Abstract
β-N-Acetylglucosaminidases (GlcNAcases) are important for many biological functions and industrial applications. In this study, a glycoside hydrolase family 20 GlcNAcase from Shinella sp. JB10 was expressed in Escherichia coli BL21 (DE3). Compared to many GlcNAcases, the purified recombinant enzyme (rJB10Nag) exhibited a higher specificity activity (538.8 µmol min−1 mg−1) or V max (1030.0 ± 82.1 µmol min−1 mg−1) toward p-nitrophenyl β-N-acetylglucosaminide and N,N′-diacetylchitobiose (specificity activity of 35.4 µmol min−1 mg−1) and a higher N-acetylglucosaminide tolerance (approximately 50% activity in 70.0 mM N-acetylglucosaminide). The degree of synergy on enzymatic degradation of chitin by a commercial chitinase and rJB10Nag was as high as 2.35. The enzyme was tolerant to most salts, especially 3.0–15.0% (w/v) NaCl and KCl. These biochemical characteristics make the JB10 GlcNAcase a candidate for use in many potential applications, including processing marine materials and the bioconversion of chitin waste. Furthermore, the enzyme has the highest proportions of alanine (16.5%), glycine (10.5%), and random coils (48.8%) with the lowest proportion of α-helices (24.9%) among experimentally characterized GH 20 GlcNAcases from other organisms.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Chitin, an unbranched β-1,4-linked polymer of N-acetylglucosamine (GlcNAc), is primarily produced by marine invertebrates, insects, fungi, and algae (Patil et al. 2000). Chitin is second only to lignocellulose in natural abundance, and every year more than 80,000 tons of chitin from marine sources goes unutilized (Patil et al. 2000). To utilize chitin, chemical reagents, e.g., hydrochloric acid, are used to degrade the chitin (Park et al. 2011). In comparison, the enzymatic degradation of chitin is more environmentally friendly and generates improved yields (Park et al. 2011). The complete enzymatic degradation of chitin depends on the synergistic actions of chitinases (EC 3.2.1.14) and β-N-acetylglucosaminidases (GlcNAcases, EC 3.2.1.52). Chitinases randomly catalyze the hydrolysis of the chitin backbone to produce N-acetyl chitooligosaccharides, and GlcNAcases further hydrolyze the N-acetyl chitooligosaccharides to produce GlcNAc, thereby diminishing the inhibition effect of N-acetyl chitooligosaccharides on chitinases (Yang et al. 2014).
GlcNAc has many important biological functions, including bacterial cell wall recycling and bacterial flagellum assembly, and has a wide range of industrial applications, such as treatment of ulcerative colitis and other gastrointestinal inflammation disorders, pharmaceutical therapies for osteoarthritis and tumors, and the production of bioethanol and single-cell protein (Inokuma et al. 2016; Yang et al. 2014; Zhou et al. 2016c).
GlcNAcases are widely distributed in animal tissues, insects, plants, bacteria, and fungi and are classified into four glycoside hydrolase (GH) families (GH 3, 20, 73, and 84) according to amino acid sequence homology (http://www.cazy.org/). To the best of our knowledge, GH 20 GlcNAcases are mainly found in microorganisms, most of which have not been characterized (http://www.cazy.org/GH20.html).
In recent years, salt-tolerant enzymes have attracted a great deal of attention, including: carbonic anhydrases (Premkumar et al. 2005; Warden et al. 2015); esterases (Wang et al. 2016); α-amylases (Qin et al. 2014); mannanases (Zhou et al. 2012); exo-inulinases (Shen et al. 2015; Zhou et al. 2015a); α-galactosidases (Zhou et al. 2016a, b); and laccases (Molina-Guijarro et al. 2009). We have previously described the molecular and biochemical characteristics of a salt-tolerant GH 3 GlcNAcase (Zhou et al. 2016c). These salt-tolerant enzymes are thought to have potential use in various applications, including the processing of various saline foods in China and marine materials with NaCl contents of 3.5–15.9% (w/w) (Zhou et al. 2016c). Regarding economic considerations, fermentation and material processing under high NaCl concentrations is helpful for reducing costs as sterilization is unnecessary (Margesin and Schinner 2001). In addition, salt-tolerant enzymes are good models for basic research on the molecular adaptation to high salt environments.
The largest deposit of phosphate rock in the country is in Yunnan Province, known in China as ‘‘the Kingdom of Nonferrous Metals”. We previously sampled slag from a phosphate rock-stacking site located in this province and discovered unique glycoside hydrolases from novel bacteria isolated from the slag (Zhou et al. 2012, 2015a, 2016b). An interesting feature of these enzymes is their salt tolerance (Zhou et al. 2012, 2015a, 2016b). In this study, a novel GH 20 GlcNAcase, designated JB10Nag, was discovered from Shinella sp. JB10, a new isolate harbored in the same slag. Molecular and biochemical characterizations of the GlcNAcase were made. Compared with the salt-tolerant GH 3 GlcNAcase that we previously reported (Zhou et al. 2016c), the GH 20 GlcNAcase JB10Nag showed higher activity and stronger GlcNAc tolerance.
Materials and methods
Vectors and reagents
The following vectors and reagents were obtained from the indicated sources: genomic DNA and plasmid isolation kits (Tiangen, Beijing, China); pEASY-E2 vector and E. coli BL21 (DE3) (TransGen, Beijing, China); Qubit protein assay kit (Invitrogen, Carlsbad, CA, USA); isopropyl-β-d-1-thiogalactopyranoside (IPTG; Amresco, Solon, OH, USA); Ni2+-NTA agarose (Qiagen, Valencia, CA, USA); dNTPs and DNA polymerases (TaKaRa, Otsu, Japan); p-nitrophenyl β-N-acetylgalactosaminide (pNPGalNAc), p-nitrophenyl β-d-gluopyranoside (pNPGlc), N,N′-diacetyl chitobiose (GlcNAc2) and N,N′,N″,N‴-tetraacetyl chitotetraose (GlcNAc4) (J&K Scientific Ltd., Beijing, China); Bacillus subtilis peptidoglycan (Ekear, Shanghai, China); silica gel G plate (Haiyang, Qingdao, China); and Streptomyces globisporus ATCC 21553 mutanolysin, p-nitrophenol (pNP), p-nitrophenyl-β-d-xylopyranoside (pNPXyl), p-nitrophenyl α-d-galactopyranoside (pNPGal), p-nitrophenyl-α-l-arabinofuranoside (pNPAra), p-nitrophenyl β-N-acetylglucosaminide (pNPGlcNAc), chitin, and chitosan (Sigma-Aldrich, St. Louis, MO, USA). All other chemicals used in this study were of analytical grade.
Microorganism isolation and preservation
The strain JB10 was isolated from the slag collected from a phosphate rock-stacking site located in Yunnan Province. The details of strain isolation are described in our previous study (Zhou et al. 2012). The pure culture was deposited in the Strains Collection of the Yunnan Institute of Microbiology under registration no. YMF 3.00678.
Gene cloning and molecular characterization
The GlcNAcase-encoding gene, jB10Nag, was identified by sequencing the genome of JB10. The methodology used to sequence the genome of JB10 is the same as that used for GN16 and is described in our previous study (Zhou et al. 2015b).
The prediction of open reading frames from the draft genome of JB10 was performed as described in our previous study (Zhou et al. 2016b). The online tools BLASTN and BLASTP (http://www.ncbi.nlm.nih.gov/BLAST/) were used to search for homologous sequences and calculate identity values. The online tools SignalP (http://www.cbs.dtu.dk/services/SignalP/) and InterPro (http://www.ebi.ac.uk/interpro/) were used to predict signal peptides and protein domains, respectively.
The tertiary structure of JB10Nag was predicted using the I-TASSER platform (http://zhanglab.ccmb.med.umich.edu/I-TASSER/). Proportions of secondary structures were determined using VADAR (http://vadar.wishartlab.com/). The charge distribution on the surface of the GlcNAcase was calculated using the Discovery Studio 2.5 software (Accelrys, San Diego, CA, USA).
Heterologous expression of GlcNAcase
The coding sequence of JB10Nag was amplified by PCR using Pyrobest DNA polymerase and the primers rjB10NagEF (5′-CCCGCCCTCGAAACCATGTT-3′) and rjB10NagER (5′-GTGTCCGGAAGATCCGTAAAGCAC-3′). After the addition of a 5′ terminal adenine using rTaq DNA polymerase, the resulting PCR product was ligated into the pEASY-E2 vector which has a single 3′-thymine overhang at the insertion site. E. coli BL21 (DE3) competent cells were transformed with the plasmid for recombinant enzyme expression. After confirmation of the correct insert sequence by DNA sequencing, a positive transformant harboring the recombinant plasmid was grown at 37 °C overnight, and then, the seed culture was used to inoculate with 1:100 dilutions into fresh Luria–Bertani medium containing 100 μg mL−1 ampicillin. When the culture reached an OD600 of approximately 0.7, IPTG at a final concentration of 0.25 mM was added to induce recombinant enzyme expression at 20 °C for 20 h.
Purification and identification of the recombinant GlcNAcase
The culture containing positive transformant cells was centrifuged and then resuspended in ice-cold buffer (20 mM Tris–HCl and 0.5 M NaCl, pH 7.2). The cells were disrupted by ultrasonication on ice, and cell debris was removed by centrifugation at 8000×g at room temperature for 6 min. The supernatant was passed over Ni2+-NTA agarose gel columns to purify the target recombinant enzyme with a linear imidazole gradient of 20–300 mM in buffer containing 20 mM Tris–HCl and 0.5 M NaCl (pH 7.2). The purified recombinant enzyme was stored at −20 °C until used for subsequent analyses.
The purity of the eluted fractions was checked by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) using 12% running gels. To identify the purified enzyme as recombinant JB10Nag (rJB10Nag), the purified protein band identified by SDS–PAGE gel was cut and analyzed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI–TOF MS) performed by Tianjin Biochip (Tianjin, China) to detect the molecular mass of an internal peptide. The protein concentration was determined by a Qubit protein assay kit using a Qubit 2.0 fluorometer (Invitrogen).
Enzyme assay and substrate specificity
The enzymatic activity of rJB10Nag toward various substrates was determined spectrophotometrically using the following methods:
-
1.
pNP method: a 500-µL assay mixture contained 50 µL of appropriately diluted rJB10Nag and 450 µL of 2.0 mM pNPGlcNAc that was prepared in McIlvaine buffer at pH 6.0. The mixture was incubated in a water bath at 50 °C for 10 min followed by the addition of 2.0 mL of 1.0 M Na2CO3 to terminate the reaction. The amount of pNP liberated from pNPGlcNAc was determined by spectrophotometrically measuring the absorption at 405 nm. One unit of GlcNAcase was defined as the amount of enzyme that released 1 µmol of pNP per minute under the assay conditions, unless otherwise noted. The activities of the enzyme toward the substrates pNPGalNAc, pNPGlc, pNPGal, pNPXyl, or pNPAra were also determined by pNP method.
-
2.
DNS method: a 500-µL assay mixture contained 50 µL of appropriately diluted rJB10Nag and 450 µL of 0.5% (w/v) substrate prepared in McIlvaine buffer at pH 6.0. The substrates include GlcNAc2, GlcNAc4, colloidal chitin, chitosan, peptidoglycan, and muropeptides (degradation products of peptidoglycan by mutanolysin treatment). The reaction was incubated in a water bath at 50 °C for 10 min followed by the addition of 2.0 mL of 3,5-dinitrosalicylic acid (DNS) reagent to terminate the reaction. The amount of reducing sugars liberated from substrate was determined by measuring the absorption at 540 nm spectrophotometrically. One unit of enzyme was defined as the amount of enzyme that released 1 µmol of reducing sugars equivalent to GlcNAc per minute under the assay conditions, unless otherwise noted.
Biochemical characterization
The pNP method, using pNPGlcNAc as a substrate, was used to determine the biochemical characteristics of purified rJB10Nag, unless otherwise noted.
The buffers, including McIlvaine buffer for pH values of 4.5–8.0 and 0.1 M glycine–NaOH for pH values of 9.0–10.0, were used to determine the pH-dependent activity and stability of purified rJB10Nag. The pH-dependent activity of rJB10Nag was assessed at 37 °C with pH values ranging from 4.5 to 8.0. The pH-dependent stability of rJB10Nag was estimated by measuring the residual enzyme activity at 37 °C, pH 6.0 after incubating the enzyme at pH values of 5.0–10.0 for 6 h.
The temperature-dependent activity of rJB10Nag was examined at temperatures ranging from 0 to 60 °C. To determine the thermostability of rJB10Nag, the enzyme was incubated at 30, 37, or 50 °C, pH 6.0 for 1–60 min, with aliquots removed to measure residual enzyme activity.
To investigate the effects of different metal ions and chemical reagents on purified rJB10Nag, the enzyme activity was measured in the presence of various metal ions and chemical reagents. The metal ions and chemical reagents assayed included: AgNO3, CaCl2, CoCl2, ZnSO4, MnSO4, FeSO4, CuSO4, NiSO4, MgSO4, Pb(CH3COO)2, HgCl2, FeCl3, EDTA, β-mercaptoethanol, and SDS [at final concentrations of 1.0 or 10.0 mM); NaCl and KCl (at final concentrations of 3.0–30.0% (w/v)]; and Tween 80 and Triton X-100 [at final concentrations of 0.5 or 1.0% (v/v)]. Any precipitation was removed by centrifugation before measuring the absorption. The enzyme stability in NaCl or KCl was estimated by measuring the residual enzyme activity following pre-incubation of the enzyme in 3.0–30.0% (w/v) of NaCl or KCl for 60 min.
The extent of GlcNAc inhibition on rJB10Nag activity was determined by the addition of 4.0–70.0 mM GlcNAc to the reaction solution.
K m, V max, and k cat values of purified rJB10Nag were determined using 0.2–1.0 mM pNPGlcNAc as a substrate. The data were plotted by nonlinear regression fit of Michaelis–Menten using software GraphPad Prism (GraphPad Software, San Diego, CA, USA).
Hydrolytic property of purified rJB10Nag
The hydrolysis mixture included 160 µL of 0.5% (w/v) GlcNAc2 or GlcNAc4 and 0.4 unit of purified rJB10Nag. The mixture was incubated in McIlvaine buffer (pH 6.0) at 37 °C for 6 h. Hydrolysis products were analyzed by thin layer chromatography (TLC) as previously described (Zhou et al. 2016b).
Synergistic action
To investigate the synergistic action of chitinase and rJB10Nag, a commercial Streptomyces griseus chitinase (CtnSg) was utilized (Shanghai YuanYe Bio-Technology, Shanghai, China). Reactions containing 0.5% (w/v) of colloidal chitin and either CtnSg (0.01 U mL−1), rJB10Nag (0.5 U mL−1) or both enzymes were incubated for 120 min. In the time-course degradation study of 0.5% (w/v) colloidal chitin, a reaction mixture containing CtnSg alone was incubated for 30, 60, or 90 min, then rJB10Nag was added and incubated for 90, 60, or 30 min. In consideration of the biochemical characteristics of CtnSg and rJB10Nag, the degradation reactions were performed at 25 °C in McIlvaine buffer (pH 6.0). The amount of reducing sugars released from colloidal chitin was measured using the DNS method. A reaction containing heat-inactivated enzyme was used as a control.
Accession numbers
The GenBank accession numbers of Shinella sp. JB10 16S rDNA and jB10Nag are KX014620 and KX014621, respectively.
Results
Strain identification
Based on the results of a BLASTN search, a nucleotide identity of greater than 97.9% between the 16S rDNA sequences of the JB10 strain and of the Shinella zoogloeoides ATCC 19623 (accession no. X74915), Shinella yambaruensis NBRC 102122 (AB681707), and Shinella fusca LMG 24714 (FM177879) type strains was determined. Phylogenetic analysis placed strain JB10 in the Shinella cluster, but not in clusters grouped by the species of other genera of Rhizobiaceae (Fig. S1). As such, JB10 was classified within the genus Shinella.
Gene cloning and sequence analysis
Sequence data of approximately 375 Mbp was generated by genome sequencing for JB10, and these data yielded a draft genomic DNA sequence of approximately 5.9 Mbp after sequence assembly. The analysis to the draft genome predicted an open reading frame designated as the gene jB10Nag. The jB10Nag gene has a length of 2001 bp and encodes the GlcNAcase JB10Nag, having a predicted molecular mass of 70.9 kDa.
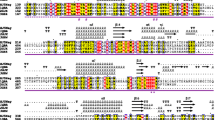
Frequencies of the amino acid residues A and G within the GlcNAcase are 16.5 and 10.5%, respectively (Table S1). Similar frequencies of other experimentally characterized GH 20 GlcNAcases are shown in Table S1. Among these GlcNAcases, JB10Nag has the highest frequencies of A and G. A signal peptide was not found in the JB10 GlcNAcase. An analysis of the GlcNAcase using the InterPro online tool showed a typical bacterial GlcNAcase N-terminal domain (from V135 to P269 of JB10Nag; domain signature: IPR015882; Fig. 1). A catalytic domain of GH 20 GlcNAcases was identified from P269 to G655 of JB10Nag (IPR015883; Fig. 1). The D444 residue was predicted as the catalytic nucleophile/base, and E445 as the catalytic proton donor/acceptor based on an amino acid sequence alignment of JB10Nag with the Vibrio harveyi 650 GlcNAcase, VhGlcNAcase (Fig. 1; Meekrathok and Suginta 2016; Suginta et al. 2010). In addition, the residues D279, R282, D311, D312, W519, W579, and W613 may also participate in ligand binding (Fig. 1; Meekrathok and Suginta 2016; Suginta et al. 2010).
Amino acid sequence alignment of JB10Nag with GH 20 GlcNAcases. Accession No. AAC44672, the experimentally characterized GH 20 GlcNAcase from V. furnissii (Keyhani and Roseman 1996); ADJ68333, the experimentally characterized GH 20 GlcNAcase from V. harveyi (Suginta et al. 2010); and KNY15668, the predicted GH 20 GlcNAcase from Shinella sp. SUS2. Asterisks and number signs show the putative catalytic residues and ligand-binding residues, respectively. Purple arrow indicates the N-terminal domain of bacterial GlcNAcases (domain signature: IPR015882). Green arrow indicates the catalytic domain of GH 20 GlcNAcases (domain signature: IPR015883)
The BLASTP result showed that JB10Nag is most similar to the GH 20 GlcNAcase from Shinella sp. SUS2 (accession no. KNY15668). The amino acid identity between the two GlcNAcases was 71.3%. However, the Shinella sp. SUS2 GH 20 GlcNAcase has not been biochemically characterized. We found that JB10Nag shared less than 30% identity with experimentally characterized GlcNAcases, such as VhGlcNAcase and a Vibrio furnissii SR1514 GlcNAcase (Fig. 1; Keyhani and Roseman 1996; Meekrathok and Suginta 2016; Suginta et al. 2010).
Structural analysis
The tertiary structure of JB10Nag was built by the threading approach, with a determined C-score of 0.39 and a TM-score of 0.77. The two values signify a high confidence and a correct topology of the model. Compared with 7 GlcNAcases that have crystal structures determined, the proportion of amino acids used to build α-helices of JB10Nag was the lowest, while the proportion of amino acids used to build random coils was the highest (Table 1). Although large negatively charged surfaces have been widely observed for salt-tolerant enzymes (Madern et al. 2000; Paul et al. 2008; Premkumar et al. 2005; Qin et al. 2014; Wang et al. 2016; Warden et al. 2015; Zhou et al. 2016a, c), Fig. S2 shows that the negatively charged surfaces of JB10Nag are not significantly greater than those of the H. sapiens GlcNAcase (PDB ID: 2GJX; Lemieux et al. 2006).
Expression, purification, and identification of rJB10Nag
The recombinant GlcNAcase was successfully expressed in E. coli BL21 (DE3) and purified by Ni2+-NTA affinity chromatography as determined by an SDS–PAGE gel showing a single band of approximately 70 kDa (Fig. S3). In addition, a MALDI-TOF MS spectrum of the band indicates that the purified enzyme is indeed rJB10Nag (Fig. S4).
Biochemical characterization
At pH 6.0 and 50 °C, the purified rJB10Nag exhibited specific activities of 538.8, 5.2, 35.4, and 26.7 μmol min−1 mg−1 toward pNPGlcNAc, pNPGalNAc, GlcNAc2, and GlcNAc4 substrates, respectively. However, no rJB10Nag activity was detected when the substrate was pNPGlc, pNPGal, pNPXyl, pNPAra, colloidal chitin, chitosan, peptidoglycan, or muropeptides. Thus, rJB10Nag is a GlcNAcase that can hydrolyze N-acetyl chitooligosaccharides, but cannot participate in bacterial cell wall turnover or recycling.
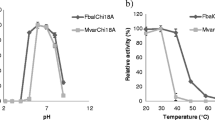
The apparent maximal activity of purified rJB10Nag occurred at pH 6.0 (Fig. 2a). Over 55% of its maximum activity was retained at pH 5.5–6.5 (Fig. 2a). The enzyme was stable across a pH range of 6–9, with more than 85% of its initial activity retained after incubating the enzyme in these buffers for 6 h (Fig. 2b).
Purified rJB10Nag was most active at 50 °C (Fig. 2c). Over 45% of its maximum activity was retained at 30–55 °C (Fig. 2c). The enzyme was stable at 30 °C, and the half-lives of the enzyme were approximately 30 and 2 min at 37 and 50 °C, respectively (Fig. 2d).
The activity of the purified rJB10Nag was completely inhibited by HgCl2, AgNO3, and SDS (Table 2). The addition of 10.0 mM CuSO4 and 1.0% (w/v) Tween 80 partially inhibited rJB10Nag, as it retained activities of 61.5 and 65.1%, respectively. No obvious effect was detected on the enzyme activity by the addition of other metal ions and chemical reagents (retaining 76.6–109.1% activity; Table 2). Furthermore, the enzyme exhibited good NaCl and KCl tolerance. The rJB10Nag enzyme remained 66.9% active in the presence of 15.0% (w/v) NaCl (Fig. 3a) and retained 99.5% of its initial activity after incubation with 15.0% (w/v) NaCl for 60 min (Fig. 3b). In 3.0–30.0% (w/v) KCl, both the activity and stability of rJB10Nag was slightly or not affected (Fig. 3c, d).
The rJB10Nag enzyme showed strong GlcNAc tolerance. The enzyme remained approximately 50% activity in the presence of 70.0 mM GlcNAc (Fig. 4).
The K m, V max, and k cat values of purified rJB10Nag toward pNPGlcNAc were 1.1 ± 0.1 mM, 1030.0 ± 82.1 µmol min−1 mg−1, and 1251.3 ± 99.8 s−1, respectively.
Hydrolytic property of purified rJB10Nag
As shown in Fig. 5, GlcNAc was the sole end-product from the hydrolysis of GlcNAc2 and GlcNAc4 by purified rJB10Nag.
Synergistic action
Although purified rJB10Nag cannot hydrolyze colloidal chitin alone, the simultaneous addition of the commercial chitinase CtnSg and rJB10Nag improved the degradation of colloidal chitin by 2.35-fold (Table 3). In addition, the sequential addition of CtnSg and rJB10Nag also improved the degradation of colloidal chitin by 1.57- to 1.88-fold (Table 3).
Discussion
Bacteria of the genus Shinella are poorly understood, with only 12 references related to Shinella being present in the Web of Science database as of Jun 1, 2016. In these references, the isolation and classification of Shinella strains have mostly been reported, such as the novel species S. yambaruensis NBRC 102122 from soil (Matsui et al. 2009). In recent years, studies on Shinella have explored their utilization for the biodegradation of environmental contaminants, such as nicotine and 4-aminobenzenesulfonate. However, to our knowledge, enzymes from Shinella strains have not been functionally characterized, except for a (S)-6-hydroxynicotine oxidase from Shinella sp. HZN7 (Qiu et al. 2014). Draft genomes of three Shinella strains have been recently sequenced and are publicly available in GenBank (Accession Nos. LGYF00000000, LGYG00000000, and AYLZ00000000). Genes encoding putative GH 20 GlcNAcases from Shinella strains have been released, including the GlcNAcases from Shinella sp. SUS2 (KNY15668), Shinella sp. GWS1 (KOC76078), and Shinella sp. DD12 (EYR78252). These Shinella GlcNAcases have been annotated based on in silico analyses without biochemical characterization. To our knowledge, this study represents the first public report on the biochemical characteristics of a Shinella GlcNAcase. The GlcNAcase has less than 30% identity with experimentally characterized GlcNAcases, suggesting that the GlcNAcase is a novel member of GH 20 GlcNAcases and may have unique enzymatic properties.
The biochemical characterization of rJB10Nag demonstrated its tolerance to salts, including NaCl, KCl, MnSO4, CuSO4, NiSO4, and FeCl3. Salt-tolerant GlcNAcases have not been widely described. To date, all available research concerning the effect of salts on GlcNAcases report that GH 20 GlcNAcases are not tolerant to many salts. For example, two GlcNAcases from Paenibacillus are strongly inhibited by 5 mM Cu2+ or Ni2+ (Sumida et al. 2009); the GlcNAcase from Aeromonas caviae is strongly inhibited by 1 mM Cu2+ or Mn2+ (Lin et al. 2006); the GlcNAcase from Lactobacillus casei is strongly inhibited by Cu2+ (Senba et al. 2000); and the GlcNAcase from Trichoderma reesei is inhibited by 1 mM Fe3+ (Chen et al. 2015). A variety of salts are extensively present or utilized in many industries (Margesin and Schinner 2001; Warden et al. 2015). The presence of salts is also helpful to retard the growth of microorganisms. The salt tolerance of rJB10Nag GlcNAcase will benefit potential applications requiring high salt concentrations.
As biocatalysts, highly active enzymes often attract a great deal of industrial attention. To date, most characterized GlcNAcases, including GH 3, 20, and 84 members, show specific activities or V max values of lower than 200 μmol min−1 mg−1 toward pNPGlcNAc (Bruce and Gounaris 2006; Chitlaru and Roseman 1996; Choi et al. 2009; da Silva Junior Sobrinho et al. 2005; Keyhani and Roseman 1996; Kubota et al. 2004; Lan et al. 2004; O’Connell et al. 2008; Ogawa et al. 2006; Ohishi et al. 1999; Park et al. 2011; Riekenberg et al. 2004; Senba et al. 2000; Slamova et al. 2014; Tomiya et al. 2006; Tsujibo et al. 1998; Yang et al. 2014; Zhou et al. 2016c). In this study, rJB10Nag exhibited a specificity activity of 538.8 μmol min−1 mg−1 and a V max of 1030.0 ± 82.1 µmol min−1 mg−1 toward pNPGlcNAc. Furthermore, rJB10Nag showed a specificity activity of 35.4 μmol min−1 mg−1 toward GlcNAc2, which is higher than the values of GlcNAcases from Trichinella spiralis (0.79 nmol min−1 mg−1; Bruce and Gounaris 2006), Rhizomucor miehei (1.06 μmol min−1 mg−1; Yang et al. 2014), Sphingobacterium sp. (0.3 μmol min−1 mg−1; Zhou et al. 2016c), and Vibrio alginolyticus (18.1 μmol min−1 mg−1; Ohishi et al. 1999).
Because GlcNAc can inhibit GlcNAcases, resulting in the accumulation of N-acetyl chitooligosaccharides that can inhibit chitinases, the tolerance of GlcNAcases to GlcNAc is an attractive feature for the bioconversion of chitin waste (Yang et al. 2014). In previous studies, 1.8 and 0.21 mM GlcNAc inhibited approximately 50% of the activities of GlcNAcases from Spodoptera frugiperda (Tomiya et al. 2006) and Trichoderma harzianum (Nieder et al. 2004), respectively. The GlcNAcase from R. miehei is a GlcNAc-tolerant GlcNAcase as approximately 50% of activity was retained in the presence of 9 mM GlcNAc (Yang et al. 2014). We determined the GlcNAc tolerance of the GH 3 GlcNAcase from Sphingobacterium sp. that we previously reported (Zhou et al. 2016c), and the result shows that the GlcNAcase also retained approximately 50% active in the presence of 9 mM GlcNAc. In this study, rJB10Nag was inhibited by approximately 50% in the presence of as high as 70.0 mM GlcNAc. To our knowledge, this value represents the highest tolerance to GlcNAc among GlcNAcases reported to date.
The statistical analysis of amino acid residues showed that JB10Nag presented the highest proportions of A, G, and random coils and the lowest proportion of α-helices than its homologs shown in Tables S1 and 1. In previous studies, salt-tolerant exo-inulinases also exhibit a higher proportion of A and G than their homologs (Shen et al. 2015; Zhou et al. 2015a) and proteins from halophiles exhibit a higher proportion of random coils, but a lower proportion of α-helices than that of non-halophiles (Paul et al. 2008). High proportions of A, G, and random coil content at the expense of α-helices probably make protein more flexible (Paul et al. 2008). There is convincing evidence that a flexible structure can enhance enzyme catalysis (Hong and Yoo 2013; Hong et al. 2014), because it is conducive to the formation of the transition state (Bhabha et al. 2011). It is worth investigating whether these molecular characteristics affect the salt tolerance and high activity of the GH 20 GlcNAcase JB10Nag in future studies.
In conclusion, a GH 20 GlcNAcase was obtained from a new isolate, Shinella sp. JB10. The enzyme showed novel biochemical characteristics, including salt tolerance, higher specificity activity or V max values than many GlcNAcases, and the highest GlcNAc tolerance of any GlcNAcase reported to date. These characteristics make the enzyme suitable for many potential applications, such as processing marine materials and the bioconversion of chitin waste. The enzyme also showed the highest proportions of A, G, and random coils and the lowest proportion of α-helices among experimentally characterized GH 20 GlcNAcases.
References
Bhabha G, Lee J, Ekiert DC, Gam J, Wilson IA, Dyson HJ, Benkovic SJ, Wright PE (2011) A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis. Science 332:234–238
Bruce AF, Gounaris K (2006) Characterisation of a secreted N-acetyl-β-hexosaminidase from Trichinella spiralis. Mol Biochem Parasit 145:84–93
Chen F, Chen XZ, Qin LN, Tao Y, Dong ZY (2015) Characterization and homologous overexpression of an N-acetylglucosaminidase Nag1 from Trichoderma reesei. Biochem Bioph Res Co 459:184–188
Chitlaru E, Roseman S (1996) Molecular cloning and characterization of a novel β-N-acetyl-d-glucosaminidase from Vibrio furnissii. J Biol Chem 271:33433–33439
Choi KH, Seo JY, Park KM, Park CS, Cha J (2009) Characterization of glycosyl hydrolase family 3 β-N-acetylglucosaminidases from Thermotoga maritima and Thermotoga neapolitana. J Biosci Bioeng 108:455–459
da Silva Junior Sobrinho I, Bataus LAM, Maitan VR, Ulhoa CJ (2005) Purification and properties of an N-acetylglucosaminidase from Streptomyces cerradoensis. Biotechnol Lett 27:1273–1276
Hong SY, Yoo YJ (2013) Activity enhancement of Candida antarctica lipase B by flexibility modulation in helix region surrounding the active site. Appl Biochem Biotech 170:925–933
Hong SY, Park HJ, Yoo YJ (2014) Flexibility analysis of activity-enhanced mutants of bacteriophage T4 lysozyme. J Mol Catal B-Enzym 106:95–99
Inokuma K, Hasunuma T, Kondo A (2016) Ethanol production from N-acetyl-d-glucosamine by Scheffersomyces stipitis strains. AMB Express 6:83
Keyhani NO, Roseman S (1996) The chitin catabolic cascade in the marine bacterium Vibrio furnissii—molecular cloning, isolation, and characterization of a periplasmic β-N-acetylglucosaminidase. J Biol Chem 271:33425–33432
Kubota T, Miyamoto K, Yasuda M, Inamori Y, Tsujibo H (2004) Molecular characterization of an intracellular β-N-acetylglucosaminidase involved in the chitin degradation system of Streptomyces thermoviolaceus OPC-520. Biosci Biotech Bioch 68:1306–1314
Lan XQ, Ozawa N, Nishiwaki N, Kodaira R, Okazaki M, Shimosaka M (2004) Purification, cloning, and sequence analysis of β-N-acetylglucosaminidase from the chitinolytic bacterium Aeromonas hydrophila strain SUWA-9. Biosci Biotech Bioch 68:1082–1090
Lemieux MJ, Mark BL, Cherney MM, Withers SG, Mahuran DJ, James MNG (2006) Crystallographic structure of human β-hexosaminidase A: interpretation of Tay-Sachs mutations and loss of G(M2) ganglioside hydrolysis. J Mol Biol 359:913–929
Lin H, Xiao X, Zeng X, Wang FP (2006) Expression, characterization and mutagenesis of the gene encoding β-N-acetylglucosaminidase from Aeromonas caviae CB101. Enzyme Microb Tech 38:765–771
Liu TA, Zhang HT, Liu FY, Wu QY, Shen X, Yang Q (2011) Structural determinants of an insect β-N-acetyl-d-hexosaminidase specialized as a chitinolytic enzyme. J Biol Chem 286:4049–4058
Madern D, Ebel C, Zaccai G (2000) Halophilic adaptation of enzymes. Extremophiles 4:91–98
Margesin R, Schinner F (2001) Potential of halotolerant and halophilic microorganisms for biotechnology. Extremophiles 5:73–83
Mark BL, Wasney GA, Salo TJS, Khan AR, Cao ZM, Robbins PW, James MNG, Triggs-Raine BL (1998) Structural and functional characterization of Streptomyces plicatus β-N-acetylhexosaminidase by comparative molecular modeling and site-directed mutagenesis. J Biol Chem 273:19618–19624
Matsui T, Shinzato N, Tamaki H, Muramatsu M, Hanada S (2009) Shinella yambaruensis sp. nov., a 3-methyl-sulfolane-assimilating bacterium isolated from soil. Int J Syst Evol Micr 59:536–539
Meekrathok P, Suginta W (2016) Probing the catalytic mechanism of Vibrio harveyi GH20 β-N-acetylglucosaminidase by chemical rescue. PLoS One 11:e0149228
Molina-Guijarro JM, Perez J, Munoz-Dorado J, Guillen F, Moya R, Hernandez M, Arias ME (2009) Detoxification of azo dyes by a novel pH-versatile, salt-resistant laccase from Streptomyces ipomoea. Int Microbiol 12:13–21
Nieder V, Kutzer M, Kren V, Gallego RG, Kamerling JP, Elling L (2004) Screening and characterization of β-N-acetylhexosaminidases for the synthesis of nucleotide-activated disaccharides. Enzyme Microb Tech 34:407–414
O’Connell E, Murray P, Piggott C, Hennequart F, Tuohy M (2008) Purification and characterization of a N-acetylglucosaminidase produced by Talaromyces emersonii during growth on algal fucoidan. J Appl Phycol 20:557–565
Ogawa M, Kitagawa M, Tanaka H, Ueda K, Watsuji T, Beppu T, Kondo A, Kawachi R, Oku T, Nishio T (2006) A β-N-acetylhexosaminidase from Symbiobacterium thermophilum; gene cloning, overexpression, purification and characterization. Enzyme Microb Tech 38:457–464
Ohishi K, Murase K, Etoh H (1999) Purification and properties of β-N-acetylglucosaminidase from Vibrio alginolyticus H-8. J Biosci Bioeng 88:98–99
Park JK, Kim WJ, Park YI (2011) Purification and characterization of an exo-type β-N-acetylglucosaminidase from Pseudomonas fluorescens JK-0412. J Appl Microbiol 110:277–286
Patil RS, Ghormade V, Deshpande MV (2000) Chitinolytic enzymes: an exploration. Enzyme Microb Tech 26:473–483
Paul S, Bag SK, Das S, Harvill ET, Dutta C (2008) Molecular signature of hypersaline adaptation: insights from genome and proteome composition of halophilic prokaryotes. Genome Biol 9:R70
Premkumar L, Greenblatt HM, Bageshwar UK, Savchenko T, Gokhman I, Sussman JL, Zamir A (2005) Three-dimensional structure of a halotolerant algal carbonic anhydrase predicts halotolerance of a mammalian homolog. P Natl Acad Sci USA 102:7493–7498
Qin YJ, Huang ZQ, Liu ZD (2014) A novel cold-active and salt-tolerant α-amylase from marine bacterium Zunongwangia profunda: molecular cloning, heterologous expression and biochemical characterization. Extremophiles 18:271–281
Qiu JG, Wei Y, Ma Y, Wen RT, Wen YZ, Liu WP (2014) A novel (S)-6-hydroxynicotine oxidase gene from Shinella sp. strain HZN7. Appl Environ Microb 80:5552–5560
Riekenberg S, Flockenhaus B, Vahrmann A, Muller MCM, Leippe M, Kiess M, Scholze H (2004) The β-N-acetylhexosaminidase of Entamoeba histolytica is composed of two homologous chains and has been localized to cytoplasmic granules. Mol Biochem Parasit 138:217–225
Senba M, Kashige N, Nakashima K, Miake F, Watanabe K (2000) Cloning of the gene of β-N-acetylglucosaminidase from Lactobacillus casei ATCC 27092 and characterization of the enzyme expressed in Escherichia coli. Biol Pharm Bull 23:527–531
Shen JD, Zhang R, Li JJ, Tang XH, Li RX, Wang M, Huang ZX, Zhou JP (2015) Characterization of an exo-inulinase from Arthrobacter: a novel NaCl-tolerant exo-inulinase with high molecular mass. Bioengineered 6:99–105
Slamova K, Kulik N, Fiala M, Krejzova-Hofmeisterova J, Ettrich R, Kren V (2014) Expression, characterization and homology modeling of a novel eukaryotic GH84 β-N-acetylglucosaminidase from Penicillium chrysogenum. Protein Expres Purif 95:204–210
Suginta W, Chuenark D, Mizuhara M, Fukamizo T (2010) Novel β-N-acetylglucosaminidases from Vibrio harveyi 650: cloning, expression, enzymatic properties, and subsite identification. BMC Biochem 11:40
Sumida T, Ishii R, Yanagisawa T, Yokoyama S, Ito M (2009) Molecular cloning and crystal structural analysis of a novel β-N-acetylhexosaminidase from Paenibacillus sp. TS12 capable of degrading glycosphingolipids. J Mol Biol 392:87–99
Tews I, Perrakis A, Oppenheim A, Dauter Z, Wilson KS, Vorgias CE (1996) Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease. Nat Struct Mol Biol 3:638–648
Tomiya N, Narang S, Park J, Abdul-Rahman B, Choi O, Singh S, Hiratake J, Sakata K, Betenbaugh MJ, Palter KB, Lee YC (2006) Purification, characterization, and cloning of a Spodoptera frugiperda Sf9 β-N-acetylhexosaminidase that hydrolyzes terminal N-acetylglucosamine on the N-glycan core. J Biol Chem 281:19545–19560
Tsujibo H, Hatano N, Mikami T, Hirasawa A, Miyamoto K, Inamori Y (1998) A novel β-N-acetylglucosaminidase from Streptomyces thermoviolaceus OPC-520: gene cloning, expression, and assignment to family 3 of the glycosyl hydrolases. Appl Environ Microb 64:2920–2924
Wang GZ, Wang QH, Lin XJ, Ng TB, Yan RX, Lin J, Ye XY (2016) A novel cold-adapted and highly salt-tolerant esterase from Alkalibacterium sp. SL3 from the sediment of a soda lake. Sci Rep-UK 6:19494
Warden AC, Williams M, Peat TS, Seabrook SA, Newman J, Dojchinov G, Haritos VS (2015) Rational engineering of a mesohalophilic carbonic anhydrase to an extreme halotolerant biocatalyst. Nat Commun 6:10278
Yang SQ, Song S, Yan QJ, Fu X, Jiang ZQ, Yang XB (2014) Biochemical characterization of the first fungal glycoside hydrolyase family 3 β-N-acetylglucosaminidase from Rhizomucor miehei. J Agr Food Chem 62:5181–5190
Zhou JP, Zhang R, Gao YJ, Li JJ, Tang XH, Mu YL, Wang F, Li C, Dong YY, Huang ZX (2012) Novel low-temperature-active, salt-tolerant and proteases-resistant endo-1,4-β-mannanase from a new Sphingomonas strain. J Biosci Bioeng 113:568–574
Zhou JP, Peng MZ, Zhang R, Li JJ, Tang XH, Xu B, Ding JM, Gao YJ, Ren JR, Huang ZX (2015a) Characterization of Sphingomonas sp. JB13 exo-inulinase: a novel detergent-, salt-, and protease-tolerant exo-inulinase. Extremophiles 19:383–393
Zhou JP, Shen JD, Zhang R, Tang XH, Li JJ, Xu B, Ding JM, Gao YJ, Xu DY, Huang ZX (2015b) Molecular and biochemical characterization of a novel multidomain xylanase from Arthrobacter sp. GN16 isolated from the feces of Grus nigricollis. Appl Biochem Biotech 175:573–588
Zhou JP, Liu Y, Lu Q, Zhang R, Wu Q, Li CY, Li JJ, Tang XH, Xu B, Ding JM, Han NY, Huang ZX (2016a) Characterization of a glycoside hydrolase family 27 α-galactosidase from Pontibacter reveals its novel salt–protease tolerance and transglycosylation activity. J Agr Food Chem 64:2315–2324
Zhou JP, Lu Q, Zhang R, Wang YY, Wu Q, Li JJ, Tang XH, Xu B, Ding JM, Huang ZX (2016b) Characterization of two glycoside hydrolase family 36 α-galactosidases: novel transglycosylation activity, lead–zinc tolerance, alkaline and multiple pH optima, and low-temperature activity. Food Chem 194:156–166
Zhou JP, Song ZF, Zhang R, Ding LM, Wu Q, Li JJ, Tang XH, Xu B, Ding JM, Han NY, Huang ZX (2016c) Characterization of a NaCl-tolerant β-N-acetylglucosaminidase from Sphingobacterium sp. HWLB1. Extremophiles 20:547–557
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant Nos. 31260215, 31660445), the Yunling Scholars (Grant No. 2015 56), the Yunling Industry Leading Talents (Grant No. 2014 1782), the Reserve Talents Project for Young and Middle-Aged Academic and Technical Leaders of Yunnan Province (Grant No. 2015HB033), and the Applied and Basic Research Foundation of Yunnan Province (Grant No. 201401PC00224).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Communicated by A. Driessen.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhou, J., Song, Z., Zhang, R. et al. A Shinella β-N-acetylglucosaminidase of glycoside hydrolase family 20 displays novel biochemical and molecular characteristics. Extremophiles 21, 699–709 (2017). https://doi.org/10.1007/s00792-017-0935-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-017-0935-1