Abstract
Various plants possess non-photosynthetic, hydrophilic chlorophyll (Chl) proteins called water-soluble Chl-binding proteins (WSCPs). WSCPs are categorized into two classes; Class I (photoconvertible type) and Class II (non-photoconvertible type). Among Class II WSCPs, only Lepidium virginicum WSCP (LvWSCP) exhibits a low Chl a/b ratio compared with that found in the leaf. Although the physicochemical properties of LvWSCP have been characterized, its molecular properties have not yet been documented. Here, we report the characteristics of the LvWSCP gene, the biochemical properties of a recombinant LvWSCP, and the intracellular localization of LvWSCP. The cloned LvWSCP gene possesses a 669-bp open reading frame. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry analysis revealed that the precursor of LvWSCP contains both N- and C-terminal extension peptides. RT-PCR analysis revealed that LvWSCP was transcribed in various tissues, with the levels being higher in developing tissues. A recombinant LvWSCP and hexa-histidine fusion protein (LvWSCP-His) could remove Chls from the thylakoid in aqueous solution and showed an absorption spectrum identical to that of native LvWSCP. Although LvWSCP-His could bind both Chl a and Chl b, it bound almost exclusively to Chl b when reconstituted in 40 % methanol. To clarify the intracellular targeting functions of the N- and C-terminal extension peptides, we constructed transgenic Arabidopsis thaliana lines expressing the Venus protein fused with the LvWSCP N- and/or C-terminal peptides, as well as Venus fused at the C-terminus of LvWSCP. The results showed that the N-terminal peptide functioned in ER body targeting, while the C-terminal sequence did not act as a trailer peptide.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chlorophylls (Chls) are essential pigments in photosynthesis. It is well known that all Chl-binding proteins functioning in photosynthesis are hydrophobic proteins embedded in the thylakoid membrane. In addition to these membranous Chl-binding proteins, highly hydrophilic Chl-binding proteins called water-soluble Chl-binding proteins (WSCPs) have been extracted from various plants classified into Brassicaceae, Chenopodiaceae, Amaranthaceae, and Polygonaceae (for a review, see Satoh et al. 2001).
The first identified WSCP, from Chenopodium album, exhibited unique photoconvertibility (Yakushiji et al. 1963). Subsequently, photoconvertible WSCPs have been extracted from Amaranthaceae and Polygonaceae plants (for a review, see Satoh et al. 2001). This photoconversion occurs in an aerobic aqueous solution (Takamiya et al. 1968) and the efficiency of photoconversion is dependent on the pH of the solution (Oku et al. 1972). The mechanism of the photoconversion has been proposed to be as follows. (a) Chls in the WSCP complex capture light energy and form a reactive triplet excited state. (b) The excited Chl reacts with molecular oxygen to produce reactive oxygen species (ROS). (c) Chl a in the complex is attacked by the ROS and then the pigment is converted to another form (Hagar and Hiyama 1979). Noguchi et al. (1999) reported that the chlorin skeleton of Chl a in WSCP is photoconverted into a bacteriochlorin-like skeleton and this conversion does not affect the protein conformation. Hirabayashi et al. (2006) isolated and characterized two kinds of photoconverted pigments that were mono-oxygenated Chl a derivatives. We recently identified a gene encoding Chenopodium album WSCP and found that this gene is a member of DUF538, a biological function unknown superfamily that distributes in Embryophyta (Takahashi et al. 2013a).
In contrast to these photoconvertible WSCPs, various non-photoconvertible WSCPs have been isolated from Brassicaceae plants. Based on their photoconvertibility, WSCPs are categorized into two classes: Class I (photoconvertible type) and Class II (non-photoconvertible type). Class II WSCPs are further categorized into two sub-classes, Class IIA and Class IIB, on the basis of their Chl a/b ratios. In comparison to the Chl a/b ratios found in vegetative leaves of Brassicaceae plants, Class IIA WSCPs exhibit higher ratios (6.3–10.0), while Class IIB WSCPs exhibit lower ones (1.0–3.5) (for a review, see Satoh et al. 2001). Class IIA WSCPs have been extracted from various plants, e.g., cauliflower (Brassica oleracea var. botrys) (Murata et al. 1971), black mustard (Brassica nigra) (Murata and Murata 1971), rapeseed (Brassica napus) (Downing et al. 1992), Brussels sprout (Brassica oleracea var. gemmifera) (Kamimura et al. 1997), Japanese radish (Raphanus sativus var. hortensis) (Shinashi et al. 2000), kale (Brassica oleracea var. acephala) (Horigome et al. 2003), Arabidopsis thaliana (Bektas et al. 2012) and Japanese wild radish (Raphanus sativus var. raphanistroides) (Takahashi et al. 2013b). On the other hand, Class IIB WSCPs have been extracted only from Virginia pepperweed (Lepidium virginicum) (Murata and Murata 1971; Murata and Ishikawa 1981; Itoh et al. 1982).
Because Chls and their metabolic intermediates (e.g., protoporphyrin IX and pheophorbide a) are photosensitizers, non-enzymatic accumulation of these pigments promotes photobleaching that ultimately leads to cell death, and thus plants strictly control both their synthetic and catabolic pathways (for reviews, see Tanaka and Tanaka 2007; Hörtensteiner and Kräutler 2011; Hörtensteiner 2013). However, it is poorly understood how plants suppress oxidative damage resulting from the excitation of Chls when a plant cell is injured.
Class II WSCPs are stress-inducible proteins [being induced in response to, for example, drought (Downing et al. 1992; Reviron et al. 1992), salinity (Reviron et al. 1992), heat (Annamalai and Yanagihara 1999), nutrient deficiency (Desclos et al. 2008; D’Hooghe et al. 2013), and methyl jasmonate treatment (Desclos et al. 2008)], and are able to reduce the singlet oxygen production from excited Chls (Schmidt et al. 2003; Horigome et al. 2007). Therefore, it has been proposed that WSCPs may act as Chl carriers under stress conditions. Recently, we demonstrated that Brussels sprout WSCP (BoWSCP) was located in the ER body (Takahashi et al. 2012), an endoplasmic reticulum (ER)-derived structure observed only in Brassicaceae plants thus far (for a review, see Yamada et al. 2011). The ER body is considered to participate in a cell defense system in response to cell disruption (for a review, see Yamada et al. 2011). In accordance with these facts, we previously proposed that BoWSCPs may act as Chl scavengers when a cell is injured (Takahashi et al. 2012).
Cauliflower WSCP (cauWSCP), the first identified Class II WSCP, was cloned as a highly homologous protein to BnD22 (a drought- and salinity-stress-induced 22 kDa protein of rapeseed), which is a member of the Kunitz-type trypsin inhibitor (KTI) family (Satoh et al. 1998). It should be noted that recombinant BnD22 was able to bind Chls, indicating that BnD22 is a rapeseed WSCP (Satoh et al. 1998). In addition, all cloned WSCPs [Arabidopsis thaliana WSCP, AtWSCP (Bektas et al. 2012); BoWSCP (Takahashi et al. 2012); Japanese wild radish WSCP, RsWSCP (Takahashi et al. 2013b)] contain a KTI motif. Despite the presence of this motif, both BnD22 and cauWSCP purified from plants did not inhibit, or only slightly inhibited, the proteinase activity of trypsin and/or chymotrypsin in vitro (Ilami et al. 1997; Nishio and Satoh 1997). However, Etienne et al. (2007) and Desclos et al. (2008) demonstrated that BnD22 exhibited trypsin inhibitor activity in vivo. Moreover, Halls et al. (2006) found that an AtWSCP precursor inhibited papain, a cysteine protease. These data indicate that proteinase inhibitor activities are another important aspect of Class II WSCPs, in addition to their Chl-binding ability.
Because Class II WSCPs contain only a few Chls without any carotenoids or other pigments (Horigome et al. 2007), they provide a suitable tool for the study of Chl–Chl and Chl–protein interactions in Chl-binding proteins (for a review, see Renger et al. 2011). Therefore, many researchers have analyzed the biochemical and spectroscopic properties of Class II WSCPs using recombinant cauWSCP (Schmidt et al. 2003; Hughes et al. 2006; Renger et al. 2007; Theiss et al. 2007; Schmitt et al. 2008; Pieper et al. 2011a, b).
Although various genes encoding Class IIA WSCPs have been identified and their recombinant proteins have been used for various studies, there has been no report describing the properties of any gene encoding a Class IIB WSCP. In the present study, we, therefore, performed molecular cloning and functional expression analysis of the WSCP from L. virginicum (LvWSCP) to clarify the molecular properties of a unique WSCP that exhibits higher affinity for Chl b than other WSCPs. In addition, we investigated the transcriptional levels of LvWSCP in various plant tissues and the intracellular localization of LvWSCP.
Materials and methods
Plant materials
Lepidium virginicum L. was collected at the Narashino campus of Toho University, Chiba, Japan. Arabidopsis thaliana L. (Col-0) was grown on 1 % agar plates containing full-strength Murashige and Skoog medium and 1 % sucrose at 24 °C under continuous light (30 μmol photons m−2 s−1). Two-week-old A. thaliana was transferred from agar plates to Jiffy-7 for further growth under the same conditions.
Purification of native LvWSCP
Water-soluble chlorophyll-binding protein was prepared from L. virginicum according to Murata and Ishikawa (1981) from 300 g of fresh leaves. The LvWSCP preparation obtained was separated by native PAGE (detergent-free) on a 10 % separation gel. A greenish band was cut out and ground in buffer (20 mM Tris–HCl, pH 8.0) with a pestle and mortar. After incubation at 25 °C for 24 h, the greenish supernatant fraction containing LvWSCP was collected by centrifugation. The recovered supernatant was concentrated with an Amicon Ultra-15 (Millipore) and subjected to gel-filtration chromatography with a Sephacryl S-300 HR (GE Healthcare). The fraction containing native LvWSCP was dialyzed with 20 mM Tris–HCl (pH 8.0). The protein concentration of the dialyzate was determined by the bicinchoninic acid method using a kit from Sigma with a BSA standard. The purified LvWSCP was kept at −20 °C until use. The molecular mass of LvWSCP was analyzed with a matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF–MS) spectrometer (Voyager-DE™; Applied Biosystems) in positive ion mode.
Determination of the N-terminal amino acid sequence of native LvWSCP
Two hundred pmol of native LvWSCP was separated by SDS-PAGE and transferred onto a PVDF membrane. The transferred proteins were visualized by Coomassie brilliant blue staining (CBB). A 19-kDa band corresponding to the LvWSCP subunit was cut from the membrane and subjected to a sequencing reaction using a gas-phase protein sequencer (Procise 491; Applied Biosystems).
Isolation of genomic DNA and RNA from plants and cDNA synthesis
Genomic DNA from leaves of L. virginicum and A. thaliana was purified using a DNeasy mini kit (Qiagen). Total RNA from various tissues of both plants was isolated using an RNeasy mini kit (Qiagen) with RNase-Free DNase (Qiagen). First-strand cDNA was synthesized with 100 ng each of total RNA and an oligo dT-adapter primer (see Supplemental Table S1) (for cDNA cloning) or an oligo (dT)20 primer (for RT-PCR analyses) with a reverse transcriptase, ReverTra Ace (Toyobo). All procedures were performed according to the instructions provided by the manufacturer.
Molecular cloning of a cDNA encoding LvWSCP
Based on the amino acid sequence (INDEEPVK) of native LvWSCP determined in this study, a degenerate primer (see Supplemental Table S1) was designed. To obtain a cDNA encoding native LvWSCP, 3′ rapid amplification of cDNA ends (RACE) was performed with the degenerate primer and an adapter-specific primer (see Supplemental Table S1). KOD-Plus-Neo DNA polymerase (Toyobo) was used for all PCR procedures. The PCR conditions were 94 °C for 5 min, followed by 30 cycles of 94 °C for 1 min, 55 °C for 2 min, and 68 °C for 2 min. The PCR product was cloned into the pGEM-T easy vector (Promega), and then the construct was introduced into E. coli (DH5α). Plasmid DNA was extracted from a culture of the transformed E. coli using a PureLink Quick Plasmid Miniprep kit (Invitrogen). The plasmid DNA was sequenced using a BigDye Terminator ver. 3.1 Cycle Sequencing kit (Applied Biosystems) and an ABI PRISM 310 Genetic Analyzer (Applied Biosystems).
For further cloning of the 5′ region of LvWSCP, 5′ RACE was performed with a 5′-phosphorylated LvWSCP-specific primer (see Supplemental Table S1) and a 5′-Full RACE Core Set (Takara Bio) with the following gene-specific primer pairs: for the 1st PCR, 5′ RACE F1 and 5′ RACE R1; for the 2nd PCR, 5′ RACE F2 and 5′ RACE R2 (see Supplemental Table S1). All procedures were performed according to the manufacturer’s instructions. The PCR product was cloned and its nucleotide sequence was determined.
Sequence analysis
Nucleotide and amino acid sequences of Class II WSCPs from cauliflower, rape, Japanese wild radish, A. thaliana and Brussels sprout (DDBJ accession numbers AB012699, AB747087, AB071430, BT025277 and AB679292) and homologous proteins, P22 (AY347537), DIP (L23554), Dr4 (AY059142), α-fucosidase (X82595), and KTI (X80039), were obtained from DNA databases. Alignment and phylogenetic analysis of the deduced amino acid sequences of the WSCPs and homologous proteins were performed with Clustal X (Thompson et al. 1997).
RT-PCR analysis
The first-strand cDNAs synthesized were used as templates for RT-PCR analysis. A constitutively expressed gene, actin, was used as an internal control. LvWSCP and actin cDNAs were amplified by PCR with specific primer pairs: for LvWSCP, LvWSCP RT F and LvWSCP RT F; for Actin of L. virginicum, Actin F and Actin R (see Supplemental Table S1). The amplification conditions were 94 °C for 2 min, followed by 30 cycles of 94 °C for 1 min, 60 °C for 30 s, and 68 °C for 1 min. The PCR products were separated on a 1.2 % agarose gel, stained with ethidium bromide and viewed under a UV transilluminator.
Expression and purification of His-tag fused recombinant LvWSCP
An expression construct of the mature form of LvWSCP was produced by fusing a hexa-histidine tag at its C-terminus (LvWSCP-His). The DNA region corresponding to the mature LvWSCP was amplified by PCR with the primers LvWSCP Exp F and LvWSCP Exp R (see Supplemental Table S1). The amplification conditions were 94 °C for 2 min, followed by 25 cycles of 94 °C for 1 min, 60 °C for 30 s, and 68 °C for 30 s. After digestion of the PCR product with NdeI and XhoI, the DNA fragment was ligated into the pET-24b vector (Novagen) predigested with the same restriction enzymes. This construct was used for transformation of E. coli BL 21 (DE3) codon plus (Novagen).
The E. coli harboring the expression construct was grown at 37 °C in LB medium containing 50 μg ml−1 kanamycin until the A600 of the culture reached 0.4. Protein expression was induced by the addition of IPTG to the cultures at a final concentration of 1 mM, and incubation was continued at 37 °C for an additional 3 h. The cells were harvested and resuspended with a buffer (20 mM Tris–HCl, pH 8.0, 500 mM NaCl), and then disrupted by sonication. Subsequently, the supernatant containing soluble LvWSCP-His was collected by centrifugation and loaded onto a Bio-Scale Mini Profinity IMAC Cartridge (Biorad) for Ni2+-chelating affinity chromatography according to the manufacturer’s protocol. The recombinant protein was recovered by elution with imidazole buffer (20 mM Tris–HCl, pH 8.0, 500 mM NaCl, 500 mM imidazole). The eluted protein fraction was dialyzed against 20 mM Tris–HCl buffer (pH 8.0) and stored at −20 °C until use.
Reconstitution of LvWSCP-His with thylakoid membranes
Methods for thylakoid membrane preparation and determination of Chls from spinach leaves were previously reported (Takahashi et al. 2012). For the reconstitution of LvWSCP-His with Chls, 50 nmol of LvWSCP-His was mixed with thylakoid membranes containing 500 nmol Chls (Chl a/b ratio = 3.3) in an aqueous solution of 20 mM Tris–HCl (pH 8.0) containing 0.5 M NaCl and 1 mM EDTA at 25 °C for 1 h. After centrifugation (16,000g for 30 min), the supernatant was collected and subjected to native PAGE (detergent-free) using a 10 % polyacrylamide gel to separate reconstituted LvWSCP-His. Subsequently, the reconstituted LvWSCP-His was recovered from the gel according to the procedure described above. The UV–visible absorption spectra of the extracted LvWSCP-His and native LvWSCP between 300 and 750 nm were measured using a U-3300 spectrophotometer (Hitachi).
Reconstitution of LvWSCP-His with purified Chl a or Chl b
Chl a and Chl b were extracted from spinach leaves according to procedures described previously (Takahashi et al. 2012). For analysis of the Chl-binding ability of LvWSCP-His under Chl excess conditions, 50 nmol of LvWSCP-His was mixed with 500 nmol of Chl a, Chl b, or mixtures containing Chl a and b in different ratios, i.e. Chl a:b = 250:250 (1:1), 750:250 (3:1), or 2500:250 (10:1) nmol, in a buffer containing 20 mM Tris–HCl (pH 8.0) and 40 % methanol. For analysis of the Chl-binding ability of LvWSCP-His under Chl b limiting conditions, 50 nmol of LvWSCP-His was added to mixtures containing Chl a and b in different ratios, i.e. Chl a:b = 500:1 (100:1), 500:15 (100:3), or 500:25 (100:5) nmol, in a buffer containing 20 mM Tris–HCl (pH 8.0) and 40 % methanol. After incubation at 25 °C for 30 min in darkness, the reconstituted LvWSCP-His was purified by native PAGE as described above. The UV–visible absorption spectra of native LvWSCP and the reconstituted LvWSCP-His series were measured between 300 and 750 nm.
Electrophoretic mobility shift assay (EMSA) for LvWSCP-His
Conformational changes of LvWSCP-His were analyzed by addition of purified Chls. After 200 pmol of LvWSCP-His was mixed with 0, 50, 100, 200, or 400 pmol Chl a or Chl b under the same conditions as described above, the resulting solutions were separated on PAGE (detergent-free) using a 10 % polyacrylamide gel. The fluorescence of Chls bound to LvWSCP-His was detected on a long wave UV transilluminator and the protein bands were visualized by CBB staining.
Construction of transgenic A. thaliana
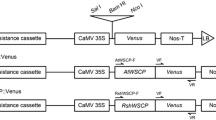
The full-length LvWSCP was amplified by PCR with the primer pair LvWSCP Full F and LvWSCP Full R (see Supplemental Table S1). After digestion of the PCR product with SalI and BglII, the DNA fragment was ligated into SalI and BamHI sites between the Cauliflower mosaic virus (CaMV) 35S promoter and Venus in a modified pBI101 vector (35SVpBI101) (Fig. 11a). The pBI101 construct was used for transformation of Agrobacterium tumefaciens (GV3101 strain). Using the floral dip method described by Clough and Bent (1998), we transformed A. thaliana (ecotype Col-0) and created 35S::LvWSCP::Venus. The N- and/or C-termini of LvWSCP were directly introduced into 35SVpBI101 with a KOD -Plus- Mutagenesis Kit (Toyobo) and the following primer pairs: for the N-signal, N-signal F and N-signal R; for the C-terminus, C-terminal F and C-terminal R (see Supplemental Table S1). These pBI101 vectors were used for the construction of three types of transgenic plants, 35S::N-signal::Venus, 35S::Venus::C-terminus, and 35S::N-signal::Venus::C-terminus. Correct insertion into the transgenic plants was confirmed by PCR using various primers: SF, CR, VF, VR, Actin8 F and Actin8 R (see Supplemental Table S1).
Fluorescence microscopy analysis of transgenic plants
Ten-day-old leaves of T2 generation transgenic plants (35S::Venus, 35S::LvWSCP::Venus, 35S::N-signal::Venus, 35S::Venus::C-terminus, and 35S::N-signal::Venus::C-terminus) were analyzed by fluorescence microscopy using procedures described previously (Takahashi et al. 2012).
Results
Amplification and molecular cloning of the LvWSCP cDNA and gene
For cDNA cloning of LvWSCP, we first determined the N-terminal amino acid sequence of native LvWSCP purified from leaves of L. virginicum. The amino acid residues spanning positions 1 through 32 of native LvWSCP were identified as NH2-INDEEPVKDTNGNPLKIETRYFIQPASDNNGG-COOH. A single amino acid peak was observed in each sequencing cycle of expected size for the protein amount (data not shown), indicating that the determined sequence corresponded to an N-terminal unblocked, single peptide of LvWSCP. This amino acid sequence exhibited homology to the N-terminal amino acid sequence of native Class IIA WSCPs (cauWSCP, BoWSCP and BnD22) with 54.8 % identity and 83.9 % similarity.
Based on the sequence 1-INDEEPVK-8, we designed a degenerated oligonucleotide primer and performed 3′ RACE PCR. The amplified DNA was cloned and sequenced. The clone contained a 769-bp cDNA with a 591-bp open reading frame encoding a polypeptide of 197 residues. The deduced amino acid sequence of the clone contained the sequence 9-DTNGNPLKIETRYFIQPASDNNGG-32 immediately downstream of 1-INDEEPVK-8, and because the latter sequence was used for primer design, we were convinced that this clone was a partial cDNA of LvWSCP. Based on the obtained cDNA sequence, we designed LvWSCP-specific primers for 5′ RACE PCR and determined its 5′-portion. Finally, we amplified and cloned a full-length cDNA encoding LvWSCP with a specific primer set for the 5′- and 3′- ends. The full-length cDNA was composed of 868 bp containing a 669-bp open reading frame encoding 223 amino acid residues (Fig. 1). The genomic gene for LvWSCP was also amplified and cloned. We found that LvWSCP was an intronless gene.
Nucleotide and deduced amino acid sequences of the open-reading frame of LvWSCP. Nucleotide and amino acid residue numbers are shown at the left and right sides of the sequence, respectively. The N-terminal amino acid sequence of the mature LvWSCP determined by Edman degradation is underlined. The region highlighted in black indicates the deduced signal peptide. The amino acid sequence with a gray background is missing in the mature LvWSCP. Black and gray arrows indicate post-translational cleavage sites for the signal peptide and C-terminal extension peptide, respectively
The cDNA encoded 26 residues forming an N-terminal extension (indicated by the black background in Fig. 1). This region was predicted to be a signal peptide by the TargetP program (ver 1.1) (Nielsen et al. 1997; Emanuelsson et al. 2000). The cleavage site deduced by the TargetP program matched the N-terminus of mature LvWSCP.
Possible post-translational processing in the C-terminus of LvWSCP
To determine the molecular mass of a subunit of native LvWSCP, we performed MALDI-TOF-MS analysis. A single major peak at 19634.9 was observed on the spectrogram (Fig. 2), although some minor peaks, considered to be contaminants, were also detected. Therefore, we concluded that the molecular mass of the mature form of LvWSCP was 19634.9 Da. This molecular mass corresponded to the deduced amino acid sequence from residue 27, which is the N-terminus of the mature form, to residue 206 with an error of 0.19 %. This indicated that 17 residues of the C-terminal region had been removed post-translationally.
Sequence comparison of LvWSCP and homologous proteins
The deduced amino acid sequences of Class IIA WSCPs from cauliflower, rapeseed, Japanese wild radish, A. thaliana and Brussels sprout, and homologous proteins, P22, DIP, Dr4, α-fucosidase and KTI (for a review, see Satoh et al. 2001), were aligned with the program ClustalX (Thompson et al. 1997) (Fig. 3), and a phylogenetic tree was generated (Fig. 4). Homology (identity and similarity) of the proteins to LvWSCP is shown in Table 1. Briefly, P22 exhibited the highest identity (43 %) and similarity (79 %) to LvWSCP. The five Class IIA WSCPs exhibited 38–40 % identity and 76–77 % similarity to LvWSCP. The remaining homologous proteins showed 24–30 % identity and 68–71 % similarity to LvWSCP. All the Class II WSCPs thus far sequenced possessed a KTI motif. This motif was conserved in LvWSCP as well. In addition, the LvWSCP precursor also possessed the conserved seven amino acid motif (L[K/R]MFPFY) at the C-terminus.
Alignment of deduced amino acid sequences of Class II WSCPs and homologous proteins. The amino acid sequences of Class II WSCPs from Brussels sprout (BoWSCP), cauliflower (cauWSCP), rape (BnD22), Japanese wild radish (RsWSCP), Arabidopsis thaliana (AtWSCP) and Lepidium virginicum (LvWSCP), and homologous proteins (P22, DIP, Dr4, α-fucosidase and KTI), were aligned using the Clustal X program (Thompson et al. 1997). Amino acid residues completely conserved among all of the proteins are highlighted in black. The amino acid residues identical to those of mature LvWSCP are shown with a gray background. Black and gray arrows indicate post-translational cleavage sites for the signal peptide and C-terminal extension peptide, respectively. A motif for the Kunitz-type proteinase inhibitor family is indicated by a black bar
Molecular phylogenetic tree of Class II WSCPs and homologous proteins. The branch lengths are proportional to the genetic distances calculated by the neighbor-joining method (Saitou and Nei 1987)
Transcriptional patterns of LvWSCP in plant tissues
To analyze the transcriptional patterns of LvWSCP in L. virginicum grown under field conditions, we performed RT-PCR analysis using single-stranded cDNA synthesized from the total RNA of various tissues of plants collected in April (roots and rosette leaves) or July (stems, cauline leaves, buds and flowers and immature seeds) at the Narashino campus of Toho University, Chiba, Japan. As shown in Fig. 5, LvWSCP was expressed in all tissues analyzed, with an especially high level of transcription detected in developing tissues. We cloned the amplified bands from all tissues tested, and determined their nucleotide sequences. The sequences of eight independent clones of the amplified products were exactly the same (data not shown), confirming that the strengths of the amplified bands were representative of the transcriptional levels of LvWSCP in these plant tissues.
RT-PCR analysis for determination of the transcriptional profile of LvWSCP. LvWSCP transcript levels were analyzed in roots, stems, rosette leaves, cauline leaves, buds and flowers, and immature seeds of Lepidium virginicum that were grown under field conditions. Actin was used as an internal control. Experiments were repeated at least three times using independent samples. The photographs represent one of the independent experiments, which all produced similar results
Expression and purification of His-tag-fused recombinant LvWSCP in E. coli
We prepared an expression construct of recombinant LvWSCP (LvWSCP-His) that was composed of the mature region of LvWSCP (without the N- and C-terminal extension peptides) and hexa-histidine.
About 60 % of the LvWSCP-His expressed in E.coli was detected in the inclusion body fraction and the remaining 40 % was found in the soluble fraction (Fig. 6a). The soluble LvWSCP-His fraction was loaded onto Ni2+ charged resin. After washing out the resin, LvWSCP-His was eluted by a buffer containing 0.5 M imidazole. Almost 95 % of the LvWSCP-His was eluted into the No. 2 fraction (Fig. 6b). This fraction was then dialyzed and used in the following experiments.
SDS-PAGE profiles of a series of experiments to obtain soluble recombinant LvWSCP (LvWSCP-His). Black arrows indicate LvWSCP-His. a Induction of LvWSCP-His expression in E. coli and subcellular fractionation. Fractions were obtained from IPTG-induced (+), or uninduced (−) E. coli. Supernatant (Sup) and pellet (Pet) fractions of sonicated E. coli were separated by 15 % SDS-PAGE. The lane with protein markers is indicated as M in the panel. b Purification of LvWSCP-His using Ni2+-chelating affinity chromatography. S supernatant fraction before chromatography, F the flow-through fraction, W washed fraction, 1–6 a series of eluted fractions
Analysis of the binding activity of LvWSCP-His to Chls in thylakoid membranes
To examine the Chl-binding ability of LvWSCP-His in aqueous solution, LvWSCP-His was mixed with purified thylakoid membranes containing a ten-fold molar excess of Chls. Subsequently, reconstituted LvWSCP-His was purified by native PAGE. The absorption spectra of reconstituted LvWSCP-His and native WSCP exhibited high identity in both the red and Soret bands (Fig. 7). The peak wavelengths of both proteins in the red band appeared to be 663 nm, and the other peak wavelengths were almost identical in both WSCP preparations (Table 2). The identity of the spectrometric properties of both proteins indicated that the Chl-binding activity of LvWSCP-His was identical to that of native LvWSCP.
Absorption spectra (300–750 nm) of native LvWSCP and LvWSCP-His reconstituted with thylakoid membranes. LvWSCP-His was reconstituted with spinach thylakoid membranes (Chl a/b = 3.3) in aqueous solution (20 mM Tris–HCl (pH 8.0), 0.5 M NaCl, 1 mM EDTA) at 25 °C for 1 h in the dark. UV–visible absorption spectra of native LvWSCP and reconstituted LvWSCP-His were measured in 20 mM Tris–HCl (pH 8.0) at 25 °C
Analysis of the binding activity of LvWSCP-His to purified Chls
The Chl a/b ratio in most vegetative plant leaves is usually around 3. Murata and Ishikawa (1981) reported that native LvWSCP extracted from leaves contained Chl a and Chl b at a ratio of 1.6–1.9. This indicates that LvWSCP exhibits a higher content of Chl b than thylakoidal Chl-binding proteins. To assess the binding specificity of LvWSCP-His to purified Chl a and/or Chl b, the following reconstitution analyses were performed. LvWSCP-His was mixed with a molar excess of Chl a, Chl b, or Chl a/b mixed at three different ratios (Chl a/b = 1.0, 3.0 or 10.0). Then, the reconstituted LvWSCP-His preparations were purified by native PAGE.
The UV–visible absorption spectra of native LvWSCP and LvWSCP-His reconstituted with different Chl a and/or b contents were measured (Fig. 8). Table 2 summarizes their absorption peaks. Seven major peaks at 339, 382, 419, 438, 470, 619 and 663 nm were observed for native LvWSCP (Fig. 8a). The shape of the absorption spectrum and the peak wavelengths of LvWSCP-His reconstituted with Chl a (Fig. 8b) were similar to those of native LvWSCP, except for the peak at 470 nm, which was attributed to Chl b. On the other hand, the absorption peaks of LvWSCP-His reconstituted with Chl b contained three major peaks at 443, 468 and 653 nm (Fig. 8c). These absorption peaks were completely different from those of native LvWSCP (Fig. 8a). The shapes of the absorption spectra and the peak wavelengths of LvWSCP-His reconstituted with three different Chl mixtures (Fig. 8d–f; Table 2) were almost identical to those of LvWSCP-His reconstituted with Chl b (Fig. 8c; Table 2), indicating that LvWSCP-His possessed much higher affinity for Chl b than Chl a in 40 % methanol.
Absorption spectra (300–750 nm) of native LvWSCP and LvWSCP-His reconstituted with purified Chls. UV–visible absorption spectra of native LvWSCP (a) and LvWSCP-His reconstituted with Chl a only (b), Chl b only (c) or three different Chl mixtures (Chl a:b = 1:1, 3:1 or 10:1) (d–f) were measured in 20 mM Tris–HCl (pH 8.0) at 25 °C. Images of disk gels of these LvWSCPs are inserted into each panel
We also performed LvWSCP-His reconstitution analyses under Chl b limiting conditions. LvWSCP-His was added to Chl mixtures at three different ratios (Chl a:b = 100:1, 100:3 or 100:5) corresponding to 1/10, 3/10, and 1/2 mol of Chl b in comparison with LvWSCP-His. The UV–visible absorption spectra of these proteins and their absorption peaks are summarized in Fig. 9; Table 2, respectively. The shape of the absorption spectrum and the absorption peaks of LvWSCP-His reconstituted with the Chl a:b = 100:1 mixture (Fig. 9a; Table 2) were similar to those of native LvWSCP (Fig. 8a; Table 2), while those of LvWSCP-His reconstituted with the Chl a:b = 100:5 mixture (Fig. 9c; Table 2) were almost identical to those of LvWSCP-His reconstituted with Chl b only (Fig. 8c; Table 2). The shape of the absorption spectrum and the absorption peaks of LvWSCP-His reconstituted with the Chl a:b = 100:3 mixture exhibited intermediate traits (Fig. 9b; Table 2). These data also supported the idea that LvWSCP-His possesses much higher affinity for Chl b than Chl a in 40 % methanol.
Absorption spectra (300–750 nm) of LvWSCP-His reconstituted with purified Chls under Chl b limiting conditions. UV–visible absorption spectra of LvWSCP-His reconstituted with three different Chl mixtures (Chl a:b = 100:1, 100:3 or 100:5) (a–c) were measured in 20 mM Tris–HCl (pH 8.0) at 25 °C. Images of disk gels of these LvWSCPs are inserted into each panel
Although Murata and Ishikawa (1981) reported using 2-butanone to extract Chls from native LvWSCP, we could not reproduce the perfect extraction of Chls from holo-LvWSCP. Moreover, the extracted Chls in 2-butanone were easily bleached in the short term. Chls in thylakoid proteins can be extracted easily with organic solvents, such as 80 % acetone. However, it is impossible to extract all of the Chls from BoWSCP using 80 % acetone (Kamimura et al. 1997; Takahashi et al. 2012). Moreover, BoWSCP exhibits resistance to proteinase K even in the presence of 1 % SDS, and therefore we could not extract Chls completely after protein digestion (Takahashi et al. 2012). Because the reconstituted LvWSCP-His also exhibited high resistance to 80 % acetone and proteinase K, we could not determine the Chl contents in the native and reconstituted LvWSCPs directly.
Analyses of the oligomerization of LvWSCP-His by EMSA
Chl-free Class II WSCP (apo-WSCP) is a monomer while Chl-binding Class II WSCP (holo-WSCP) forms a tetramer. These WSCPs could be easily separated by native (detergent-free) PAGE. To investigate the number of Chl molecules required for LvWSCP-His to form a tetramer, we performed an electrophoretic mobility shift assay (EMSA) of reconstituted LvWSCP-His with a series of different concentrations of Chl a or Chl b (Fig. 10). When LvWSCP-His was incubated with a quarter or half molar amount of Chl a or Chl b in 40 % methanol, most of the LvWSCP-His formed tetramers. This result indicated that one to two Chls were sufficient to reconstitute apo-LvWSCP (i.e., the monomer) to holo-LvWSCP (i.e., the tetramer).
Conformational changes of LvWSCP-His on binding with purified Chl a or Chl b. LvWSCP-His (200 pmol) was mixed with 0, 50, 100, 200 or 400 pmol Chl a or Chl b in a buffer containing 20 mM Tris–HCl (pH 8.0) and 40 % methanol. After reconstitution, these mixtures were subjected to 10 % native PAGE. M monomer of LvWSCP-His corresponding to the Chl-free form (apo-form), T tetramer of LvWSCP-His corresponding to the Chl-bound form (holo-form). a Image of the gel after native PAGE. b Profile of the CBB stained gel. c Image of Chl fluorescence from reconstituted LvWSCP-His under UV irradiation
Intracellular localization of LvWSCP
From sequence analysis of LvWSCP, we predicted that the N-terminal extension of LvWSCP was a signal peptide targeting the ER. Recently, we demonstrated that the N-terminal signal peptide of BoWSCP targeted the ER body (Takahashi et al. 2012). However, the homology between the N-terminal peptides of LvWSCP and BoWSCP was quite low; they showed only 19.2 % identity and 34.6 % similarity (Fig. 3). To assess the intracellular localization of LvWSCP, we constructed transgenic A. thaliana (35S::N-signal::Venus) expressing a chimeric protein that was composed of the N-terminal signal peptide of LvWSCP and Venus, a yellow fluorescent protein (Fig. 11a). In addition, Class II WSCPs, including LvWSCP, contain a C-terminal extension sequence that was predicted to be a trailer peptide (Ilami et al. 1997). To reveal the function of the C-terminus in intracellular targeting, we constructed more transgenic A. thaliana (35S::Venus::C-terminus and 35S::N-signal::Venus::C-terminus), as well as transgenic A. thaliana expressing Venus fused at the C-terminus of the LvWSCP precursor (35S::LvWSCP::Venus). All constructs used for this experiment are shown in Fig. 11a. Correct insertion into the transformants was confirmed by PCR analysis with specific primer pairs (Fig. 11a, b). Fluorescence images of the leaf cells of ten-day-old transformants at different scales are shown in Fig. 11c. In three of the transgenic plants (35S::LvWSCP::Venus, 35S::N-signal::Venus, and 35S::N-signal::Venus::C-terminus), yellow fluorescence emitted from the Venus fusions was detected in the ER body, a spindle-shaped structure (Fig. 11c). On the other hand, yellow fluorescence in 35S::Venus::C-terminus was detected in the cytosol, as in the case of free Venus in the 35S::Venus control (Fig. 11c). These observations clearly indicated that the N-terminal extension peptide was a signal peptide targeting the ER body, but the C-terminal extension peptide might not be a trailer peptide. Because the ER body is an organelle unique to Brassicaceae plants, and the ER body was found in L. sativum (Iversen 1970), we concluded that LvWSCP is located in the ER body in L. virginicum.
Intracellular localization of LvWSCP in Arabidopsis thaliana. a Schematic representation of the control and modified Venus constructs for intracellular localization analysis of LvWSCP. b PCR analysis to confirm correct insertion into transgenic plants, 35S::Venus, 35S::LvWSCP::Venus, 35S::N-signal::Venus, 35S::Venus::C-terminus, and 35S::N-signal::Venus::C-terminus. The sites of primers (SF, CR, VF and VR) are shown in a. Actin8 was used as a control. c Fluorescence images of ten-day-old leaves of the transgenic plants. Bar 20 μm
Discussion
In this study, we cloned a cDNA and a gene encoding a Class IIB WSCP from L. virginicum (Fig. 1). Murata and Ishikawa (1981) previously extracted two different kinds of WSCPs (CP661 and CP663, named after the wavelengths (nm) of their red absorption peaks) from L. virginicum. The amino acid compositions and N-terminal residues of CP661 (glycine) and CP663 (isoleucine) are different (Murata and Ishikawa 1981), indicating that the two WSCPs are chemically distinct from each other. It should be noted that CP661 has only been extracted from plants collected in California, while CP663 has only been isolated from plants harvested in Japan (Murata and Murata 1971; Murata and Ishikawa 1981; Itoh et al. 1982; Horigome et al. 2007). The N-terminal amino acid residue of the mature form of native LvWSCP used in this study was determined to be isoleucine (Fig. 1). In addition, the absorption peaks of native LvWSCP and LvWSCP-His reconstituted with thylakoid membranes appeared at 663 nm (Fig. 7). Thus, we concluded that the LvWSCP identified in this study corresponded to the former CP663.
Recently, we demonstrated that the N-terminal signal peptides that are conserved among Class IIA WSCPs (BoWSCP, cauWSCP, and BnD22) target the proteins into the ER body. In the present study, we demonstrated that the N-terminal signal peptide of LvWSCP was also responsible for transportation into the ER body (Fig. 11c). Previous studies reported that the C-terminal 22 ± 1 amino acid residues of BnD22 (Ilami et al. 1997) and 21 amino acid residues of BoWSCP undergo post-translational cleavage (Takahashi et al. 2012). In this study, we found that the C-terminal 17 amino acid residues of the precursor LvWSCP were also removed in the mature LvWSCP. Ilami et al. (1997) speculated that the C-terminal extension peptide might play a role in targeting towards a specific cellular compartment. However, we found that the LvWSCP C-terminal fusion did not affect the intracellular localization of Venus (Fig. 11c), indicating that the C-terminus is not a trailer peptide. Although the physiological function of the C-terminal peptide is still unclear, both recombinant BoWSCP-His (Takahashi et al. 2012) and LvWSCP-His lacking this region are able to bind Chls, and BnD22 is also able to act as a trypsin inhibitor without this region (Desclos et al. 2008), indicating that this region is not essential for these activities.
Water-soluble Chl-binding proteins have been suggested to function as Chl carriers in the initial step of the Chl catabolic pathway together with chlorophyllase (CLH), which removes a phytol chain from Chls (Matile et al. 1997; Kamimura et al. 1997). However, several studies have revealed that CLH is not involved in Chl breakdown (Schenk et al. 2007; Schelbert et al. 2009). Most recently, Sakuraba et al. (2012) demonstrated that Chl breakdown occurred in a protein complex at the thylakoid, indicating that the removal of chlorophylls by WSCPs would not be required. Furthermore, cauWSCP overexpression in Nicotiana tabacum did not affect Chl metabolism (Damaraju et al. 2011). In fact, both Class IIA and Class IIB WSCPs are located in the ER body but not in chloroplasts. These observations strongly indicate that WSCPs do not play a role in the enzymatic Chl breakdown pathway in chloroplasts. Although the ER body is a subdomain of the rough ER, the biological function of the ER body is thought to be different from that of the rough ER, which is involved in the synthesis of proteins. The ER body is considered to be a defense system against pathogens (for a review, see Yamada et al. 2011) and metal stress (Yamada et al. 2013). The ER body accumulates a large amount of PYK10, a β-glucosidase (Matsushima et al. 2003), which is important for resistance against Piriformospora indica infection as it produces toxic compounds (Sherameti et al. 2008). PYK10 localizes to the ER body, but its substrates and activator are stored in different compartments (the vacuole and cytosol, respectively) in non-damaged cells. PYK10, therefore, does not produce toxic compounds in non-damaged cells. Pest and/or pathogen attacks cause plant cell disruption accompanied by organelle disruption. Therefore, it was proposed that PYK10 produces toxic compounds when a plant cell is damaged (for a review, see Yamada et al. 2011). Based on this PYK10 activation model, we produced a Chl-binding model for LvWSCP (shown in Fig. 12). In non-damaged cells, apo-LvWSCPs are located in ER bodies. When cells are subjected to severe damage causing organelle disruption, apo-LvWSCPs are released from burst ER bodies and transferred to disrupted chloroplasts, where they bind Chls. Because Class II WSCPs suppress the generation of reactive oxygen species derived from excited Chls (Schmidt et al. 2003; Horigome et al. 2007), we hypothesize that LvWSCP is an urgent Chl scavenger that protects healthy cells around damaged cells.
Since Chls are synthesized in chloroplasts and Class II WSCPs were extracted from leaves and stems (for a review, see Satoh et al. 2001), we would expect LvWSCP to be transcribed in greenish plant tissues if LvWSCP only functions as a Chl scavenger. However, LvWSCP was transcribed not only in greenish tissues, but also in roots (Fig. 5). Recently, Bektas et al. (2012) reported that AtWSCP is located in the transmitting tract in the gynoecium and silique. In addition, Halls et al. (2006) found that the AtWSCP precursor could inhibit papain. Moreover, Desclos et al. (2008) demonstrated that BnD22 possessed trypsin inhibitor activity in vivo. Because all Class II WSCPs possess a KTI motif in the mature protein region (Fig. 2), Bektas et al. (2012) proposed that Class II WSCPs might be bifunctional molecules (acting as both Chl carriers and protease inhibitors). It may be plausible that LvWSCP acts as a protease inhibitor in roots. However, the target proteinases and proteinase inhibitor activities of Class II WSCPs have not been consistent in previous reports. Elucidation of the relationship between the Chl-binding and protease inhibitor activities of Class II WSCPs will be one of our next goals.
Although native LvWSCP exhibits a lower Chl a/b ratio (1.6–1.9 (Murata and Ishikawa 1981)) than that in vegetative leaves, native LvWSCP nonetheless contains more Chl a than Chl b. It is interesting that, in 40 % methanol, apo-LvWSCP-His bound Chl b almost exclusively (Fig. 8d–f), and only a few Chl a molecules were incorporated into LvWSCP-His when the reconstitution analyses were performed with a molar excess of Chl a and Chl b. Furthermore, LvWSCP-His preferentially bound Chl b rather than Chl a under Chl b limiting conditions (Fig. 9a–c). Although native BoWSCP exhibits the highest Chl a/b ratio among Brassica-type WSCPs, i.e., 10 (Kamimura et al. 1997), recombinant BoWSCP-His bound Chl a and Chl b with almost the same affinity in 40 % methanol (Takahashi et al. 2012). These observations indicate that the Chl-selectivity of the Class II WSCPs might depend on reconstitution conditions or the substrate Chls, e.g., free molecules or Chl–protein complexes in the thylakoid. Previously, Itoh et al. (1982) reported that the Chl a/b ratio of LvWSCP extracted from leaves (CP663L) was 1.5–1.7, while that of LvWSCP extracted from stems (CP663S) was 3.4–3.5. As shown in Fig. 5, LvWSCP was transcribed not only in leaves but also stems, indicating that the difference in the Chl a/b ratio between CP663L and CP663S might have been caused by a difference in extraction conditions, e.g., a difference in the tissues, Chl content, or expression levels of WSCP.
Cauliflower WSCP requires a half molar ratio of Chls to protein to form a tetramer (Satoh et al. 1998), while BoWSCP needs an equal molar amount of Chls (Takahashi et al. 2012). Horigome et al. (2007) reported that the native LvWSCP tetramer complex contained four Chls. However, most of the apo-LvWSCP-His could be reconstituted in a low molar ratio of Chl a and/or Chl b to protein (Fig. 10), suggesting that the holo-LvWSCP-His reconstituted with purified Chls may contain few Chls in the complex. The UV–visible absorption spectrum of LvWSCP-His reconstituted with the Chl a:b = 100:5 mixture (Fig. 9c) was similar to that of LvWSCP-His reconstituted with Chl b only (Fig. 8c), indicating that almost all of the Chls bound in the reconstitution were Chl b, not Chl a, even though the amount of Chl a was 20 times higher than that of Chl b. The Chl a:b = 100:5 mixture contained a half mole of Chl b compared with the LvWSCP-His used for the reconstitution, also supporting the idea that a low molar ratio of Chl is enough to reconstitute apo-LvWSCP-His to holo-LvWSCP-His in a solution containing 40 % methanol. It should be noted that native LvWSCP was prepared in a solution without organic solvent while reconstitution of LvWSCP-His for the EMSA assay was performed in a solution containing 40 % methanol. We therefore consider the discrepancy in Chl contents between native LvWSCP and reconstituted LvWSCP-His to be due to Chl-binding conditions.
Compared with Chl-binding proteins functioning in photosynthesis, Class II WSCPs are highly water-soluble, stable and simple (they contain only Chls without any carotenoids), and thus many researchers have used Class II WSCPs, especially recombinant cauWSCP, to analyze Chl–Chl and Chl–protein interactions in Chl-binding proteins (for a review, see Renger et al. 2011). Because Class II WSCPs bind Chls so tightly, extraction procedures with strong organic solvent are required to remove Chls from the holo-form of LvWSCP to obtain apo-LvWSCP from native LvWSCP. We could not extract Chls completely from LvWSCP even using 2-butanone. Therefore, LvWSCP has not been used for these studies even though it is the only Class IIB WSCP yet discovered. In this study, we succeeded in expressing functional recombinant apo-LvWSCP as a His-tagged fusion protein in E. coli. The reconstituted LvWSCP exhibited almost the same spectral features as the native LvWSCP. Based on the simple procedures established in the present study, researchers should be able to easily prepare apo-LvWSCP-His from E. coli harboring the expression construct, and to readily obtain holo-LvWSCP-His containing only Chl a or only Chl b.
Abbreviations
- CaMV:
-
Cauliflower mosaic virus
- Chl:
-
Chlorophyll
- C-terminal/terminus:
-
Carboxy terminal/terminus
- EMSA:
-
Electrophoretic mobility shift assay
- ER:
-
Endoplasmic reticulum
- IPTG:
-
Isopropyl β-d-1-thiogalactopyranoside
- KTI:
-
Kunitz-type trypsin inhibitor
- MALDI-TOF–MS:
-
Matrix-assisted laser desorption ionization time-of-flight mass spectrometry
- NOS:
-
Nopaline synthase
- N-terminal/terminus:
-
Amino terminal/terminus
- WSCP:
-
Water-soluble chlorophyll-binding protein
- WT:
-
Wild type
References
Annamalai P, Yanagihara S (1999) Identification and characterization of heat-stress induced gene in cabbage encodes a Kunitz type protease inhibitor. J Plant Physiol 155:226–233
Bektas I, Fellenferg C, Paulsen H (2012) Water-soluble chlorophyll protein (WSCP) of Arabidopsis is expressed in the gynoecium and developing silique. Planta 236:251–259
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:736–743
D’Hooghe P, Escamez S, Trouverie J, Avice JC (2013) Sulphur limitation provokes physiological and leaf proteome changes in oilseed rape that lead to perturbation of sulphur, carbon and oxidative metabolisms. BMC Plant Biol 13:23. doi:10.1186/1471-2229-13-23
Damaraju S, Schlede S, Eckhardt U, Lokstein H, Grimm B (2011) Functions of the water soluble chlorophyll-binding protein in plants. J Plant Physiol 168:1444–1451
Desclos M, Dubousset L, Etienne P, Le Caherec F, Satoh H, Bonnefoy J et al (2008) A proteomic profiling approach to reveal a novel role of Brassica napus drought 22 kD/water-soluble chlorophyll-binding protein in young leaves during nitrogen remobilization induced by stressful conditions. Plant Physiol 147:1830–1844
Downing WL, Mauxion F, Fauvarque MO, Reviron MP, de Vienne D, Vartanian N et al (1992) A Brassica napus transcript encoding a protein related to the Künitz protease inhibitor family accumulates upon water stress in leaves, not in seeds. Plant J 2:685–693
Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300:1005–1016
Etienne P, Desclos M, Le Gou L, Gombert J, Bonnefoy J, Maurel K et al (2007) N-protein mobilisation associated with the leaf senescence process in oilseed rape is concomitant with the disappearance of trypsin inhibitor activity. Funct Plant Biol 34:895–906
Hagar WG, Hiyama T (1979) Characterization of the light-induced transient states of the chlorophyll proteins 668 and 743 from Atriplex rosea. Plant Physiol 63:1182–1186
Halls CE, Rogers SW, Oufattole M, Ostergard O, Sevensson B, Rogers JC (2006) A Kunitz-type cysteine protease inhibitor from cauliflower and Arabidopsis. Plant Sci 170:1102–1110
Hirabayashi H, Amakawa M, Kamimura Y, Shino Y, Satoh H, Itoh S et al (2006) Analysis of photooxidized pigments in water-soluble chlorophyll protein complex isolated from Chenopodium album. J Photochem Photobiol 183:121–125
Horigome D, Satoh H, Uchida A (2003) Purification, crystallization and preliminary X-ray analysis of a water-soluble chlorophyll protein from Brassica oleracea L. var. acephala (kale). Acta Crystallogr D Biol Csystallogr 59:2283–2285
Horigome D, Satoh H, Itoh N, Mitsunaga K, Oonishi I, Nakagawa A et al (2007) Structural mechanism and photoprotective function of water-soluble chlorophyll-binding protein. J Biol Chem 282:6525–6531
Hörtensteiner S (2013) Update on the biochemistry of chlorophyll breakdown. Plant Mol Biol 82:505–517
Hörtensteiner S, Kräutler B (2011) Chlorophyll breakdown in higher plants. Biochim Biophys Acta 1807:977–988
Hughes JL, Razeghifard R, Logue M, Oakle A, Wydrzynski T, Krausz E (2006) Magneto-optic spectroscopy of a protein tetramer binding two exciton-coupled chlorophylls. J Am Chem Soc 128:3649–3658
Ilami G, Nespoulous C, Huet JC, Vartanian N, Pernollet JC (1997) Characterization of BnD22, a drought-induced protein expressed in Brassica napus leaves. Phytochemistry 45:1–8
Itoh R, Itoh S, Sugawa M, Oishi O, Tabata K, Okada M et al (1982) Isolation of crystalline water-soluble chlorophyll proteins with different chlorophyll a and b contents from stems and leaves of Lepidium virginicum. Plant Cell Physiol 23:557–560
Iversen TH (1970) The morphology, occurrence, and distribution of dilated cisternae of the endoplasmic reticulum in tissues of plants of the Cruciferae. Protoplasma 71:467–477
Kamimura Y, Mori T, Yamasaki T, Katoh S (1997) Isolation, properties and a possible function of a water-soluble chlorophyll a/b-protein from brussels sprouts. Plant Cell Physiol 38:133–138
Matile P, Schellenberg M, Vicentini F (1997) Localization of chlorophyllase in the chloroplast envelope. Planta 201:96–99
Matsushima R, Kondo M, Nishimura M, Hara-Nishimura I (2003) A novel ER-derived compartment, the ER body, selectively accumulates a β-glucosidase with an ER-retention signal in Arabidopsis. Plant J 33:493–502
Murata T, Ishikawa C (1981) Chemical, physicochemical and spectrophotometric properties of crystalline chlorophyll-protein complexes from Lepidium virginicum L. Biochim Biophys Acta 635:341–347
Murata T, Murata N (1971) Water-soluble chlorophyll-proteins from Brassica nigra and Lepidium virginicum. Carnegie Inst Wash Yearb 70:504–507
Murata T, Toda F, Uchino K, Yakushiji E (1971) Water-soluble chlorophyll protein of Brassica oleracea var. botrys (cauliflower). Biochim Biophys Acta 245:208–215
Nielsen H, Engelbrecht J, Brunak S, von Heijne G (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Prot Eng 10:1–6
Nishio N, Satoh H (1997) A water-soluble chlorophyll protein in cauliflower may be identical to BnD22, a drought-induced, 22-kilodalton protein in rapeseed. Plant Physiol 115:841–846
Noguchi T, Kamimura Y, Inoue Y, Itoh S (1999) Photoconversion of a water-soluble chlorophyll protein from Chenopodium album: resonance Raman and Fourier transform infrared study of protein and pigment structures. Plant Cell Physiol 40:305–310
Oku T, Yoshida M, Tomita G (1972) The photoconversion of heat-treated Chenopodium chlorophyll protein and its pH dependence. Plant Cell Physiol 13:773–782
Pieper J, Rätsep M, Trostmann I, Paulsen H, Renger G, Freiberg A (2011a) Excitonic energy level structure and pigment-protein interactions in the recombinant water-soluble chlorophyll protein. I. Difference fluorescence line-narrowing. J Phys Chem B 115:4042–4052
Pieper J, Rätsep M, Trostmann I, Schmitt FJ, Theiss C, Paulsen H et al (2011b) Excitonic energy level structure and pigment-protein interactions in the recombinant water-soluble chlorophyll protein. II. Spectral hole-burning experiments. J Phys Chem B 115:4053–4065
Renger T, Trostmann I, Theiss C, Madjet ME, Richter M, Paulsen H et al (2007) Refinement of a structural model of a pigment-protein complex by accurate optical line shape theory and experiments. J Phys Chem B 111:10487–10501
Renger G, Pieper J, Theiss C, Trostmann I, Paulsen H, Renger T et al (2011) Water soluble chlorophyll binding protein of higher plants: a most suitable model system for basic analyses of pigment–pigment and pigment-protein interactions in chlorophyll protein complexes. J Plant Physiol 168:1462–1472
Reviron MP, Vartanian N, Sallantin M, Huet JC, Pernollet JC, de Vienne D (1992) Characterization of a novel protein induced by progressive or rapid drought and salinity in Brassica napus leaves. Plant Physiol 100:1486–1493
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sakuraba Y, Schelbert S, Park SY, Han SH, Lee BD, Andrès CB et al (2012) STAY-GREEN and chlorophyll catabolic enzymes interact at light-harvesting complex II for chlorophyll detoxification during leaf senescence in Arabidopsis. Plant Cell 24:507–518
Satoh H, Nakayama K, Okada M (1998) Molecular cloning and functional expression of a water-soluble chlorophyll protein, a putative carrier of chlorophyll molecules in cauliflower. J Biol Chem 273:30568–30575
Satoh H, Uchida A, Nakayama K, Okada M (2001) Water-soluble chlorophyll protein in Brassicaceae plants is a stress-induced chlorophyll-binding protein. Plant Cell Physiol 42:906–911
Schelbert S, Aubry S, Burla B, Agne B, Kessler F, Krupinska K et al (2009) Pheophytin pheophorbide hydrolase (pheophytinase) is involved in chlorophyll breakdown during leaf senescence in Arabidopsis. Plant Cell 21:767–785
Schenk N, Schelbert S, Kanwischer M, Goldschmidt EE, Dörmann P, Hörtensteiner H (2007) The chlorophyllase AtCLH1 and AtCLH2 are not essential for senescence-related chlorophyll breakdown in Arabidopsis thaliana. FEBS Lett 581:5517–5525
Schmidt K, Fufezan C, Krieger-Liszkay A, Satoh H, Paulsen H (2003) Recombinant water-soluble chlorophyll protein from Brassica oleracea var. Botrys binds various chlorophyll derivatives. Biochemistry 42:7427–7433
Schmitt FJ, Trostmann I, Theiss C, Pieper J, Renger T, Fuesers J et al (2008) Excited state dynamics in recombinant water-soluble chlorophyll proteins (WSCP) from cauliflower investigated by transient fluorescence spectroscopy. J Phys Chem B 112:13951–13961
Sherameti I, Venus Y, Drzewiecki C, Tripathi S, Dan VM, Nitz I et al (2008) PYK10, a β-glucosidase located in the endoplasmatic reticulum, is crucial for the beneficial interaction between Arabidopsis thaliana and the endophytic fungus Piriformospora indica. Plant J 54:428–439
Shinashi K, Satoh H, Uchida A, Nakayama K, Okada M, Oonishi I (2000) Molecular characterization of a water-soluble chlorophyll protein from main veins of Japanese radish. J Plant Physiol 157:255–262
Takahashi S, Yanai H, Nakamaru Y, Uchida A, Nakayama K, Satoh H (2012) Molecular cloning, characterization and analysis of the intracellular localization of a water-soluble chlorophyll-binding protein from Brussels sprouts (Brassica oleracea var. gemmifera). Plant Cell Physiol 53:879–891
Takahashi S, Yoshikawa M, Kamada A, Ohtsuki T, Uchida A, Nakayama K, et al (2013a) The photoconvertible water-soluble chlorophyll-binding protein from Chenopodium album is a member of DUF538, a superfamily that distributes in Embryophyta. J Plant Physiol (in press)
Takahashi S, Ono M, Uchida A, Nakayama K, Satoh H (2013b) Molecular cloning and functional expression of a water-soluble chlorophyll-binding protein from Japanese wild radish. J Plant Physiol 170:406–412
Takamiya A, Kamimura Y, Kira A (1968) A transient form of chlorophyll produced by flash photolysis of Chenopodium chlorophyll protein, CP668. In: Shibata K, Takamiya A, Jagendorf AT, Fuller RC (eds) Comparative biochemistry and biophysics of photosynthesis. University of Tokyo Press, Tokyo, pp 229–239
Tanaka R, Tanaka A (2007) Tetrapyrrole biosynthesis in higher plants. Annu Rev Plant Biol 58:321–346
Theiss C, Trostmann I, Andree S, Schmitt FJ, Renger T, Eichler HJ et al (2007) Pigment-pigment and pigment-protein interactions in recombinant water-soluble chlorophyll proteins (WSCP) from cauliflower. J Phys Chem B 111:13325–13335
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Yakushiji E, Uchino K, Sugimura Y, Shiratori I, Takamiya F (1963) Isolation of water-soluble chlorophyll protein from the leaves of Chenopodium album. Biochim Biophys Acta 75:293–298
Yamada K, Hara-Nishimura I, Nishimura M (2011) Unique defense strategy by the endoplasmic reticulum body in plants. Plant Cell Physiol 52:2039–2049
Yamada K, Nagano AJ, Nishina M, Hara-Nishimura I, Nishimura M (2013) Identification of two novel endoplasmic reticulum body-specific integral membrane proteins. Plant Physiol 161:108–120
Acknowledgments
We thank Dr. Soichiro Watanabe for his kind help in the preparation of Chls a and b.
Author information
Authors and Affiliations
Corresponding author
Additional information
The nucleotide sequence reported in this paper has been submitted to DDBJ under accession number AB002589.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Takahashi, S., Yanai, H., Oka-Takayama, Y. et al. Molecular cloning, characterization and analysis of the intracellular localization of a water-soluble chlorophyll-binding protein (WSCP) from Virginia pepperweed (Lepidium virginicum), a unique WSCP that preferentially binds chlorophyll b in vitro. Planta 238, 1065–1080 (2013). https://doi.org/10.1007/s00425-013-1952-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-013-1952-7