Abstract
Background
The survival rate of patients with pancreatic cancer is low; therefore, continuous discovery and development of novel pancreatic cancer drugs are required. Functional network analysis is an integrated bioinformatics approach based on gene, target, and disease networks interaction, and it is extensively used in drug discovery and development.
Objective
This study aimed to identify if atenolol, a selective adrenergic inhibitor, can be repurposed for the treatment of pancreatic cancer using functional network analysis.
Methods
Direct target proteins (DTPs) and indirect target proteins (ITPs) were obtained from STITCH and STRING databases, respectively. Atenolol-mediated proteins (AMPs) were collected from DTPs and ITPs and further analyzed for gene ontology, KEGG pathway enrichment, genetic alterations, overall survival, and molecular docking.
Results
We obtained 176 AMPs that consisted of 10 DTPs and 166 ITPs. Among the AMPs involved in the pancreatic cancer pathways, several AMPs such as MAPK1, RELA, MAPK8, STAT1, and STAT3 were identified. Genetic alterations in seven AMPs were identified in 0.9%–16% of patients. Patients with high mRNA levels of MAPK1, RELA, STAT3, GNB1, and MMP9 had significantly worse overall survival rates compared with patients with low expression. Molecular docking studies showed that RELA and MMP9 are potential target candidates of atenolol in the treatment of patients with pancreatic cancer.
Conclusion
In conclusion, atenolol can potentially be repurposed to target pancreatic cancer cells by modulating MMP9 and NF-κB signaling. The results of this study need to be further validated in vitro and in vivo.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In 2018, pancreatic cancer was estimated to be the seventh leading cause of cancer deaths in the world [1]. The overall five-year survival rate for this disease is only 2%–9% [2]. The mortality rate of pancreatic cancer is high, partly because of inaccurate diagnosis and limited treatment options [3]. Current treatments for pancreatic cancer are surgery and adjuvant chemotherapy [3]. However, the response of patients to chemotherapy is low because of chemoresistance, metabolic aberrations, invasion, and metastasis [4]. Therefore, targeted therapies need to be developed for the effective treatment of pancreatic cancer.
Only a few effective and safe anticancer agents have been successfully developed worldwide. Furthermore, the research, development, and marketing of these drugs are expensive and time consuming [5]. Drug repurposing, which is the use of approved drugs for new medical indications, is a strategy to accelerate the discovery and development of anticancer drugs. The development of information technology and the use of big data enable drug development to be efficient and cost-effective [6]. The use of omics-based technology, improvements in data storage, data mining, and machine learning have provided adequate sources of information about diseases, molecular mechanisms, and drugs [7]. Functional network analysis is an integrated bioinformatics approach based on gene, target, and disease networks interaction, and it is extensively used in drug discovery and development [8].
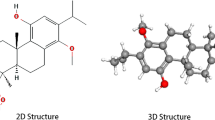
Atenolol (Fig. 1a) may be develop as anticancer agent on the basis of the drug re-purposing approach using functional network analysis. Atenolol is a β-blocker that inhibits β-adrenergic 1 and 2 receptors (ADRB1 and ADRB2) [9]. Atenolol is used to control blood pressure in patients with angina pectoris, hypertension [6], and ischemic heart disease [10]. A previous study have shown that atenolol is also effective as an anticancer agent against infantile hemangioma, which is a benign vascular tumor derived from blood vessel cell types [11]. In addition, the combination of atenolol with metformin effectively eradicates breast cancer tumors and their cellular microenvironment [12].
Activation of ADRB2 signaling promotes the progression of pancreatic ductal carcinoma [13] and pancreatic cancer microenvironment [14], thus, blocking ADRB2 signaling is a promising strategy for pancreatic cancer therapy. Atenolol is known to block β-1 and β-2 adrenergic receptor [15]. Propranolol, an ADRB antagonist, was found to inhibit pancreatic cancer cell proliferation [16]. Nevertheless, the use of atenolol, which has longer action and fewer side effects than propranolol [17], for the treatment of patients with pancreatic cancer has never been explored yet.
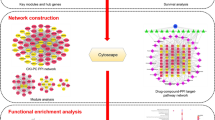
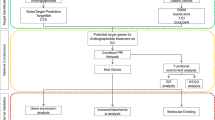
Using functional network analysis, we explored the potential of repurposing atenolol in pancreatic cancer. Direct target proteins (DTPs) and indirect target proteins (ITPs) were obtained from the STITCH and STRING databases, respectively. Atenolol-mediated proteins (AMPs) were collected from DTPs and ITPs. AMPs were further analyzed for gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment. A protein–protein interaction (PPI) network of AMPs was constructed using the STRING database, and hub proteins were selected based on their degree score. Genetic alterations were analyzed using cBioportal. Molecular docking studies were performed to identify the potential interaction between atenolol and the target proteins. We identified a possible molecular mechanism of atenolol using integrated bioinformatics analysis, which suggested that RELA and MMP9 could be potential targets of atenolol in pancreatic cancer therapy.
Material and methods
DTP acquisition
DTPs are the target protein of a compound that possesses as agonist, antagonist, or inhibitor towards the DTPs. DTPs of atenolol were obtained from the STITCH database (http://stitch.embl.de) [18]. The results were then used for subsequent analysis.
ITP acquisition
The proteins that are interacted with the DTPs were called Indirect target proteins (ITPs). The ITPs of each DTP were obtained from the STRING database (https://string-db.org) [19], with a minimum interaction score of 0.4 and maximum number of interactors of 20. The ITPs of all DTPs were generated after removing repetitive proteins. A total of DTPs and ITPs were further considered as atenolol-mediated proteins (AMPs).
PPI network
PPI networks were constructed with STRING-DB v11.0 [19], with confidence scores >0.7 and visualized by Cytoscape software [20]. Genes with a degree score of more than 10 analyzed by CytoHubba plugin were selected as hub genes [21].
GO and KEGG pathway enrichment
GO and KEGG pathway enrichment analyses were conducted by The DAVID v6.8 [22]. p < 0.05 was selected as the cutoff value.
Analysis of genetic alterations among hub genes
The genetic alterations in selected genes were analyzed using cBioPortal (http://www.cbioportal.org) [23, 24]. In the present study, the genes MAPK1, RELA, STAT3, ADCY8, GNB1, and MMP9 were screened for genetic alterations in all pancreatic studies available in the cBioportal database. The pancreatic cancer study with the highest frequency of genetic alterations was chosen for further connectivity analysis.
Gene expression profile and Kaplan–Meier survival analysis
Gene expression profiles and the prognostic value of the hub genes across pancreatic adenocarcinoma samples were evaluated using GEPIA [25] and Kaplan–Meier survival curves (http://kmplot.com) [26], respectively, by log-rank test. p < 0.05 was selected as the cutoff value.
Molecular docking
Docking simulation was conducted to predict the binding properties of atenolol with MMP9 and IKK. All computational simulations were generated on the Windows 10 Operating System, with Intel Core i5-7th Gen as a processor and 4 GB of RAM. PDB ID 4H3X contained a non-selective MMP hydroxamic acid derivative representing the model of a compound bound to the catalytic site of MMP9, while PDB ID 2OVX embedded barbiturate inhibitor RO-206-0222, depicting the model of a compound bound to MMP9 with the inactive E402Q mutant [27, 28]. The model of IKK was represented by the non-canonical NFκB pathway IKKα (5EBZ) and canonical NFκB pathway IKKβ (4KIK) on the basis of the presence of the known inhibitors, including IKK inhibitor XII and staurosporine, respectively [29, 30]. MOE-Dock program on MOE 2010 (licensed from the Faculty of Pharmacy, UGM) was used for docking simulation, RMSD calculation, and visualization of the binding interaction. The structure of atenolol was drawn in ChemDraw software and subjected to a conformational search that was minimized in MOE using the energy minimization module. The calculation allowed an induced fit using the rigid backbone for the conformation of the template protein with Amber10: EHT force field, triangle matcher as placement, and GBVI/WSA dG (kcal/mol) as the scoring function. The default settings were used in each application unless any further explanation was available. The results of the analysis were used to identify the conformation that produced the lowest energy state when atenolol was bound to the target protein.
Results and discussion
DTP and ITP acquisition
This study explored the potential of repurposing atenolol for the treatment of pancreatic cancer. We searched the DTPs of atenolol in the STITCH database, which identified 10 DTPs of atenolol (Table 1). The interactions between atenolol and DTPs were also analyzed (Fig. 1b).
PPI networks
We then generated DTP-related proteins using the STRING database, and the results are summarized in Supplementary Table 1. Using STITCH and STRING, we retrieved 176 AMPs that consisted of 10 DTPs and 166 ITPs (Supplementary Table 1). The AMPs were then constructed into a PPI network, which consisted of 173 nodes, 1470 edges, an average node degree of 17, and a high confidence interaction (Fig. 2)change to Fig. 1c. Furthermore, hub proteins were selected from the PPI network based on a specific degree score (Table 2).
a Overview of the changes in MAPK1, RELA, STAT3, ADCY8, GNB1, ADRB2, and MMP9 in genomic datasets from nine studies of pancreatic cancer. b Summary of the alterations in MAPK1, RELA, STAT3, ADCY8, GNB1, ADRB2, and MMP9 across pancreatic cancer samples (based on a study by Witkiewicz et al., 2015). c Gene network and d drug–gene network connected to MAPK1, RELA, STAT3, ADCY8, GNB1, and MMP9 across pancreatic cancer samples (based on a study by Witkiewicz et al., 2015)
Among the hub proteins, ADRB2 was the only DTP with the highest degree score of 40. This result indicated that the biological effect of atenolol was strongly correlated with ADRB2. GO analysis showed that AMPs were involved in the adenylate cyclase-activating G protein-coupled receptor signaling pathway, peptidyl-threonine phosphorylation, and negative regulation of apoptosis. The β-2 adrenergic receptor is a member of GPCR, which is involved in prostate cancer progression [31]. The AMPs are located in the cytosol, extracellular matrix, and cytoplasm. Matrix metalloproteinases (MMPs) are key extracellular matrix enzymes and targets for anticancer drugs [32]. Moreover, the AMPs play a molecular function in modulating MAP kinase activity and DNA binding. Activation of the MAPK signaling pathway plays an important role in human pancreatic cancer [33].
GO and KEGG pathway enrichment analysis
GO analysis of AMPs was classified into three groups, namely, biological process, cellular component, and molecular function (Supplementary Table 2). Pathway enrichment by KEGG of the AMPs (Supplementary Table 3) showed the regulation of ∼94 pathways. Many of the proteins, such as MAPK1, RELA, MAPK8, STAT1, and STAT3, were found to be involved in pancreatic cancer. Hub protein selection based on degree score showed that ADRB2 was the only DTP with a high degree score. These findings highlight the potential importance of ADRB2 in pancreatic cancer progression.
Analysis of genetic alterations among hub proteins
Seven AMPs were analyzed using cBioportal to explore their genomic alterations in pancreatic cancer. These AMPs consisted of three genes involved in the pancreatic cancer pathway (MAPK1, RELA, and STAT3), three genes with the highest degree scores (ADCY8, GNB1, and MMP9), and ADRB2, which is the only DTP with a high degree score. Among ten pancreatic cancer studies, the highest frequency of genetic alterations (40%) was found in a study by Witkiewicz et al. (2015) [34], Fig. 3should be Fig. 2a), which was selected for further analysis. Oncoprint analysis showed genetic alterations in seven AMPs ranging from 0.9% to 16% (Fig. 4change to Fig. 2b). Furthermore, amplification was the most common gene alteration.
This result was supported by previous studies on ADCY8, GNB1, MMP9, and RELA genetic alterations in cancer progression. For example, a mutation in ADCY8 is associated with decreased expression of tumoral PD-L1 in lung squamous cell carcinoma [35]. An activating mutation in GNB1 is associated with resistance to tyrosine kinase inhibitors in ETV6-ABL1-positive leukemia cells [36]. Genetic polymorphisms in MMP9 are associated with breast cancer risk in the Chinese Han population [37]. In addition, high expression of RELA is associated with the activation of NF-κB signaling and poor prognosis in patients with pancreatic cancer [38].
MAPK1 is a member of the MAPK family, which regulates various cellular processes, such as proliferation, differentiation, invasion, and metastasis [39]. Pancreatic cancer is characterized by constitutive activation of the MAPK1 pathway [40]. Moreover, increased MAPK1 activation is found in patients with pancreatic ductal adenocarcinoma with liver metastasis [41]. Thus, targeting MAPK1 may be a strategic way to treat pancreatic cancer.
Signal transducer and activator of transcription 3 (STAT3) is a member of the STAT family, which is activated by phosphorylation, regulates transcriptional activity, and is involved in various human tumors [42]. Activation of the STAT3 signaling pathway enhances the migration and invasion of pancreatic ductal adenocarcinoma cells [43]. Furthermore, suppression of STAT3 by emodin increases the sensitivity of human pancreatic cancer cells to EGFR inhibitors, such as erlotinib, gefitinib, afatinib, and cetuximab [44].
ADCY8 encodes adenylate cyclase, an enzyme that catalyzes cyclic AMP formation from ATP. ADCY8 is expressed in pancreatic β-cells and plays an important role in insulin secretion [45]. Another study revealed that genetic polymorphism of ADCY8 is associated with glioma risk in female patients with type I neurofibromatosis [46]. In addition, ADCY8 has been shown to play a role as a tumor suppressor gene in cervical cancer in which promoter methylation of ADCY8 is correlated with poor prognosis in patients [47].
Guanine nucleotide-binding protein beta 1 (GNB1) or the β subunit of heterotrimeric G proteins is a regulator of neurodevelopment [48]. GNB1 regulates the mTOR-induced antiapoptotic pathway in human breast cancer [49]. Furthermore, the downregulation of GNB1 is associated with poor prognosis in patients with clear cell renal cell carcinoma and is related to the VEGF signaling pathway [50]. Therefore, targeted therapy against GNB1 may be a promising candidate in pancreatic cancer treatment.
ADRB2 encodes the β-2 adrenergic receptor (also known as ADRB2), a member of the G protein-coupled receptor family, which regulates the cardiovascular system [51]. To date, the role of ADRB2 in carcinogenesis is not well understood. Adrenergic signaling plays an important role in tumor development [52]. The ADRB2 blocker propranolol inhibits migration in MDA-MB 231 breast cancer cells [53]. Another study showed that blocking of ADRB2 reduces pancreatic nerve growth factor expression, which accelerates tumor development in mice [54]. Thus, the study of ADRB2 as a target of atenolol in pancreatic cancer cells is an interesting topic.
Mutual exclusivity analysis showed that only three gene pairs (RELA-MMP9, RELA-STAT3, and GNB1-MMP9) exhibited significant (p < 0.05) co-occurrence in pancreatic samples from the UTSW study (Table 3). Subsequently, we explored the interactive relationship between seven selected genes and altered genes in the UTSW study. The results showed that a network contained six query and 45 neighbor genes (Fig. 5Achange to Fig. 2c). To reduce network complexity, we filtered out neighbor genes with 25% alterations. The results showed that only HRAS with the highest alterations remained among neighbor genes (Fig. 5B). Moreover, ADRB2, MAPK1, and MMP9 were the main targets of most cancer drugs, which indicated the potential of those proteins to be a potential atenolol target in pancreatic cancer treatment.
The co-occurrence of MMP9, RELA, and three other genes revealed an essential role of MMP9 and RELA in the mechanisms involved in atenolol treatment. MMPs play an important role in cancer initiation, tumor growth, and metastasis [55]. A previous study showed that the expression of MMP9 is increased and associated with vascular invasion, lymph node invasion, liver metastases, and TNM stage in patients with pancreatic cancer [56]. The results of this study highlighted the potency of atenolol as an inhibitor of MMP9 in pancreatic cancer cells. These results were supported by a recent study, which demonstrated that inhibition of MMP9 with an antibody leads to increased tumor-associated IL-28 and decreased stromal markers and vimentin, thereby enhancing the efficacy of chemotherapy in pancreatic cancer [57].
Gene expression profile and Kaplan–Meier survival analysis
We explored the gene expression level of seven AMPs among patients with pancreatic adenocarcinoma using the KM Plotter database. The expression of MAPK1, RELA, STAT3, GNB1, and MMP9 was significantly higher in pancreatic cancer samples than in normal samples (Fig. 6change to Fig. 3a). Additionally, ADRB2 levels were higher in pancreatic cancer samples than in normal samples. There was no difference in the level of ADCY8 between normal and pancreatic cancer samples. A Kaplan–Meier plot for the overall survival of patients with pancreatic cancer was obtained based on the low and high expression levels of each gene. Patients with high mRNA expression of MAPK1 (p = 0.0084), RELA (p = 0.062), STAT3 (p = 0.22), GNB1 (p = 0.0071), and MMP9 (p = 0.046) had significantly worse overall survival rates than those in the low expression level group (Fig. 7change to Fig. 3b). Moreover, patients with increased mRNA levels of ADCY8 (p = 0.032) and ADRB2 (p = 0.055) had better overall survival rates than those in the low expression level group.
Molecular docking
The inhibition of RELA/NFκB signaling and MMPs can be used as a treatment strategy for pancreatic cancer cell therapy. In this study, we performed molecular docking analysis to predict the possible atenolol-mediated inhibition of regulatory proteins in pancreatic cancer cells. Docking simulation and ligand–protein binding visualization were generated by Molecular Operating Environment (MOE) software. The protein targets of IKK and MMP9 were selected from KEGG pathway enrichment analysis, the top 50 genes with the highest degree score, the Kaplan–Meier plot, and networks of atenolol-associated genes in pancreatic cancer. We used two PDB IDs for each protein to validate the binding properties of atenolol. Our molecular docking study using a 4H3X model found that atenolol exhibited a lower docking score than a hydroxamic acid derivative, indicating potent binding affinity (Table 4). The visualization of the binding interaction data revealed that the hydrophilic amide group of atenolol formed an H-bond with His236, rather than forming a direct metal bond with Zn2+ and an H-bond with Leu188, which was observed with the hydroxamic acid derivative (Fig. 8)change to Fig. 4. Using a 2OVX model, atenolol had a comparable docking score with RO-206-0222. The comparable binding affinity was mediated by forming an H-bond between the hydroxyl group and Arg424 and an Arene-H bond between the Cα atom of Atenolol and His401. These bonds were stabilized by another Arene-H bond with Leu418, Arg424, and Tyr423, resulting in the hydrophobic binding properties of RO-206-0222 (Fig. 8change to Fig. 4).
Given the instability of full-length MMP9, every molecular docking approach requires several types of crystal structure models to validate the binding properties of a certain compound [28]. In our study, we used two different PDB IDs representing the wild type and mutant catalytic sites in MMP9 for validation. Our analysis of the wild-type catalytic site in MMP9 revealed that the hydrophilic amide group of atenolol contributed to the H-bond binding with His236 and one of three histidine residues coordinated with the catalytic ion Zn2+ responsible for substrate proteolysis in the active enzyme [58]. The interruption of the catalytic ion Zn2+ also appeared between Cα of atenolol and His401 with the E402Q mutant, highlighting the importance of the hydrophilic amide group on the binding interaction. In addition, a similar hydrophobic binding pattern was observed with the strong MMP9 inhibitor RO-206-0222, suggesting the strong inhibitory activity of atenolol on the MMP9-E402Q mutant [28]. Atenolol bound to the catalytic site of MMP9 by interrupting the ion Zn2+ catalytic site and was predicted to possess strong inhibitory activity.
The molecular docking study of the ATP-binding site of IKKα and IKKβ demonstrated that atenolol had comparable binding affinity with IKK inhibitor XII but lower binding affinity than staurosporine (Table 4). The three H-bonds represented by the binding between the amide group with Glu19, hydroxyl group with Thr23, and ether group with Gly22 contributed to the comparable binding affinity of atenolol with IKKα (Fig. 8)should be Fig. 4. The low affinity of atenolol compared with that of staurosporine and IKKβ was caused by a reduced number of amino acid bindings, which only formed one H-bond between the amide group and Asp166 and one Arene-H bond between benzene and Val29. Meanwhile, the staurosporine interaction possessed three H-bonds with a short distance and four Arene-H bonds (Fig. 8)should be Fig. 4. In summary, atenolol interacted with MMP9 on its wild type or E402Q mutant catalytic site and with IKK through the ATP-binding site.
NF-κB is a transcription factor that contributes to cancer development [59]. RELA (also known as p65) is a subunit of the NF-κB transcription factor complex, together with p50 [60]. Activation of NF-κB signaling leads to phosphorylation of IkBα and subsequent translocation of the p65/p50 complex into the nucleus [61]. NF-κB signaling contributes to pancreatic cancer development. Furthermore, high expression of RELA is associated with the activation of NF-κB signaling and poor prognosis in patients with pancreatic cancer [38]. Activation of NF-κB/p65 signaling stimulates anti-apoptotic responses in pancreatic cancer cells [62]. The inhibition of NF-kB signaling induces apoptosis [63] and inhibits angiogenesis in pancreatic cancer cells [64]. Taken together, targeting NF-κB/RELA may be a promising strategy in pancreatic cancer therapy.
IKK is a component of NF-κB signaling proteins and a multiprotein complex consisting of two kinase subunits. The first is IKKα, which is responsible for adaptive immunity, and the second is IKKβ, which is essential for innate immunity and inflammation [65]. Our molecular docking data on IKKα demonstrated the important role of the hydroxyl and ether groups beside the amide group in forming an H-bond. The hydroxyl group interacted with Thr23, the target binding site of Akt for phosphorylation, whereas the ether group bound to Gly22, an important residue on the glycine-rich loop [66]. The amide group and the benzene ring were two important functional groups for atenolol binding to IKKβ. Although the binding affinity of atenolol was lower than that of staurosporine, the interaction of the amide group from atenolol with Asp166 was similar to compound NSC-719177, a strong IKKβ inhibitor that is effective at low concentrations [67]. The hydrophobic binding between the benzene ring with Val29 was also similar to the other selective IKKβ kinase inhibitors, such as 4-phenyl-7-azaindoles, thiophenecarboxamide, and 2-amino-3-cyano-4-alkyl-6-(2-hydroxyphenyl) pyridine derivatives [68,69,70]. This study highlighted the key structure of atenolol responsible for its interaction with the ATP-binding site of IKK. The findings suggested that atenolol possibly inhibited its activity through the canonical or non-canonical pathway of NF-kB signaling.
The results of this study highlighted the potential of atenolol for pancreatic cancer therapy. RELA and MMP9 are potential target candidates of atenolol in the treatment of patients with pancreatic cancer. These target candidates greatly complement other genomics data and provide essential information for further research on atenolol, as well as the development of pancreatic cancer drugs. This study used bioinformatics approaches, and the results must be further validated in vitro and in vivo. In addition, the potential drug repurposing of atenolol for use in other types of cancer should be explored.
Conclusion
In conclusion, we performed functional network analysis using the databases STITCH; STRING; Database for Annotation, Visualization and Integrated Discovery (DAVID); cBioportal; and KMPlotter to investigate the potential of atenolol for drug repurposing in pancreatic cancer. A molecular docking study revealed that atenolol may inhibit MMP9 and NF-κB signaling in pancreatic cancer cells. Therefore, atenolol has the potential to be repurposed in pancreatic cancer therapy by targeting MMP9 and NF-κB signaling. The results of this study need to be further validated in vitro and in vivo.
Data availability
All data generated or analysed during this study are included in the Additional Files of this article.
Abbreviations
- ADRB1:
-
β-Adrenergic 1 Receptor
- ADRB2:
-
β-Adrenergic 2 Receptor
- AMPs:
-
Atenolol-mediated proteins
- DAVID:
-
Database for Annotation, Visualization and Integrated Discovery
- DTPs:
-
Direct target proteins
- GO:
-
Gene Ontology
- ITPs:
-
Indirect target proteins
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- MMP:
-
Matrix metalloproteinase
- PPI:
-
Protein–Protein Interaction
- SNP:
-
Single-Nucleotide Polymorphism
- STAT3:
-
Signal Transducer and Activator of Transcription 3
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. https://doi.org/10.3322/caac.21492.
Ilic M, Ilic I. Epidemiology of pancreatic cancer. World J Gastroenterol. 2016;22(44):9694–705. https://doi.org/10.3748/wjg.v22.i44.9694.
Rawla P, Sunkara T, Gaduputi V. Epidemiology of pancreatic cancer: global trends, etiology and risk factors. World J Oncol. 2019;10(1):10–27. https://doi.org/10.14740/wjon1166.
Grasso C, Jansen G, Giovannetti E. Drug resistance in pancreatic cancer: impact of altered energy metabolism. Crit Rev Oncol Hematol. 2017;114:139–52. https://doi.org/10.1016/j.critrevonc.2017.03.026.
Sleire L, Forde HE, Netland IA, Leiss L, Skeie BS, Enger PO. Drug repurposing in cancer. Pharmacol Res. 2017;124:74–91. https://doi.org/10.1016/j.phrs.2017.07.013.
Heel RC, Brogden RN, Speight TM, Avery GS. Atenolol: a review of its pharmacological properties and therapeutic efficacy in angina pectoris and hypertension. Drugs. 1979;17(6):425–60. https://doi.org/10.2165/00003495-197917060-00001.
Mottini C, Napolitano F, Li Z, Gao X, Cardone L. Computer-aided drug repurposing for cancer therapy: approaches and opportunities to challenge anticancer targets. Semin Cancer Biol. 2019:S1044-579X(19)30139-7. https://doi.org/10.1016/j.semcancer.2019.09.023.
Shi J, Dong B, Zhou P, Guan W, Peng Y. Functional network analysis of gene-phenotype connectivity associated with temozolomide. Oncotarget. 2017;8(50):87554–67. https://doi.org/10.18632/oncotarget.20848.
Nuttall SL, Routledge HC, Kendall MJ. A comparison of the beta1-selectivity of three beta1-selective beta-blockers. J Clin Pharm Ther. 2003;28(3):179–86. https://doi.org/10.1046/j.1365-2710.2003.00477.x.
Cruickshank JM, McAinsh J. Atenolol and ischaemic heart disease: an overview. Curr Med Res Opin. 1991;12(8):485–96. https://doi.org/10.1185/03007999109111659.
Bayart CB, Tamburro JE, Vidimos AT, Wang L, Golden AB. Atenolol versus propranolol for treatment of infantile Hemangiomas during the proliferative phase: a retrospective noninferiority study. Pediatr Dermatol. 2017;34(4):413–21. https://doi.org/10.1111/pde.13177.
Talarico G, Orecchioni S, Dallaglio K, Reggiani F, Mancuso P, Calleri A, et al. Aspirin and atenolol enhance metformin activity against breast cancer by targeting both neoplastic and microenvironment cells. Sci Rep. 2016;6:18673. https://doi.org/10.1038/srep18673.
Wan C, Gong C, Zhang H, Hua L, Li X, Chen X, et al. beta2-adrenergic receptor signaling promotes pancreatic ductal adenocarcinoma (PDAC) progression through facilitating PCBP2-dependent c-myc expression. Cancer Lett. 2016;373(1):67–76. https://doi.org/10.1016/j.canlet.2016.01.026.
Kim-Fuchs C, Le CP, Pimentel MA, Shackleford D, Ferrari D, Angst E, et al. Chronic stress accelerates pancreatic cancer growth and invasion: a critical role for beta-adrenergic signaling in the pancreatic microenvironment. Brain Behav Immun. 2014;40:40–7. https://doi.org/10.1016/j.bbi.2014.02.019.
Baker JG. The selectivity of beta-adrenoceptor antagonists at the human beta1, beta2 and beta3 adrenoceptors. Br J Pharmacol. 2005;144(3):317–22. https://doi.org/10.1038/sj.bjp.0706048.
Zhang D, Ma Q, Shen S, Hu H. Inhibition of pancreatic cancer cell proliferation by propranolol occurs through apoptosis induction: the study of beta-adrenoceptor antagonist's anticancer effect in pancreatic cancer cell. Pancreas. 2009;38(1):94–100. https://doi.org/10.1097/MPA.0b013e318184f50c.
Larsen TA, Teravainen H, Calne DB. Atenolol vs. propranolol in essential tremor. A controlled, quantitative study. Acta Neurol Scand. 1982;66(5):547–54. https://doi.org/10.1111/j.1600-0404.1982.tb03141.x.
Kuhn M, von Mering C, Campillos M, Jensen LJ, Bork P. STITCH: interaction networks of chemicals and proteins. Nucleic Acids Res. 2007;36(suppl_1):D684–8.
Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447–52. https://doi.org/10.1093/nar/gku1003.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. https://doi.org/10.1101/gr.1239303.
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8(Suppl 4):S11. https://doi.org/10.1186/1752-0509-8-s4-s11.
da Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. https://doi.org/10.1093/nar/gkn923.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discovery. 2012;2(5):401–4. https://doi.org/10.1158/2159-8290.cd-12-0095.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. https://doi.org/10.1126/scisignal.2004088.
Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102.
Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123(3):725–31. https://doi.org/10.1007/s10549-009-0674-9.
Antoni C, Vera L, Devel L, Catalani MP, Czarny B, Cassar-Lajeunesse E, et al. Crystallization of bi-functional ligand protein complexes. J Struct Biol. 2013;182(3):246–54. https://doi.org/10.1016/j.jsb.2013.03.015.
Tochowicz A, Maskos K, Huber R, Oltenfreiter R, Dive V, Yiotakis A, et al. Crystal structures of MMP-9 complexes with five inhibitors: contribution of the flexible Arg424 side-chain to selectivity. J Mol Biol. 2007;371(4):989–1006. https://doi.org/10.1016/j.jmb.2007.05.068.
Liu S, Misquitta YR, Olland A, Johnson MA, Kelleher KS, Kriz R, et al. Crystal structure of a human IκB kinase β asymmetric dimer. J Biol Chem. 2013;288(31):22758–67. https://doi.org/10.1074/jbc.M113.482596.
Polley S, Passos DO, Huang D-B, Mulero MC, Mazumder A, Biswas T, et al. Structural basis for the activation of IKK1/α. Cell Rep. 2016;17(8):1907–14. https://doi.org/10.1016/j.celrep.2016.10.067.
Kulik G. ADRB2-targeting therapies for prostate cancer. Cancers (Basel). 2019;11(3). https://doi.org/10.3390/cancers11030358.
Jabłońska-Trypuć A, Matejczyk M, Rosochacki S. Matrix metalloproteinases (MMPs), the main extracellular matrix (ECM) enzymes in collagen degradation, as a target for anticancer drugs. J Enzyme Inhib Med Chem. 2016;31(sup1):177–83. https://doi.org/10.3109/14756366.2016.1161620.
Lee KH, Hyun MS, Kim JR. Growth factor-dependent activation of the MAPK pathway in human pancreatic cancer: MEK/ERK and p38 MAP kinase interaction in uPA synthesis. Clin Exp Metastasis. 2003;20(6):499–505.
Witkiewicz AK, McMillan EA, Balaji U, Baek G, Lin WC, Mansour J, et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun. 2015;6:6744. https://doi.org/10.1038/ncomms7744.
Choi M, Kadara H, Zhang J, Parra ER, Rodriguez-Canales J, Gaffney SG, et al. Mutation profiles in early-stage lung squamous cell carcinoma with clinical follow-up and correlation with markers of immune function. Ann Oncol: official journal of the European Society for Medical Oncology. 2017;28(1):83–9. https://doi.org/10.1093/annonc/mdw437.
Zimmermannova O, Doktorova E, Stuchly J, Kanderova V, Kuzilkova D, Strnad H, et al. An activating mutation of GNB1 is associated with resistance to tyrosine kinase inhibitors in ETV6-ABL1-positive leukemia. Oncogene. 2017;36(43):5985–94. https://doi.org/10.1038/onc.2017.210.
Wang K, Zhou Y, Li G, Wen X, Kou Y, Yu J, et al. MMP8 and MMP9 gene polymorphisms were associated with breast cancer risk in a Chinese Han population. Sci Rep. 2018;8(1):13422. https://doi.org/10.1038/s41598-018-31664-3.
Weichert W, Boehm M, Gekeler V, Bahra M, Langrehr J, Neuhaus P, et al. High expression of RelA/p65 is associated with activation of nuclear factor-kappaB-dependent signaling in pancreatic cancer and marks a patient population with poor prognosis. Br J Cancer. 2007;97(4):523–30. https://doi.org/10.1038/sj.bjc.6603878.
Li X-W, Tuergan M, Abulizi G. Expression of MAPK1 in cervical cancer and effect of MAPK1 gene silencing on epithelial-mesenchymal transition, invasion and metastasis. Asian Pac J Trop Med. 2015;8(11):937–43. https://doi.org/10.1016/j.apjtm.2015.10.004.
Furukawa T. Impacts of activation of the mitogen-activated protein kinase pathway in pancreatic cancer. Front Oncol. 2015;5:23.
Sahu N, Chan E, Chu F, Pham T, Koeppen H, Forrest W, et al. Cotargeting of MEK and PDGFR/STAT3 pathways to treat pancreatic ductal adenocarcinoma. Mol Cancer Ther. 2017;16(9):1729–38.
Morichika K, Karube K, Kayo H, Uchino S, Nishi Y, Nakachi S, et al. Phosphorylated STAT3 expression predicts better prognosis in smoldering type of adult T-cell leukemia/lymphoma. Cancer Sci. 2019;110(9):2982–91.
Chen Q, Zhang JJ, Ge WL, Chen L, Yuan H, Meng LD, et al. YY1 inhibits the migration and invasion of pancreatic ductal adenocarcinoma by downregulating the FER/STAT3/MMP2 signaling pathway. Cancer Lett. 2019;463:37–49. https://doi.org/10.1016/j.canlet.2019.07.019.
Wang Z, Chen H, Chen J, Hong Z, Liao Y, Zhang Q, et al. Emodin sensitizes human pancreatic cancer cells to EGFR inhibitor through suppressing Stat3 signaling pathway. Cancer Manag Res. 2019;11:8463–73. https://doi.org/10.2147/cmar.s221877.
Kong X, Li B, Deng Y, Ma X. FXR mediates adenylyl cyclase 8 expression in pancreatic β-cells. J Diabetes Res. 2019;2019.
Warrington NM, Sun T, Luo J, McKinstry RC, Parkin PC, Ganzhorn S, et al. The cyclic AMP pathway is a sex-specific modifier of glioma risk in type I neurofibromatosis patients. Cancer Res. 2015;75(1):16–21. https://doi.org/10.1158/0008-5472.can-14-1891.
Shen-Gunther J, Wang CM, Poage GM, Lin CL, Perez L, Banks NA, et al. Molecular pap smear: HPV genotype and DNA methylation of ADCY8, CDH8, and ZNF582 as an integrated biomarker for high-grade cervical cytology. Clin Epigenetics. 2016;8:96. https://doi.org/10.1186/s13148-016-0263-9.
Szczałuba K, Biernacka A, Szymańska K, Gasperowicz P, Kosińska J, Rydzanicz M, et al. Novel GNB1 de novo mutation in a patient with neurodevelopmental disorder and cutaneous mastocytosis: clinical report and literature review. Eur J Med Genet. 2018;61(3):157–60. https://doi.org/10.1016/j.ejmg.2017.11.010.
Wazir U, Jiang WG, Sharma AK, Mokbel K. Guanine nucleotide binding protein beta 1: a novel transduction protein with a possible role in human breast cancer. Cancer Genomics Proteomics. 2013;10(2):69–73.
Chen C, Chi H, Min L, Junhua Z. Downregulation of guanine nucleotide-binding protein beta 1 (GNB1) is associated with worsened prognosis of clearcell renal cell carcinoma and is related to VEGF signaling pathway. J BUON: official journal of the Balkan Union of Oncology. 2017;22(6):1441–6.
Li Y, Yuan H, Sun L, Zhou Q, Yang F, Yang Z, et al. β2-adrenergic receptor gene polymorphisms are associated with cardiovascular events but not all-cause mortality in coronary artery disease patients: a meta-analysis of prospective studies. Genet Test Mol Biomarkers. 2019;23(2):124–37.
Wu FQ, Fang T, Yu LX, Lv GS, Lv HW, Liang D, et al. ADRB2 signaling promotes HCC progression and sorafenib resistance by inhibiting autophagic degradation of HIF1alpha. J Hepatol. 2016;65(2):314–24. https://doi.org/10.1016/j.jhep.2016.04.019.
Rivero EM, Pinero CP, Gargiulo L, Entschladen F, Zanker K, Bruzzone A, et al. The beta 2-adrenergic agonist salbutamol inhibits migration, invasion and metastasis of the human breast cancer MDA-MB- 231 cell line. Curr Cancer Drug Targets. 2017;17(8):756–66. https://doi.org/10.2174/1568009617666170330151415.
Renz BW, Takahashi R, Tanaka T, Macchini M, Hayakawa Y, Dantes Z, et al. beta2 adrenergic-neurotrophin feedforward loop promotes pancreatic cancer. Cancer Cell. 2018;33(1):75–90.e77. https://doi.org/10.1016/j.ccell.2017.11.007.
Knapinska AM, Estrada CA, Fields GB. The roles of matrix metalloproteinases in pancreatic cancer. Prog Mol Biol Transl Sci. 2017;148:339–54. https://doi.org/10.1016/bs.pmbts.2017.03.004.
Xu Y, Li Z, Jiang P, Wu G, Chen K, Zhang X, et al. The co-expression of MMP-9 and Tenascin-C is significantly associated with the progression and prognosis of pancreatic cancer. Diagn Pathol. 2015;10:211. https://doi.org/10.1186/s13000-015-0445-3.
Awasthi N, Mikels-Vigdal AJ, Stefanutti E, Schwarz MA, Monahan S, Smith V, et al. Therapeutic efficacy of anti-MMP9 antibody in combination with nab-paclitaxel-based chemotherapy in pre-clinical models of pancreatic cancer. J Cell Mol Med. 2019;23(6):3878–87. https://doi.org/10.1111/jcmm.14242.
Jacobsen JA, Major Jourden JL, Miller MT, Cohen SM. To bind zinc or not to bind zinc: an examination of innovative approaches to improved metalloproteinase inhibition. Biochim Biophys Acta (BBA) – Mol Cell Res. 2010;1803(1):72–94. https://doi.org/10.1016/j.bbamcr.2009.08.006.
Lu X, Yarbrough WG. Negative regulation of RelA phosphorylation: emerging players and their roles in cancer. Cytokine Growth Factor Rev. 2015;26(1):7–13.
Tian B, Widen SG, Yang J, Wood TG, Kudlicki A, Zhao Y, et al. The NFκB subunit RELA is a master transcriptional regulator of the committed epithelial-mesenchymal transition in airway epithelial cells. J Biol Chem. 2018;293(42):16528–45.
Mitchell S, Vargas J, Hoffmann A. Signaling via the NFκB system. Wiley Interdiscip Rev Syst Biol Med. 2016;8(3):227–41.
Wang H, Cai J, Du S, Guo Z, Xin B, Wang J, et al. Fractalkine/CX3CR1 induces apoptosis resistance and proliferation through the activation of the AKT/NF-kappaB cascade in pancreatic cancer cells. Cell Biochem Funct. 2017;35(6):315–26. https://doi.org/10.1002/cbf.3278.
Li W, Wu J, Li Z, Zhou Z, Zheng C, Lin L, et al. Melatonin induces cell apoptosis in Mia PaCa-2 cells via the suppression of nuclear factor-kappaB and activation of ERK and JNK: a novel therapeutic implication for pancreatic cancer. Oncol Rep. 2016;36(5):2861–7. https://doi.org/10.3892/or.2016.5100.
Saito K, Matsuo Y, Imafuji H, Okubo T, Maeda Y, Sato T, et al. Xanthohumol inhibits angiogenesis by suppressing nuclear factor-kappaB activation in pancreatic cancer. Cancer Sci. 2018;109(1):132–40. https://doi.org/10.1111/cas.13441.
Claudio E, Brown K, Park S, Wang H, Siebenlist U. BAFF-induced NEMO-independent processing of NF-κB2 in maturing B cells. Nat Immunol. 2002;3(10):958–65. https://doi.org/10.1038/ni842.
Sala E, Guasch L, Iwaszkiewicz J, Mulero M, Salvadó M-J, Pinent M, et al. Identification of human IKK-2 inhibitors of natural origin (part I): modeling of the IKK-2 kinase domain, virtual screening and activity assays. PLoS One. 2011;6(2):e16903. https://doi.org/10.1371/journal.pone.0016903.
Noha SM, Atanasov AG, Schuster D, Markt P, Fakhrudin N, Heiss EH, et al. Discovery of a novel IKK-β inhibitor by ligand-based virtual screening techniques. Bioorg Med Chem Lett. 2011;21(1):577–83. https://doi.org/10.1016/j.bmcl.2010.10.051.
Baxter A, Brough S, Cooper A, Floettmann E, Foster S, Harding C, et al. Hit-to-lead studies: the discovery of potent, orally active, thiophenecarboxamide IKK-2 inhibitors. Bioorg Med Chem Lett. 2004;14(11):2817–22. https://doi.org/10.1016/j.bmcl.2004.03.058.
Liddle J, Bamborough P, Barker MD, Campos S, Cousins RPC, Cutler GJ, et al. 4-Phenyl-7-azaindoles as potent and selective IKK2 inhibitors. Bioorg Med Chem Lett. 2009;19(9):2504–8. https://doi.org/10.1016/j.bmcl.2009.03.034.
Murata T, Shimada M, Kadono H, Sakakibara S, Yoshino T, Masuda T, et al. Synthesis and structure–activity relationships of novel IKK-β inhibitors. Part 2: improvement of in vitro activity. Bioorg Med Chem Lett. 2004;14(15):4013–7. https://doi.org/10.1016/j.bmcl.2004.05.040.
Acknowledgments
The authors thank Badan Penerbit dan Publikasi Universitas Gadjah Mada for their assistance in writing.
Funding
This research did not receive any specific grants from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
AH—conception and design of the study, acquisition, analysis and interpretation of data, drafting and revising the article, HP and RYU—acquisition and analysis of data, drafting the article. All authors had final approval of the submitted manuscript.
Corresponding author
Ethics declarations
Competing of interests
The authors declare that they have no no competing interests.
Ethical approval
Not applicable.
Consent for publication
All authors agree to publish our manuscript.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(XLSX 86 kb)
Rights and permissions
About this article
Cite this article
Hermawan, A., Putri, H. & Utomo, R.Y. Functional network analysis reveals potential repurposing of β-blocker atenolol for pancreatic cancer therapy. DARU J Pharm Sci 28, 685–699 (2020). https://doi.org/10.1007/s40199-020-00375-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40199-020-00375-4