Abstract
A chitinolytic Streptomyces strain isolated from an alkaline habitat produced six different isozymes of chitinases. PCR amplification of DNA with chitinase domain specific primers yielded six amplicons of which, a 0.8 Kb fragment was cloned in pQE-30UA direct cloning vector and transformed into E. coli M15 cells (pREP4). The recombinant homodimer protein had a molecular mass of 44 kDa, and the 22 kDa monomers displayed 45 and 60 % activity in the presence of reducing agents. The size of the monomers is close to the predicted putative ORF of 17.8 kDa. The enzyme exhibits extreme pH and temperature optima of 10.0 and 70 °C respectively making it favorable for industrial applications. Its gene sequence revealed no homology with the reported N-acetylglucosaminidases suggesting that it could have novel attributes. This enzyme could be useful in the large scale production of N-acetylglucosamine which is currently having numerous therapeutic and commercial applications.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chitin is a long chain homopolymer composed of repeating N-acetylglucosamine (GlcNAc) units which are covalently linked by β-1,4 glycosidic bonds. It is a major structural component of fungal cell walls, arthropod exoskeleton and crustacean shells [1]. This tough polymer can be biologically broken down by a class of hydrolases known as chitinases which find major applications in biocontrol of plant pathogens and pests [2–4] and management of seafood waste [5]. Chitinases (E.C 3.2.1.14) are produced by fungi, bacteria, insects, plants and animals. According to their mode of action they are classified as endo- and exochitinases [6]. Endochitinases cleave within the chitin polymer and release chitin oligomers which are further broken down by exochitinases. An important category of enzymes that release N-acetylglucosamine from the non-reducing end of the chitin or from chitin oligomers are called N-acetylglucosaminidase (E.C.3.2.1.30). Chitin oligomers have tremendous biomedical applications [7] and their production by chemical methods is hazardous to the environment. Due to this rationale, their enzymatic resurgence is favored [8]. N-acetylglucosamine (GlcNAc) has recently found several applications in the treatment of arthritic diseases, osteoarthritis, inflammatory bowel disease, etc. It also has extensive usage as a valuable ingredient in several cosmetics and as a food additive [9].
Many chitinases have been cloned for their industrial application for the production of GlcNAc and its oligomers [10]. But a few have been successfully applied in industries. This study demonstrates the alkalophilic and thermophilic properties of chitinases from Streptomyces sp. NK52 isolated from an alkaline habitat. Streptomyces sp. NK52 produced six isozymes of chitinases. The gene encoding N-acetylglucosaminidase was cloned in E. coli for the rapid and targeted recovery of N-acetylglucosamine from chitin and marine waste.
Material and Methods
Isolation of Streptomyces sp. NK 52
Streptomyces sp. NK52 was isolated from soil at a location near Pune, India where laundry activity has been prevalent since several decades. Chitinase activity was detected on a medium containing colloidal chitin 10 g l−1; yeast extract 0.2 g l−1; (NH4)2SO4 0.25 g l−1; K2HPO4 0.7 g l−1; KH2PO4 0.3 g l−1; MgSO4 0 g l−1; FeSO4 0.01 g l−1; ZnSO4 0.01 g l−1; MnCl2 0.01 g l−1. Colloidal chitin was prepared as described by Hsu and Lockwood [11].
Amplification of 16S rDNA and Chitinase Fragments
Streptomyces sp. NK52 was grown in Luria–Bertani broth (LB), pH 7.0 at 40 °C for 3 days with shaking at 150 rpm and the genomic DNA was isolated from the harvested cells as described by Pospiech and Newmann [12]. Amplification of the 16S rDNA was carried out using domain specific primers [13]. The chitinase genes were amplified using conserved catalytic domain specific primers [14] with slight modifications. DNA sequencing and analysis was done on Avant 3100 gene analyzer, USA.
Cloning, Expression and Purification of the Putative Chitinase Gene
The amplicons obtained using chitinase domain specific primers were purified and the PCR product (0.8 Kb) was ligated with pQE-30UA vector overnight at 15 °C. The ligation mixture was used for the transformation of E. coli M15 cells by standard techniques [15] and plated on Luria Agar (LA) containing 100 μ g ml−1 of ampicillin and 25 μ g ml−1 of kanamycin. The recombinants were screened for chitinolytic activity on LA containing above mentioned antibiotics, 1 mM IPTG and 0.1 % colloidal chitin. Colonies displaying clearance zones were chosen for further studies. Induction of the recombinant protein was carried out with 3 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) for 6 h and cell pellet obtained after centrifugation was sonicated for 5 min (2″ pulse, 5″ standby, 4 °C, 36 %, Ultrasonic processor-Sigma Braun Biotech, USA) in 5 ml native purification lysis buffer as per the manufacturer’s instructions. The proteins bound to the Nickel NTA column were eluted in a buffer containing 50 mM Na2HPO4, 300 mM NaCl and 90 mM imidazole (pH 8.0) with flow rate of 1 ml min−1. Absorbance of the eluates was measured at 280 nm and protein containing fractions were checked for chitinase activity.
Determination of Chitinase Activity and Protein Content
During purification of chitinase, the enzyme activity in fractions was initially determined with swollen chitin as substrate [13]. Protein content was measured as previously described [16] with bovine serum albumin as standard.
To determine the nature of the chitinase under study, p-nitrophenyl-N-acetyl-β-d-glucosaminide (pNP-GlcNAc), p-nitrophenyl-β-d-N,N′-diacetylchitobiose [pNP-(GlcNAc)2] and p-nitrophenyl-β-d-N,N′,N″-triacetylchitotriose [pNP-(GlcNAc)3] were used as substrates [17]. One unit of enzyme activity was defined as the amount of enzyme that released 1 μ mol of p-nitrophenol per minute under the mentioned assay conditions.
Characterization of the Recombinant Protein
The molecular weight and purity of protein fractions was determined by SDS-PAGE carried out by the method of Laemmli [18] and protein bands were detected by silver staining [19]. Size exclusion chromatography of pure native protein was carried out on Sephadex G-100 calibrated using Gel Filtration Molecular Weight Markers Kit (molecular weights 12,000–200,000 Da MWGF200, Sigma–Aldrich, MO, USA). Chitinase activity of proteins was detected on glycol chitin gels as described earlier [13]. Isoelectric focusing (IEF) was carried out on a 17.0 cm, pH 3.0–10.0 Readystrip™ IPG strip and run in a protean–IEF cell (Biorad laboratories Inc, USA) along with standard pI marker (3.6–9.3).
Optimum pH was measured at different pH values using pNP-GlcNAc as the substrate. The pH of the reaction mixtures was made varied by using 100 mM citrate phosphate (pH 2.0–7.0), Tris–glycine (pH 8.0–9.0) and glycine–NaOH (pH 10.0–12.0) [13]. The enzyme activity was determined under the standard assay conditions. The enzyme activity was determined in the presence of metal ions and denaturants (refer Table 1) at a final concentration of 1 mM under the standard assay conditions.
Hydrolysis Products from Chitin Oligomers and Derivatives
The purified enzyme (0.05 unit, hereafter referred to as CHIT_52) was incubated with 0.5 mM each of (GlcNAc)1–6, swollen chitin and colloidal chitin for 1 h at pH 10.0 and 70 °C. Aliquots of the products were withdrawn at various time intervals and purified on HPLC (Gilson, USA) on a Sugar-D column (2.5 × 25 cm, Nacalai–Tesque, Japan) using acetonitrile:water (70:30) as mobile phase at flow rate of 4.0 ml min−1 and detected at 210 nm. The purified end products were compared with standard chitin oligosaccharides (GlcNAc)1–6 for identification. All experiments were carried out in triplicates.
Results and Discussion
The isolate NK52 was identified as Streptomyces sp. based on morphological and biochemical properties and 16S rDNA sequence (GenBank accession number GQ221681). Streptomyces sp. NK52 is publicly available at KCTC Korea (KCTC 19739). The strains of genus Streptomyces have inherent multiple chitinases which enable them to breakdown chitin more efficiently due to their collective action [20]. Streptomyces sp. NK52 reported in this study was isolated from conditions favoring alkalophiles. Alkalophilic actinomycetes have been previously isolated from soil samples having both alkaline and neutral pH [21] which shows the predominance of Streptomyces sp. in varied environmental conditions.
The DNA fragments obtained after amplification with domain specific chitinase gene primers are shown in Fig. 1. Ligation of the 0.8 Kb DNA fragment with pQE-30UA vector and its transformation in E. coli M15 gave colonies displaying clearance on LA with 0.1 % colloidal chitin. Maximum expression of the recombinant protein (here afterwards referred to as CHIT_52) was achieved after 6 h incubation in the presence of 3.0 mM IPTG thus, the expression was carried out under these conditions.
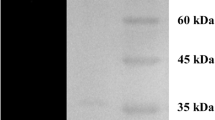
The elution profile of CHIT_52 was loaded on Ni–NTA agarose and eluted in the presence of increasing concentration of imidazole (10 to 200 mM). Fractions 8–11 eluted with 90 mM imidazole displayed activity with swollen chitin. These fractions were pooled, concentrated and analyzed by SDS-PAGE. The large scale purification of CHIT_52 was carried out using Ni–NTA agarose affinity chromatography as the vector had a hexa-histidine tag. Metal affinity chromatography has been earlier used for rapid small scale purification of recombinant chitinases as reported previously [22]. A 41 % yield and two fold purity of CHIT_52 in a single step purification is similar to the yield and fold purification of an N-acetylglucosaminidase reported earlier [23]. This would be helpful in the large scale purification of CHIT_52 making it more applicable for industries. The fractions displayed a single band of 44 kDa in the absence of any reducing agent (Fig. 2, lane A) and a single band of 22 kDa in the presence of 100 mM dithiothreitol (DTT) (Fig. 2, lane B), indicating the protein to be a homodimer. Chitinase activity was seen on glycol chitin gel in the presence (Fig. 2, lane C) and absence (Fig. 2, lane D) of reducing agent. This indicates that the enzyme is active in the presence of reducing agents. Figure 2, lane E shows the SDS-PAGE profile (without reducing agent) of crude protein extract from Streptomyces sp. NK52 grown in the presence of chitin. Figure 2, lane F shows the activity staining of a 24 kDa band in the crude protein extract with 4-methylumbelliferyl-N-acetyl-d-glucosaminide, a substrate selective for the detection N-acetylglucosaminidase activity on acrylamide gels. The molecular weight of pure native protein was 44 kDa as confirmed by size exclusion chromatography.
SDS-PAGE and zymogram of CHIT_52 and crude chitinase from Streptomyces sp. NK52 lane (M) molecular weight marker (10–200 kDa), lane (A) SDS-PAGE CHIT_52 in non-reducing conditions, lane (B) SDS-PAGE of CHIT_52 in reducing conditions, lane (C) zymogram of CHIT_52 in reducing conditions, lane (D) zymogram of CHIT_52 in non-reducing conditions, lane (E) SDS-PAGE of crude protein extract from Streptomyces sp. NK52 grown on chitin medium, lane (F) zymogram of crude protein extract of Streptomyces sp. NK52 with 4-methylumbelliferyl-N-acetyl-d-glucosaminide as substrate demonstrating presence of 24 kDa N-acetylyglucosaminidase
CHIT_52 exhibited maximum activity at pH 10.0 (Fig. 3) and 70 °C (Fig. 4). The enzyme was most stable at pH 6.0 and 40 °C with a wide range of stability between pH 4.0–8.0 and 40–70 °C. Alkalophilic chitinases are reported from Thermoascus aurantiacus var. levisporus [24] and Aeromonas caviae [25] where the optimum pH was 8.0 and 9.0 respectively. Thermophilic N-acetylglucosaminidases are reported from Streptomyces thermoviolaceus OPC-520 [26], Bacillus sp. NCIM 5120 [27] and Bacillus stearothermophilus [28] with optimum activity at 60, 70 and 75 °C respectively. The extreme pH and temperature optima make CHIT_52 more robust for industrial applications.
CHIT_52 released p-nitrophenol from pNP-GlcNAc and GlcNAc from chitin oligomers like (GlcNAc)2–6, colloidal chitin and swollen chitin; confirming its N-acetylglucosaminidase activity (Fig. 5). CHIT_52 produced N-acetylglucosamine as the end product on reaction with different chitin substrates. CHIT_52 could release N-acetylglucosamine from (GlcNAc)2–3 in 15 min exhibiting 100 % conversion to GlcNAc however, from (GlcNAc)5 and (GlcNAc)6 it released N-acetylglucosamine only on prolonged incubation.CHIT_52 could convert (GlcNAc)5 into smaller oligomers at 20 min giving 40 % (GlcNAc)3 and 10 % GlcNAc. Whereas at 30 min, CHIT_52 released 50 % (GlcNAc)2 and 25 % GlcNAc. It released 50 % each (GlcNAc)2 and GlcNAc at 40 min. This indicates the rapid conversion of oligomers into GlcNAc. The crude chitinase as well as CHIT_52 from Streptomyces sp. NK52 also released GlcNAc from swollen chitin after an incubation period of 1 h. This suggests that N-acetylglucosaminidase plays an important role in the complete breakdown of swollen chitin to GlcNAc. The large scale production of N-acetylglucosamine currently involves the employment of high temperature (40–80 °C) and strong acids (15–36 % HCl) which yields lower quantity of GlcNAc along with O-acetylated and di-acetylated products which give a bitter taste making it unfit for consumption [10]. The biological production of GlcNAc is more environmental friendly along with improved yield and sweet taste owing to the mild conditions employed. N-acetylglucosamine is non-toxic and can be administered easily for therapeutic applications [10]. This enzyme finds appreciable value for the selected large scale production of GlcNAc from chitin derivatives.
The isoelectric point (pI) of CHIT_52 was 3.6. The activity of CHIT_52 was inhibited by Hg2+ and enhanced in presence of Mg2+, Mn2+and Ag+. EDTA did not inhibit the enzyme activity indicating that it did not require any cofactors for activity. CHIT_52 retained 86 and 55 % activity in the presence of 1 and 10 M urea respectively. In the presence of 10–100 mM DTT, the enzyme exhibited relative activity of 80 and 60 % respectively as shown in Table 1. Previous reports indicate the addition of various detergents for loosening of the chitin structure to enable action of chitinases for the production of GlcNAc [10]. This makes CHIT_52 more valuable since it is not inactivated by any of these detergents.
The gene sequence of chit_52 was determined with specific primers and it spanned a region of 788 bp encoding two open reading frames of 162 and 127 amino acids. The amino acid sequence from the ORF encoding 162 amino acids was deduced using CLC Sequence Viewer version 6.0. The molecular weight determined by SDS-PAGE (22 kDa) correlated well with the predicted molecular weight of 17.8 kDa from the ORF encoding 162 amino acids. The chit_52 gene sequence is submitted in the GenBank database with the accession number GQ851949 and the deduced protein sequence is submitted with accession number ACX29957. The translation of annotated chit_52 gene sequence revealed the presence of glutamate and glutamine residues that are topographically conserved in family 85 glycosyl hydrolases as reported earlier [29]. The gene sequence also exhibited numerous glutamate and aspartate residues which are required for activity [30]. Four ORF’s were found in the 8 Kb fragment encoding chitinase from Aeromonas sp. No.10S-24 with 535, 474, 538 and 287 amino acids which is dissimilar to the size of the ORF’s found here [31]. The second and the fourth domains in the ORF1 from the same fragment were found to be rich in threonine and proline residues, which is similar to the ORF encoding the N-acetylglucosaminidase enzyme in the present study. The different domains of the ORFs from Aeromonas sp. No.10S-24 also exhibited homology to domains of xylanases and cellulases. However the chit_52 gene or translated amino acid sequence did not exhibit appreciable homology with any of the reported glycosyl hydrolases or chitinases. Figure 6 shows the phylogenic relation of putative N-acetylglucosaminidase with related protein sequences in the database.
N-acetylglucosamine has numerous applications as a dietary supplement and anti-inflammatory drug. Recently, novel anti-HIV drugs were synthesized from glucosamine and N-acetylglucosamine containing dipeptides and polypeptides [32]. This opens new vistas for the use of N-acetylglucosamine in the synthesis of novel glycopeptides which may find important medical and industrial applications. Besides, Streptomyces sp. NK52 harbors other chitinases which need to be studied at the protein and molecular level throwing some light on the diversity of chitinases present in this system.
Conclusions
CHIT_52 is an important industrial N-acetylglucosaminidase owing to its extreme alkalophilic and thermophilic nature. In addition, it is suitable for rapid recovery of N-acetylglucosamine at an industrial scale owing to other robust characters like low molecular weight, stability over wide range of pH, temperature and detergents. A large scale, rapid and low cost production of N-acetylglucosamine using the recombinant strain developed in this study will be beneficial for industries involved in the production of this medicinally important saccharide. Studies on utility of this strain at an industrial level are in progress. Thus, CHIT_52 is different in terms of gene sequence, low molecular weight, extreme pH and temperature optima along with thermal and chemical stability, than other reported N-acetylglucosaminidases, making it an interesting enzyme for further studies.
References
Majeti N, Kumar R (2000) A review of chitin and chitosan applications. React Funct Polym 46:1–27
Driss F, Kallassy-Awad M, Zouari N, Jaoua S (2005) Molecular characterization of a novel chitinase from Bacillus thuringiensis subsp. kurstaki. J Appl Microbiol 99:945–953
dela Vega LM, Barboza-Corona JE, Aguilar-Uscanga MG, Ramírez-Lepe M (2006) Purification and characterization of an exochitinase from Bacillus thuringiensis subsp. aizawai and its action against phytopathogenic fungi. Can J Microbiol 52:651–657
Huang CJ, Wang TK, Chung SC, Chen CY (2005) Identification of an antifungal chitinase from a potential biocontrol agent, Bacillus cereus 28–9. J Biochem Mol Biol 38:82–88
Wang S, Lin B-S, Liang T, Wang C, Wu P, Liu J (2010) Purification and characterization of chitinase from a new species strain, Pseudomonas sp. TKU008. J Microbiol Biotechnol 20:1001–1005
Sahai AS, Manocha MS (1993) Chitinases of fungi and plants: their involvement in morphogenesis and host-parasite interaction. FEMS Microbiol Rev 11:317–338
Patil RS, Ghormade V, Deshpande MV (2000) Chitinolytic enzymes: an exploration. Enzyme Microb Technol 26:473–483
Il’ina AV, Zueva OY, Lopatin SA, Varlamov VP (2005) Enzymatic hydrolysis of α-chitin. Appl Biochem Microbiol 35:1154–1156
Chen JK, Shen CR, Liu CL (2010) N-acetylglucosamine: production and applications. Mar Drugs 8:2493–2516
Kuk JH, Jung WJ, Jo GH, Kim YC, Kim KY, Park RD (2005) Production of N-acetyl-β-d-glucosamine from chitin by Aeromonas sp. GJ-18 crude enzyme. Appl Microbiol Biotechnol 68:384–389
Hsu SC, Lockwood JL (1975) Powdered chitin agar as a selective medium for enumeration of actinomycetes in water and soil. Appl Microbiol 29:422–426
Pospiech A, Neumann B (1995) A versatile quick prep of genomic DNA from gram-positive bacteria. Trends Genet 11:217–218
Nawani NN, Kapadnis BP, Das AD, Rao AS, Mahajan SK (2002) Purification and characterization of a thermophilic and acidophilic chitinase from Microbispora sp. V2. J Appl Microbiol 93:965–975
Xiao X, Yin X, Lin J, Sun L, You Z, Wang P, Wang F (2005) Chitinase genes in lake sediments of Ardley islands, Antartica. Appl Environ Microbiol 71:7904–7909
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, New York
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the folin-phenol reagent. J Biol Chem 193:265–275
Matsuo Y, Kurita M, Park JK, Tanaka K, Nakagawa T, Kawamukai M, Matsuda H (1999) Purification, characterization and gene analysis of N-acetylyglucosaminidase from Enterobacter sp. G1. Biosci Biotechnol Biochem 63:1261–1268
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Merril CR, Goldman D, Vankeuren ML (1983) Silver staining methods for polyacrylamide gel electrophoresis. Methods Enzymol 96:230–239
Tsujibo H, Okamoto T, Hatano N, Miyamoto K, Watanabe T, Mitsutomi M, Inamori Y (2000) Family 19 chitinases from Streptomyces thermoviolaceus OPC-520: molecular cloning and characterization. Biosci Biotechnol Biochem 64:2445–2453
Selyanin VV, Oborotov EG, Zenova GM, Zvyagintsev DG (2005) Alkaliphilic soil actinomycetes. Microbiology 74:729–734
Christodoulou E, Duffner F, Vorgias CE (2001) Overexpression, purification and characterization of a thermostable chitinase (Chi40) from Streptomyces thermoviolaceus OPC-520. Protein Expr Purif 23:97–105
Reddy A, Grimwood BG, Plummer TH, Tarentino AL (1998) High-level expression of the endo-β-N-acetylglucosaminidase F2 gene in E.coli: one step purification to homogeneity. Glycobiology 8:633–636
Li A, Yu K, Liu H, Zhang J, Li H, Li D (2010) Two novel thermostable chitinase genes from thermophilic fungi: cloning, expression and characterization. Bioresour Technol 101:5546–5551
Lin J, Xiao X, Zeng X, Wang F (2006) Expression, characterization and mutagenesis of the gene encoding β-N-acetylgluosaminidase from Aeromonasa caviae CB101. Enzyme Microb Technol 38:765–771
Kubota T, Miyamoto K, Yasuda M, Inamori Y, Tsujibo H (2004) Molecular characterization of an intracellular β-N-acetylglucosaminidase involved in the chitin degradation system of Streptomyces thermoviolaceus OPC-520. Biosci Biotechnol Biochem 68:1306–1314
Amutha B, Khire JM, Khan MI (1998) Characterization of a novel exo-N-acetyl-β-d-glucosaminidase from the thermotolerant Bacillus sp. NCIM 5120. Biochim Biophys Acta 1425:300–310
Sakai K, Narihara M, Kasama Y, Wakayama M, Moriguchi M (1994) Purification and characterization of thermostable β-N-acetylhexoaminidase of Bacillus stearothermophilus CH-4 isolated from chitin-containing compost. Appl Environ Microbiol 60:2911–2915
Abbott DW, Macauley MS, Vocadlo DJ, Boraston AB (2009) Streptococcus pneumoniae endohexosaminidase D, structural and mechanistic insight into substrate-assisted catalysis in family 85 glycoside hydrolases. J Biol Chem 284:11676–11689
Okazaki K, Yamashita Y, Noda M, Sueyoshi N, Kameshita I, Hayakawa S (2004) Molecular cloning and expression of the gene encoding family 19 chitinase from Streptomyces sp.J13-3. Biosci Biotechnol Biochem 68:341–351
Shiro M, Ueda M, Kawaguchi T, Arai M (1996) Cloning of a cluster of chitinase genes from Aeromonas sp. 10S–24. Biochim Biophys Acta 1305:44–48
Huang W, Groothuys S, Heredia A, Kuijpers BH, Rutjes FP, van Delft FL, Wang LX (2009) Enzymatic glycosylation of triazole-linked GlcNAc/Glc-peptides: synthesis, stability and anti-HIV activity of triazole-linked HIV-1 gp41 glycopeptide C34 analogues. Chem Biochem 10:1234–1242
Acknowledgments
The authors are thankful to Department of Science and Technology, Government of India for financial support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Prakash, D., Nawani, N. & Kapadnis, B. Cloning, Expression and Characterization of Thermophilic and Alkalophilic N-acetylglucosaminidase from Streptomyces sp. NK52 for the Targeted Production of N-acetylglucosamine. Proc. Natl. Acad. Sci., India, Sect. B Biol. Sci. 83, 431–437 (2013). https://doi.org/10.1007/s40011-013-0158-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40011-013-0158-x