Abstract
The type-1 water soluble chlorophyll binding proteins (WSCP1) have been generally known as chlorophyll extractors and transporters from the thylakoid membrane to the chloroplast envelope, where the membrane bound chlorophyllase catabolizes the chlorophyll. Despite the type-2 WSCP, WSCP1 has been known to be located in the chloroplasts of the green plants. In the present study, the non-chloroplastic protein superfamily containing domain of unknown function 538 (DUF538) as functional homologue of type-1 WSCP has been identified in plants. The structural similarities/differences and the cellular locations of Celosia cristata DUF538 and Chenopodium album WSCP1 were predicted by using computational tools and the chlorophyll binding abilities of their purified maltose binding protein-fused forms were estimated by maltose binding affinity method. It was predicted that despite CaWSCP1, CcDUF538 is a non-chloroplastic protein. The chlorophyll binding abilities of the recombinant fusion forms of test WSCP1 and DUF538 were estimated to be about 58 and 56%, respectively. Considering DUF538 as stress-induced protein, it was speculated that they may form complex with chlorophyll molecules or their hydrolyzed products out of chloroplasts to proceed the chlorophyll breakdown and nitrogen/carbon recycling in stress-challenged plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Most of the chlorophyll binding proteins of plants have been known to be hydrophobic types embedded in the thylakoid membrane of the chloroplasts. Besides this, the non-membranous highly hydrophilic water-soluble types of chlorophyll binding proteins have also been numerously isolated and identified from green plants. These proteins have been isolated from Amaranthaceae, Chenopodiaceae, and Polygonaceae as class I type of WSCP (WSCP1), and from Brassicaceae as class II type (WSCP2). These two classes of chlorophyll binding proteins differ in their photoconvertibilties. Class I proteins are generally photoconvertible, while class II types do not show such ability. In addition, the amino acid sequences of these two protein classes exhibit no significant similarities to each other [1–4].
The water soluble protein of Chenopodium album is the first characterized member of class I type WSCP whose photoconverted form generates light wavelengths of 668 and 742 nm, respectively [2]. The encoding gene of this protein has been recently identified and cloned [4]. But, the biological function of this protein is still unclear and not studied well. It has been generally speculated that WSCP1 acts as the scavenger of free chlorophyll molecules, by transporting them from the thylakoid membrane to the chloroplast envelope, where the membrane bound chlorophyllase enzyme catabolizes the chlorophyll [1, 2]. Despite class I types of WSCP, WSCP2 has generally received much attention. The numerous genes of class II WSCP have been reported and their potential functional properties have been analyzed using recombinant technologies [4]. Recent studies on the subcellular localization of the water soluble chlorophyll binding proteins demonstrated that some of the class II types of WSCP including Brassica oleracea WSCP (BoWSCP), Lepidium virginicum WSCP (LvWSCP), Arabidopsis thaliana WSCP (AtWSCP) and Raphanus sativus WSCP (RsWSCP) are located in the endoplasmic reticulum (ER) [5, 6]. These endoplasmic homologues of WSCP2 proteins have been reported to contribute to the unique defense system in Brassicaceae plants [7, 8].
In a recent study, based on the amino acid sequence analysis, it has been indicated that photoconvertable WSCP1 of Chenopodium album possesses the structural signature of DUF538 protein superfamily in plants [4]. DUF538 members have been mostly found as putative hypothetical proteins in wide ranges of monocotyledonous and dicotyledonous plant species. These proteins have often been identified from plants challenged with various environmental stresses such as nutrient deficiency, crown gall, mixed elicitors, and mild drought [9, 10]. They have molecular weights of about 19–21 kDa encoding around 170 amino acids and having a molecular structure dominated by ß-strands (PDB ID: D1ydua1).
In a different study, it has been suggested that DUF538 proteins are functional homologues of BPI proteins in the innate immune system of mammalia [11]. They have been suggested to affect the bacterial growth rate through the binding to the lipid A moiety of lipopolysaccharides on the outer leaflet of the bacterial membranes is like BPI in mammalians immune system. Based on this report and considering chlorophyll molecules containing lipid moiety in their structure, the structural similarities between DUF538 and WSCP proteins were furthermore confirmed and led to the conduction of the present research to investigate more about the functional properties of DUF538 domain containing proteins in plants.
Keeping this in view, an attempt was made to predict the subcellular location of DUF538 proteins and to examine their complex formation ability with chlorophyll molecules in comparison to WSCP1, in vitro. By using computational and experimental tools including recombination methods, it is predicted that DUF538 proteins might be the non-chloroplastic functional homologues of WSCP1 in the chloroplasts of plants. Considering DUF538 as stress-induced proteins, it was speculated that DUF538 bind to the chlorophyll molecules or their hydrolyzed products out of the chloroplasts to proceed or help increase the chlorophyll breakdown process and nitrogen/carbon recycling in stress-challenged plants.
Material and Methods
Computational Material and Analysis
The amino acid sequences of Celosia cristata DUF538 (AJ535713) and Chenopodium album (K4PX49) were extracted from EMBL database. Primary sequence alignments were performed using CLUSTALW server at http://www.genome.ad.jp/. Conserved domains were identified using the NCBI conserved domain architectural tool (CDART). The online PSIPRED v 2.0 server was used for secondary structure analysis. Protein tertiary structure predictions were done by using the internet-based Phyre v 2.0 server. Predictions of protein phosphorylation sites were performed by NetPhos v 2.0 Server. The presence of signal peptides and subcellular localization prediction was analyzed using Signal P online server.
Experimental Material
Trizol reagent was used for total RNA extraction (Cat. no. RN7713C; RNX™; Fermentas), mRNA purification was carried out by using mRNA mini prep kit (Cat. no. 70022; QIAGEN). AcessQuick™ RT-PCR System was provided for reverse-transcription PCR reaction (Cat. no. A1701; Promega). pGEM-T easy plasmid vector used for the cloning of RT-PCR end product was from Promega. Purification of the DNA fragments from the agarose gel DNA was done by using extraction kit (Cat. no. K0513; Fermentas). Bacterial transformation by E. coli strain TB1 and plasmid pMALc2X vector for recombinant protein expression studies provided in protein fusion and purification system kit (Cat. no. E8000S; NEW ENGLAND, Bio Lab). Materials and columns for the fusion protein isolation and purification were provided in protein fusion and purification system kit. Antibody against MBP was provided in protein fusion and purification system kit. The leaf extract of Celosia cristata plant was considered as test material for chlorophyll assay.
Molecular Cloning of DUF538 and WSCP cDNAs
The cloning of DUF538 cDNA from Celosia cristata leaves had been carried out in previous work [10]. The same procedure was repeated to clone the WSCP1 cDNA from Chenopodium album leaves using the following primer pair: Forward (5′actcgaattcaacaaccactacctccctcg3′) and Reverse (5′tctaggatccgaagattggtgtttgtgttgg3′). Briefly, RT-PCR based cloning method was used. For total RNA isolation, about 0.2 g of leaf material was fine homogenized, 2 ml of Trizol reagent and the RNA was collected after addition of 200 μl of chloroform and an equal volume of isopropanol. Poly (A+) RNA was purified from total RNA using oligo dT-columns according to the provided kit protocol. RT-PCR reaction was performed with 0.5 μg of each mRNA sample in 25 μl of Master Mix (2x) and 1 μl of primer set. The reaction mixture was incubated at 45 °C for 45 min and followed by PCR cycling. PCR was carried out after a pre-denaturation stage at 95 °C for 3 min with 25 cycles of 1 min denaturation at 95 °C, 1.5 min annealing at 56 °C, 2 min extension at 72 °C, ending with 10 min final extension at 72 °C. In the next step, the amplified products were extracted from the agarose gel, cloned in pGEM-T easy cloning vector. The cloned fragments proceeded for the partial sequencing in Microsynth DNA sequencing center at Switzerland.
Recombinant Fusion Expression of DUF538 and WSCP
The expression of DUF538 cDNA as MBP-fused product using E. coli system had been described in the previous work [10]. The same protocol was used for the bacterial expression of WSCP1 cDNA. Briefly the RT-PCR amplification product of WSCP1 after digestion with EcoRI and BamHI restriction enzymes, ligated into the pMALc2X expression vector which had already been linearized at the EcoRI and BamHI sites within the multiple cloning region. The ligation mixture was transferred into competent E. coli TB1 cells. The transformed cells were plated on LB-agar medium (supplemented with Ampicillin and X-gal) at 37 °C and a recombinant clone was selected for protein extraction and purification.
Purification and Western Blotting of Fused Products
For the extraction of fused products from recombinant bacteria, the cells were separately grown in 500 mL of rich broth medium (supplemented with glucose and ampicillin). The fused products were induced by the addition of IPTG (0.3 mM final concentration) for an incubation period of 8 h. The cells were then harvested and precipitated by centrifugation at 4000×g for 10 min, followed by dissolving the pellet in 25 mL of extraction buffer (consisted of 20 mM of Tris-Cl, 200 mM of NaCl, 1 mM of EDTA, 1 mM of sodium azide, and 10 mM of BME. For sonication purpose), freezing in the same extraction buffer at −20 °C and then sonicating in short pulses of 15 s. The sonicated samples were finally centrifuged at 10,000×g at 4 °C for 20 min, and the obtained supernatants were used as crude protein extracts of fused proteins. The purification processes were carried out by using the single-step affinity column chromatography. For this purpose, columns having the dimensions of 2.5 cm × 45 cm were packed with amylase resins (specific for the maltose-binding protein). The bound MBP-fused proteins were eluted out from the amylase column by using the specific elution buffer consisting of the crude protein extraction buffer plus 10 mM maltose. The homogeneities of the eluted products were analyzed by separating them on 10% polyacrylamide gel using SDS-PAGE [12].
Western analysis was carried out on the induced protein extracts using anti-body against maltose-binding protein. For this, about 10–15 µg of total protein extracts were separated on 10% SDS-PAGE and transferred onto nitrocellulose membrane using blotting buffer containing 0.025 M Tris-Cl pH 8.3, 0.192 M glycine and 20% ethanol. The blots were then kept in TBS–BSA buffer (0.02 M Tris-Cl, pH 7.5, 0.5 M NaCl, 1% BSA) at 4 °C overnight and then incubated for 2 h with specific antiserum (anti-maltose binding protein) at 1:500 ratios in the same buffer at 37 °C for 1 h. After washing with TBS-T (TBS + 0.05% Tween 20), the membrane was incubated for 1 h with alkaline phosphatase conjugated goat-anti rabbit antibody (GARXAP) diluted at the rate of 1:20,000 in TBS buffer containing bovine serum albumin. After extensive washes, the signal bands were visualized using a substrate solution consisting of 0.33 mg/ml NBT and 0.165 mg/ml BCIP in 0.1 M Tris-Cl buffer (pH 9.5) containing 0.1 M NaCl and 5 mM MgCl2.
Chlorophyll Extraction and Estimation
The chlorophyll content of the test leaf extract of Celosia cristata plant was detected according to method of Moran [13] with some modifications. The harvested fresh leaf material (about 200 mg) were chopped and treated with 4 ml of ethanol and DMF (at the rate 1/100) at 4 °C for 48 h. DMF was then removed from the mixture by using a rotary evaporator. The dried sample was dissolved in 100% methanol and its chlorophyll (a, b, pchl and total) contents were quantified according to the following equations:
where Ca, Cb, Cp and CT represent the concentrations of a, b, proto and total chlorophylls, respectively.
Data on the graph shows the changes of total chlorophyll contents.
Reconstitution of Fused DUF538/WSCP with Chlorophyll Molecules
Each purified fused product of 10 μg/ml was separately dissolved in aqueous buffer (consisted of 20 mM of Tris-Cl, 200 mM of NaCl, 1 mM of EDTA) and then mixed with chlorophyll extracts in 20% methanol. The mixture was incubated under laboratory temperature and light conditions for 2 h. The total chlorophyll content of each test sample was measured before and after mixing with fused protein and passing through maltose affinity column (as used for fusion protein purification). Changes in the chlorophyll contents of test samples were estimated and presented as its approximate binding capacity towards chlorophyll molecules. To elucidate the possible effect of MBP partner, a different solution containing purified MBP was considered as control sample.
Statistics
Each experiment was carried out with three replications using the same starting materials and experimental conditions. Data points on the graphs represent the mean values ± SD of the replicates of each test.
Results and Discussion
Comparative Computational Studies
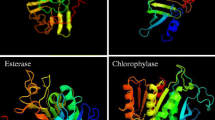
The protein sequence of WSCP1 (K4PX49) and deduced amino acid sequence of DUF538 (AJ535713) were extracted from universal EMBL database. Comparison of their amino acid sequences by CLASTALW server revealed that these proteins are not significantly identical in their primary structures (Appendix 1). The homology score between these two proteins were predicted to be 22%. However, conserved domain analysis by CDART confirmed that WSCP protein has the signature of DUF538 protein domain (output data not presented). It is because of this that conserved domain analysis by CDART is based on protein similarities across significant evolutionary distances using sensitive domain profiles rather than direct sequence similarity. The secondary structure prediction results by PSIPRED online software indicated that despite primary structures, the secondary folds do much more support to the presence of the homologous regions between WSCP1 and DUF538 molecules. These regions are shown to be confidently dominated by 8 ß-strands (Appendix 1). These ß-strands were found to have the same organizations and orientations in both the proteins. Similarly, the three-dimensional molecular structures of WSCP1 and DUF538 were predicted by Phyre v. 2 server. Comparison of the output data revealed that the ß-stranded regions between WSCP1 and DUF538 proteins have the similar patterns and orientations (Appendix 1).
The second comparative computational study, by using TMHMM trans-membrane helix prediction server, showed that despite DUF538 proteins, the N-terminal helical region of WSCP1 has the trans-membrane spanning (Fig. 1). Based on the obtained results, the N-terminus peptide sequence “MSPKTTTTSLALLAITLTLSSAHAH” is predicted to contain a chloroplastic transmembrane helix and a cleavage site in WSCP1 in comparison to the N-terminus peptide of DUF538. According to the secondary and tertiary structural predictions, the N-terminal part of WSCP1 is shown to be more helical and spiral than DUF538. Prediction data with high probabilities confirmed the differential cellular locations of selected proteins, indicating the chloroplastic and non-chloroplastic localizations of WSCP1 and DUF538, respectively.
The third computational based evidence was carried out on the comparative analysis of phosphorylation potentials in selected WSCP1 and DUF538 proteins. The obtained results showed that despite DUF538, WSCP1 has no considerable phosphorylation capacity (Fig. 2). As it was shown, DUF538 domain containing proteins is more likely predicted to be thoroughly phosphorylated by serine, tyrosine and threonine. While, WSCP1 proteins are not considerably detected as phosphorylable molecules. In comparison to WSCP1, four serine phosphorylation sites including “KEFNSVGDD”, “SMGSSIGDG”, “YKDSSVLRF”, and “KKTRSREAY”, two tyrosine phosphorylation sites including “GEVGYKDSS” and “SREAYQVLR” and one threonine phosphorylation site “PRDATHYEF” were detected to have high potential for phosphorylation in DUF538 protein.
Based on the present computational results, the primary structural analysis revealed that WSCP1 domain is not an extra domain, but it is the potential substitute of architectural unknown DUF538 domain in widely known DUF538 domain containing protein superfamily. As a strong clue about the functions or the mechanisms of the actions of the selected test proteins, the tertiary structure prediction data highlighted that DUF538 domain containing proteins might be the functional homologue of WSCP1 in plant system. Computational predicted data with the high probabilities confirmed the differential cellular localizations of two test proteins, indicating the chloroplastic and non-chloroplastic localizations of WSCP1 and DUF538, respectively. This result may apparently be in contrast with the function of WSCP1 that is suggested to bind to the chlorophyll molecules in the thylakoid membranes and chloroplasts stroma [1, 14]. Despite the secondary and tertiary structural identifications, the differences of trans-membrane capacities between two test proteins do not support their possible bio-functional similarities. Like trans-membrane capacities, the considerable difference between the phosphorylation potentials of WSCP1 and DUF538 may also not support their potential functional homologues. It has been generally suggested that phosphorylated proteins belong to the key elements in plants. They were known to play regulatory roles under various cellular or environmental conditions [15]. According to this, DUF538 protein superfamily may regulate chlorophyll metabolism out of chloroplast in plant system. But to how extent these computational predictions support the functional similarities between the two proteins will depend on experimental analysis. In a previous work, by using computational amino acid sequence similarity data, it had been initially suggested that WSCP1 proteins of plant chloroplasts posses the structural signature of DUF538 proteins in plants [4]. But the present computational data predicted that despite WSCP1 protein of Chenopodium, DUF538 protein of Celosia is located out of chloroplast and has more significant phosphorylation capacity that may be in contrast with their functional similarities. Therefore, due to this controversy the authors attempted to extend their analysis by using experimental tools, too.
Molecular Cloning and Expression Analysis
The cDNA clones containing DUF538 and WSCP1 domains were amplified from C. cristata and C. album leaf cDNA populations using specific primer sets based on the previously reported sequences.. The presence and the sizes of the amplified products were analyzed on agarose gel and the partial nucleotide sequences were carried out at Microsynth DNA sequencing center. The results showed that the sizes and the sequences of the amplified cDNA clones were in perfect agreement with the expected size and the sequence of the previously reported cDNAs (sequence data and the gel photographs not presented). To clone the primed cDNAs in E. coli expression vector pMALc2X, the cDNAs and vector were restricted with EcoRI and BamHI enzymes and directionally ligated and transformed into TB1 strain of E. coli. The cloned map of DUF538/WSCP1on MBP-fusion expression vector is presented (Fig. 3).
The MBP-fused products of the selected test proteins were separately and homogenously prepared from expressing recombinant bacteria by using maltose-affinity chromatography method. The MBP fusion tag was chosen due to its efficient expression and purification procedure as well as its role in the production of correctly folded protein structures in heterologous prokaryotic expression systems such as E. coli. This was important requirement for the functional studies on DUF538 and WSCP1. To confirm the purities of the products, SDS-PAGE as well as Western blot analysis were carried out using anti-body against maltose-binding protein (Fig. 2). Both the WSCP and DUF538 proteins are expressed as recombinant proteins with the molecular weight of about 66 kDa. However, WSCP1 has got 2 extra amino acids in comparison to DUF538.
Chlorophyll Binding Ability Assay
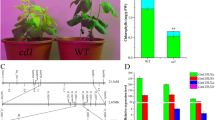
In order to assess the chlorophyll binding abilities, two test solutions containing the same amounts of chlorophyll and purified DUF538 or WSCP1 were separately considered as the starting experimental materials. The free chlorophyll contents of the test solutions were measured after (Fig. 4A) and before mixing with the purified fusion proteins and passing through the maltose-affinity column (Fig. 4B). The results showed that the amount of the chlorophyll is sharply decreased to about 42 and 44% in the solutions containing fused WSCP1 and DUF538, respectively. To test the possible effect of MBP protein on the absorption of chlorophyll molecules, a different solution containing purified MBP (instead of the purified recombinant DUF538 or WSCP1) was considered as a control sample. The obtained data (not presented) showed that MBP has no absorption effect on chlorophyll molecules and has not interfered with the binding abilities of fused DUF538 and WSCP1. The overall results of this experiment revealed that about 58 and 56% of the chlorophyll molecules are absorbed by WSCP1and DUF538 proteins, respectively. The results not only confirmed the chlorophyll binding ability of WSCP1, but also clarified the same ability for DUF538 superfamily. Since the difference (about 2%) between their chlorophyll capturing abilities were found not to be significant, they were most probably suggested as functional homologues of each other in the plant system.
Chlorophyll binding ability assessment. Upper Changes in the chlorophyll contents of the test solution before (B) and after (A) mixing with WSCP1 and passing through the maltose affinity column. Lower Changes in the chlorophyll contents of the test solution before (B) and after (A) mixing with DUF538 and passing through the maltose affinity column
For further clarifications, the time course experiments were also followed. Analysis of the chlorophyll assessment data showed that the chlorophyll contents of the WSCP1 and DUF538 containing solutions are decreased with time. Based on the obtained data, the similar decrease patterns were observed for both the test solutions. Data revealed that the amount of the chlorophyll in both samples declined to the level of about 40% after 10 h of experimental period (Fig. 5). Like chlorophyll binding capacity, the time course experiment result also confirmed the potent functional homology between WSCP1 and DUF538 proteins.
The present experimental data strongly support the bio-functional identities between WSCP1 and DUF538 proteins in photosynthetic apparatus in plant system. According to the previous literature reviews, DUF538 proteins are the functional homologues of bactericidal permeability increasing proteins in the innate immune system of mammalia [11]. They do affect the bacterial growth rates through the binding to the lipid A moiety of lipopolysaccharides on the outer leaflet of the bacterial membranes like BPI in mammalian immune system. Based on this report and considering chlorophyll molecules containing lipid moiety in their structures, the chlorophyll binding ability of DUF538 proteins might be acceptable.
Although, the exact bio-physiological functions of WSCP1 proteins have not been clearly understood, but it has been speculated that they act as scavengers of free chlorophyll molecules to transport them to the chloroplast envelop, where chlorophyllase enzyme initiates chlorophyll catabolism [14]. In view of this, it can be expected that the chlorophyll binding abilities of WSCP1 proteins are crucial for chlorophyll breakdown and catabolism in plants. The present data revealed that the chlorophyll binding ability not only plays an important catabolic role (by WSCP1) inside the chloroplast environment, but also it might have the same level of significance out side the chloroplast, through DUF538 protein members. The present data may also reveal that chlorophyll catabolism is completed out of chloroplasts, where the nitrogen and carbon recycling has occurred. In this case, DUF538 proteins are speculated to get intermediate role between chloroplasts and cytosol of photosynthesizing cells.
In many cases, WSCP1 have been reported to be expressed in response to various environmental stresses such as drought, detachments and heat stresses as well as in senescence and developing tissues [16–19]. The similar stress conditions have also been included for DUF538 protein superfamily expression in plant system [9]. From this point of view, the bio-functional homologies between WSCP1 and DUF538 can also be predictable. Considering DUF538 as stress-induced protein, it is speculated that DUF538 bind to the chlorophyll molecules or their hydrolytic products to proceed or help increase the chlorophyll breakdown process and nitrogen/carbon recycling rates out of the chloroplasts of stress-challenged plants.
Chlorophyll catabolism has been mostly reported to be carried out by chloroplastic chlorophyllase. Although some reports speculated about different cellular locations of this enzyme [20, 21], it is not very likely that chlorophyllase isoforms are located outside the plastids. Recently, two chlorophyllase isoforms have been reported to be localized in cytosol and involved in stress responses of Arabidopsis plant [22]. This extra plastidial localization (cytosolic) of chlorophyllase is in accordance with the assumption of Takamiya group that points a novel chlorophyll catabolism pathway in vacuoles, distinct from that of plastids [20]. Keeping this information in view, the out of chloroplastic functional homologue of water soluble chlorophyll binding proteins can not be away from thinking.
Recently, it has been demonstrated that some of the class II types of WSCP target the endoplasmic reticulum and contribute to the unique defense system in plants [5–8, 23]. The present investigation as a first report showed that the C. album WSCP1 homologue “DUF538” may also be located out of chloroplasts and like WSCP2 homologues it may help to increase the chlorophyll breakdown process in response to stress stimuli. The results of the present work will open the new gate to study the chlorophylls non-chloroplastic catabolic pathway that is thought to be related to the various stress responses in plant system.
Conclusions
Despite WSCP1, DUF538 is non-chloroplastic protein. It was predicted to be the functional homologue of WSCP1 in plant cells. Considering plants WSCP1 and DUF538 both as stress induced proteins, it was most likely proposed that they do co-function to catabolize and breakdown chlorophyll molecules in response to various stress responses in plants.
Abbreviations
- WSCP:
-
Water soluble chlorophyll binding proteins
- DUF:
-
Domain of unknown function
- BPI:
-
Bactericidal permeability increasing
- IPTG:
-
Isopropyl-1-thio-β-d-galactoside
- NBT:
-
Nitro blue tetrazolium
- BCIP:
-
5-bromo-1-chloro-3-indolyl phosphate
- DMF:
-
Dimethyl formamide
- Pchl:
-
Proto chlorophyll
- EDTA:
-
Ethylene diamine tetra acidic acid
- MBP:
-
Maltose-binding protein
References
Horigome D, Satoh H, Itoh N, Mitsunaga K, Oonishi I, Nakagawa A, Uchida A (2007) Structural mechanism and photoprotective function of water-soluble chlorophyll-binding protein. J Biol Chem 282:6525–6531
Noguchi T, Kamimura Y, Inoue Y, Itoh S (1999) Photoconversion of a water-soluble chlorophyll protein from Chenopodium album: resonance raman and fourier transform infrared study of protein and pigment structures. Plant Cell Physiol 40:305–310
Satoh H, Uchida A, Nakayama K, Okada M (2001) Water-soluble chlorophyll protein in Brassicaceae plants is a stress-induced chlorophyll-binding protein. Plant Cell Physiol 42:906–911
Takahashi S, Yoshikawa M, Kamada A, Ohtsuki T, Uchida A, Nakayama K, Satoh H (2013) The photoconvertible water-soluble chlorophyll-binding protein of Chenopodium album is a member of DUF538, a superfamily that distributes in Embryophyta. J Plant Physiol 170:1549–1552
Takahashi S, Aizawa K, Nakayama K, Satoh H (2015) Water soluble chlorophyll binding proteins from Arabidopsis thaliana and Raphanus sativus target the endoplasmic reticulum body. BMC Res Notes 8:365
Takahashi S, Yanai H, Oka-Takayama Y, Zanma-Sohtome A, Fujiyama K, Uchida A, Nakayama K, Satoh H (2013) Molecular cloning, characterization and analysis of the intracellular localization of a water-soluble chlorophyll binding protein (WSCP) from Virginia pepperweed (Lepidium virginicum), a unique WSCP that preferentially binds chlorophyll b in vitro. Planta 238:1065–1080
Yamada K, Hara-Nishimura I, Nishimura M (2011) Unique defense strategy by the endoplasmic reticulum body in plants. Plant Cell Physiol 52:2039–2049
Yamada K, Nagano AJ, Nishina M, Hara-Nishimura I, Nishimura M (2013) Identification of two novel endoplasmic reticulum body-specific integral membrane proteins. Plant Physiol 161:108–120
Gholizadeh A, BaghbanKohnehrouz B (2010) Identification of DUF538 cDNA clone from Celosia cristata expressed sequences of none stressed and stressed leaves. Russ J Plant Physiol 57:247–252
Gholizadeh A (2011) Heterologous expression of stress-responsive DUF538 domain containing protein and its morpho-biochemical consequences. Protein J 30:351–358
Gholizadeh A, Baghbankohnehrouz S (2013) DUF538 protein super family is predicted to be the potential homologue of bactericidal/permeability-increasing protein in plant system. Protein J 32:163–171
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Moran R (1982) Formulae for determination of chlorophyllous pigments extracted with N, N-dimethylformamide. Plant Physiol 69:1376–1381
Fang Z, Bouwkamp JC, Solomos T (1998) Chlorophyllase activities and chlorophyll degradation during leaf senescence in non-yellowing mutant and wild type of Phaseolus vulgaris L. J Exp Bot 49:503–510
Nakagami H, Sugiyama N, Mochida K, Daudi A, Yoshida Y, Toyoda T, Tomita M, Ishihama Y, Shirasu K (2010) Large-scale comparative phosphoproteomics identifies conserved phosphorylation sites in plants. Plant Physiol 153:1161–1174
Annamalai P, Yanagihara S (1999) Identification and characterization of a heat-stress induced gene in cabbage encodes a kunitz type protease inhibitor. J Plant Physiol 155:226–233
Downing WL, Mauxion F, Fauvarque MO, Reviron MP, de Vienne D, Vartanian N, Giraudat J (1992) A Brassica napus transcript encoding a protein related to the Künitz protease inhibitor family accumulates upon water stress in leaves, not in seeds. Plant J 2:685–693
Nishio N, Satoh H (1997) A water-soluble chlorophyll protein in cauliflower may be identical to BnD22, a drought-induced, 22-kilodalton protein in rapeseed. Plant Physiol 115:841–846
Satoh H, Nakayama K, Okada M (1998) Molecular cloning and functional expression of a water-soluble chlorophyll protein, a putative carrier of chlorophyll molecules in cauliflower. J Biol Chem 273:30568–30575
Takamiya KI, Tsuchiya T, Ohta H (2000) Degradation pathway (s) of chlorophyll: what has gene cloning revealed? Trends Plant Sci 5:426–431
Tsuchiya T, Ohta H, Masuda T, Mikami B, Kita N, Shioi Y, Takamiya K (1997) Purification and characterization of two isozymes of chlorophyllase from mature leaves of Chenopodium album. Plant Cell Physiol 38:1026–1031
Schenk N, Schelbert S, Kanwischer M, Goldschmidt EE, Dörmann P, Hörtensteiner S (2007) The chlorophyllases AtCLH1 and AtCLH2 are not essential for senescence-related chlorophyll breakdown in Arabidopsis thaliana. FEBS Lett 27:5517–5525
Takahashi S, Yanai H, Nakamaru Y, Uchida A, Nakayama K, Satoh H (2012) Molecular cloning, characterization and analysis of the intracellular localization of a water-soluble chlorophyll-binding protein from Brussels sprouts (Brassica oleracea var. gemmifera). Plant Cell Physiol 53:879–891
Acknowledgements
The author of this paper is thankful to the Research Institute for Fundamental Sciences (RIFS), University of Tabriz for the financial support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
There is no conflict of interest in this study.
Appendix
Appendix

Prediction and comparison of primary, secondary and tertiary structures of WSCP1 and DUF538 proteins
Rights and permissions
About this article
Cite this article
Gholizadeh, A. Chlorophyll Binding Ability of Non-chloroplastic DUF538 Protein Superfamily in Plants. Proc. Natl. Acad. Sci., India, Sect. B Biol. Sci. 88, 967–976 (2018). https://doi.org/10.1007/s40011-016-0834-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40011-016-0834-8