dynamics, and progression-free survival at the patient level, while accounting for microenvironment adaptations including those of the CSC-like state, has confirmed an integral role that the entire population of interactive cancer cells contribute to resistance [47]. This latest example highlights the emerging view that resistance is perhaps not a phenomenon of only the single-cell level. Rather, it may require an understanding of the entire population.
Drug Resistance in the Context of the Entire Tumor Population
At the cell population level, plasticity is usually understood as adaptability and is also understood as “fitness” in a changing environment. As such, it does not assume mechanisms relying on mutations or epimutations, but it may be due to endogenous local regulatory responses, e.g., of protein synthesis such as the lac operon in Escherichia coli populations, that use it for adaptation to metabolic changes in their environment. It may also be due to more sophisticated mechanisms at the single-cell level that are considered below, but the global result in a tumor is as such only assessed as preservation of the cell population and its capacities of proliferation in a changing environment.
The complexity of population-based dynamics requires the novel integration of mathematics and computational simulation, since experimental evidence alone is insufficient [48]. This is exemplified by the use of mathematical frameworks that incorporate clonal interference and heterogeneity, which interrogate growth behavior. For example, by estimating the clone-specific exponential growth rates for each cell population in different heterogeneous contexts, Marusyk et al. recently determined that heterogeneity and drug resistance are maintained in a cell non-autonomous fashion, integrating population-based dynamics at the single-cell level [49]. Results from these studies in population dynamics might play a substantial role in driving new therapeutic strategies. Indeed, when considering drug resistance, others have determined that transient evolution through non-Darwinian dynamics is operational, which directly opposes the established dogma of natural selection and somatic mutational evolution [26]. Another fundamental question at the cell population level is—given a phenotypic and/or spatial snapshot of a cancer cell population, can we reconstitute the history of its development? This would, of course, have consequences at the single-cell level as well. Some methods of phylogenetic studies can give such answers with respect to a historical hierarchy of the development of coexistent clones [50, 51], but these answers are scarce.
The first heterogeneity that can be investigated is of a spatial nature, and it has been investigated from the point of view of successive genetic mutations giving rise by branching to different clones in different regions (spatially isolated) of the cancer cell population. It is thus of both genetic and spatial nature, at least if one assumes, following Gause’s ecological competitive exclusion principle [52], that genetically different cancer clones do not mix, as only one will prevail in the long term. The 2013 article from Gerlinger and colleagues [53] was one of the first to document such spatial and genetic heterogeneity as exemplified by sampling tissue in different parts of the same renal carcinoma and its metastatic sites. Predictable as it seemed, this report showed, to the surprise of many, that one single tumor biopsy was not enough to identify a tumor and adapt a treatment to a supposed fixed genotype.
Biological Models to Study Therapy Resistance
Our current approach to experimentation is to develop biological models, which can be exploited to study and predict therapy response in true clinical scenarios. Indeed, pre-clinical models that can be employed to study response and resistance to therapy are penultimate to translating drugs into the clinic, or understanding their clinical efficacy and utility. However, one of the major challenges to study resistance is the lack of available biological models that recapitulate the physiologic human context. It is possible that in vitro 3D cultures could partially fill the gap between conventional 2D in vitro testing and animal models [54]. Emerging studies are harnessing co-cultured organoids that comprise multiple components within the microenvironment, including cells of the endothelium and stroma with tumor cells. For example, Nyga et al. created a biomimetic in vitro model of colorectal cancer that combines tumor cells with connective stroma in 3D [55]. Emerging tools are now making 3D organotypic models more simplified by removing the need for extracellular matrices such as Matrigel that can modify the signaling tumor-stroma contexture. For example, nanofabricated scaffolds that enable complex organoid structures can serve as a platform to interrogate the effect of drugs in real time, using high-throughput screens [56]. The question remains, however, how close can these current strategies come to the complex, heterogeneous, and individual complexity that exists within patients?
Despite the exceptional utility of in vitro and in vivo models, the current paradigms fail to completely mimic the complexity of the entire tumor ecosystem, which is extraordinarily dynamic and heterogeneous. Indeed, the recent advances in immunotherapy and the necessity for a fully human model have continued to elude current biological strategies. Addressing these limitations, more recent efforts are challenging the simplicity of the binary view and are harnessing the understanding that a continuum of phenotypes predisposes the individual to resistance development. For example, utilizing mathematical algorithms that employ machine learning technology provides a computational platform to explain therapy response and resistance using 3D human tumor explanted tissue, which maintains the native architecture, heterogeneity, and immune context within the stroma [57]. Not only has this complex model achieved the goal of being derived entirely from human components, it simultaneously integrates mathematics to seamlessly describe the role of the biological system and explains drug resistance in a comprehensive manner. How can we leverage these technologies, what more can we do to develop a personalized approach to therapy that both informs and explains resistance in the context of the heterogeneous microenvironment? Many open-ended questions exist that require the use of both math and biology.
Conclusions and Future Directions
It is clearer now, more than ever, that science needs an injection of novel experimentation to unmask the mechanisms of drug resistance. Here, we have demonstrated, through emerging research, that the fastest route to discovery of novel phenomena, and unmasking the reason for cancer therapy failure, is the integration of both math and biology. From a purely biological approach to more complex models that incorporate the entire tumor ecosystem, it is increasingly clear that we need to integrate scientific disciplines. How can math inform biology, and how can we leverage our biological understanding of drug resistance to inform computational approaches? We are at an exciting inflection point for these two distinct, yet synergistic scientific disciplines to emerge as a powerhouse of discovery.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance
Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer. 2002;2(1):48–58.
Okabe S, Tauchi T, Ohyashiki K. Characteristics of dasatinib- and imatinib-resistant chronic myelogenous leukemia cells. Clin Cancer Res. 2008;14(19):6181–6.
Gorman MF, et al. Outcome for children treated for relapsed or refractory acute myelogenous leukemia (rAML): a Therapeutic Advances in Childhood Leukemia (TACL) Consortium study. Pediatr Blood Cancer. 2010;55(3):421–9.
de Rooij JD, Zwaan CM, van den Heuvel-Eibrink M. Pediatric AML: from biology to clinical management. J Clin Med. 2015;4(1):127–49.
Marjanovic ND, Weinberg RA, Chaffer CL. Cell plasticity and heterogeneity in cancer. Clin Chem. 2013;59(1):168–79.
Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501(7467):328–37.
American Cancer Society. Early history of cancer. 2014. https://www.cancer.org/cancer/cancer-basics/history-of-cancer/what-is-cancer.html.
Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer. 2003;3(6):453–8.
Christakis P. The birth of chemotherapy at Yale. Bicentennial lecture series: surgery grand round. Yale J Biol Med. 2011;84(2):169–72.
Luria SE, Delbruck M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics. 1943;28(6):491–511.
Skipper HE. The forty-year-old mutation theory of Luria and Delbruck and its pertinence to cancer chemotherapy. Adv Cancer Res. 1983;40:331–63.
Law LW. Origin of the resistance of leukaemic cells to folic acid antagonists. Nature. 1952;169(4302):628–9.
Goldie JH, Coldman AJ. A mathematic model for relating the drug sensitivity of tumors to their spontaneous mutation rate. Cancer Treat Rep. 1979;63(11–12):1727–33.
Riordan JR, Ling V. Purification of P-glycoprotein from plasma membrane vesicles of Chinese hamster ovary cell mutants with reduced colchicine permeability. J Biol Chem. 1979;254(24):12701–5.
Flintoff WF, et al. Overproduction of dihydrofolate reductase and gene amplification in methotrexate-resistant Chinese hamster ovary cells. Mol Cell Biol. 1982;2(3):275–85.
Gambacorti-Passerini C. Part I: milestones in personalised medicine—imatinib. Lancet Oncol. 2008;9(6):600.
Mahon FX, et al. Selection and characterization of BCR-ABL positive cell lines with differential sensitivity to the tyrosine kinase inhibitor STI571: diverse mechanisms of resistance. Blood. 2000;96(3):1070–9.
Berrieman HK, Lind MJ, Cawkwell L. Do beta-tubulin mutations have a role in resistance to chemotherapy? Lancet Oncol. 2004;5(3):158–64.
Talpaz M, et al. Imatinib induces durable hematologic and cytogenetic responses in patients with accelerated phase chronic myeloid leukemia: results of a phase 2 study. Blood. 2002;99(6):1928–37.
Hirschmann-Jax C, et al. A distinct “side population” of cells with high drug efflux capacity in human tumor cells. Proc Natl Acad Sci U S A. 2004;101(39):14228–33.
Gupta PB, et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138(4):645–59.
Wan X, et al. Rapamycin induces feedback activation of Akt signaling through an IGF-1R-dependent mechanism. Oncogene. 2007;26(13):1932–40.
• Sharma SV, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141(1):69–80. A milestone paper that elicits transient, epigenetically controlled, drug resistance.
Gupta PB, et al. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146(4):633–44.
Goldman A, et al. Temporally sequenced anticancer drugs overcome adaptive resistance by targeting a vulnerable chemotherapy-induced phenotypic transition. Nat Commun. 2015;6:6139.
Pisco AO, et al. Non-Darwinian dynamics in therapy-induced cancer drug resistance. Nat Commun. 2013;4:2467.
O'Brien CA, Kreso A, Jamieson CH. Cancer stem cells and self-renewal. Clin Cancer Res: Off J Am Assoc Cancer Res. 2010;16(12):3113–20.
Patel P, Chen EI. Cancer stem cells, tumor dormancy, and metastasis. Front Endocrinol (Lausanne). 2012;3:125.
Trumpp A, Wiestler OD. Mechanisms of disease: cancer stem cells—targeting the evil twin. Nat Clin Pract Oncol. 2008;5(6):337–47.
Kern SE, Shibata D. The fuzzy math of solid tumor stem cells: a perspective. Cancer Res. 2007;67(19):8985–8.
Klonisch T, et al. Cancer stem cell markers in common cancers—therapeutic implications. Trends Mol Med. 2008;14(10):450–60.
Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5(4):275–84.
Wichmann HE, Loeffler M. Mathematical modeling of cell proliferation: stem cell regulation in hemopoiesis. Vol. 1. Boca Raton: CRC Press; 1985.
Ganguly R, Puri IK. Mathematical model for the cancer stem cell hypothesis. Cell Prolif. 2006;39(1):3–14.
Turner C, et al. Characterization of brain cancer stem cells: a mathematical approach. Cell Prolif. 2009;42(4):529–40.
Michor F. Mathematical models of cancer stem cells. J Clin Oncol. 2008;26(17):2854–61.
Boman BM, et al. Symmetric division of cancer stem cells—a key mechanism in tumor growth that should be targeted in future therapeutic approaches. Clin Pharmacol Ther. 2007;81(6):893–8.
van Leeuwen IM, et al. Crypt dynamics and colorectal cancer: advances in mathematical modelling. Cell Prolif. 2006;39(3):157–81.
Merrell AJ, Stanger BZ. Adult cell plasticity in vivo: de-differentiation and transdifferentiation are back in style. Nat Rev Mol Cell Biol. 2016;17(7):413–25.
Heerboth S, et al. Use of epigenetic drugs in disease: an overview. Genet Epigenet. 2014;6:9–19.
Turner C, Kohandel M. Investigating the link between epithelial-mesenchymal transition and the cancer stem cell phenotype: a mathematical approach. J Theor Biol. 2010;265(3):329–35.
Chaffer CL, et al. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci U S A. 2011;108(19):7950–5.
Jilkine A, Gutenkunst RN. Effect of dedifferentiation on time to mutation acquisition in stem cell-driven cancers. PLoS Comput Biol. 2014;10(3):e1003481.
Kaveh K, Kohandel M, Sivaloganathan S. Replicator dynamics of cancer stem cell: selection in the presence of differentiation and plasticity. Math Biosci. 2016;272:64–75.
Goldman A. Tailoring combinatorial cancer therapies to target the origins of adaptive resistance. Mol Cell Oncol. 2016;3(1):e1030534.
Goldman, A., et al. Rationally designed 2-in-1 nanoparticles can overcome adaptive resistance in cancer. ACS Nano. 2016;10(6):5823–34.
Sun X, Bao J, Shao Y. Mathematical modeling of therapy-induced cancer drug resistance: connecting cancer mechanisms to population survival rates. Sci Rep. 2016;6:22498.
Tovar JD. Supramolecular construction of optoelectronic biomaterials. Acc Chem Res. 2013;46(7):1527–37.
Marusyk A, et al. Non-cell-autonomous driving of tumour growth supports sub-clonal heterogeneity. Nature. 2014;514(7520):54–8.
Moore LR, Rocap G, Chisholm SW. Physiology and molecular phylogeny of coexisting prochlorococcus ecotypes. Nature. 1998;393(6684):464–7.
Valaskova V, et al. Phylogenetic composition and properties of bacteria coexisting with the fungus Hypholoma fasciculare in decaying wood. ISME J. 2009;3(10):1218–21.
Hardin G. The competitive exclusion principle. Science. 1960;131(3409):1292–7.
Gerlinger M, Norton L, Swanton C. Acquired resistance to crizotinib from a mutation in CD74-ROS1. N Engl J Med. 2013;369(12):1172–3.
Zanoni M, et al. 3D tumor spheroid models for in vitro therapeutic screening: a systematic approach to enhance the biological relevance of data obtained. Sci Rep. 2016;6:19103.
Nyga A, et al. A novel tissue engineered three-dimensional in vitro colorectal cancer model. Acta Biomater. 2013;9(8):7917–26.
Arai K, et al. A novel high-throughput 3D screening system for EMT inhibitors: a pilot screening discovered the EMT inhibitory activity of CDK2 inhibitor SU9516. PLoS One. 2016;11(9):e0162394.
Majumder B, et al. Predicting clinical response to anticancer drugs using an ex vivo platform that captures tumour heterogeneity. Nat Commun. 2015;6:6169.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Aaron Goldman, Mohammad Kohandel, and Jean Clairambault declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
This article is part of the Topical Collection on Mathematical Models of Stem Cell Behavior
Rights and permissions
About this article
Cite this article
Goldman, A., Kohandel, M. & Clairambault, J. Integrating Biological and Mathematical Models to Explain and Overcome Drug Resistance in Cancer. Part 1: Biological Facts and Studies in Drug Resistance. Curr Stem Cell Rep 3, 253–259 (2017). https://doi.org/10.1007/s40778-017-0097-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40778-017-0097-1