Abstract
Thirty-five isolates of actinomycetes were characterized for their antagonistic potential against phyto-pathogens of chickpea by dual-culture and metabolite production assays. The seven most promising isolates of Streptomyces were evaluated for their physiological and plant growth-promoting traits under in vitro and in vivo conditions. All the seven isolates exhibited good growth at temperatures between 20 and 40 °C, pH between 7 and 11 and saline concentrations up to 4%; all the isolates were highly tolerant to fungicide Bavistin, three isolates were moderately tolerant to Captan and all the isolates were susceptible to Thiram, Benlate and Ridomil. All the seven isolates of Streptomyces produced siderophore, chitinase (except isolate CAI-133), cellulase, lipase, protease (except isolates BCA-689 and CAI-133), hydrocyanic acid (except isolate CAI-133), indole acetic acid and β-1,3-glucanase. The greenhouse studies revealed that the isolates of Streptomyces enhanced the plant growth by promoting root length and weight, nodule numbers, shoot weight, pod numbers and pod weight over the un-inoculated control. Under field conditions, the Streptomyces treated plots increased the nodule numbers, root weight, stover yield and grain yield over the un-inoculated control plots. In the rhizosphere, the Streptomyces were also found to enhance the total nitrogen, available phosphorus and organic carbon compared to un-inoculated control. The colonizing capability of the Streptomyces on the roots of chickpea was confirmed by scanning electron microscopic analysis. All the isolates were identified as Streptomyces species by 16S rDNA analysis; five of the seven isolates clustered in one clade, whereas the other two belonged to two different clades in phylogenetic analysis.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Chickpea (Cicer arietinum L.) is the third most important leguminous crop after bean and soybean. India is the largest chickpea-producing country accounting for 64% of the global chickpea production [45]. Chickpea is important because of its high protein content and thus widely used in human and animal diet. It is also used as fodder and green manures [25]. The production of chickpea is affected by many biotic and abiotic factors. The biotic factors include insect pests, fungi, bacteria, viruses, nematodes and mycoplasma which result in severe economic loss. Among the fungal pathogens, Fusarium wilt, dry root rot, collar rot, Ascochyta blight and botrytis grey mould (BGM) are important [1, 40]. Of the various management strategies, the biological control is recognized as the best alternative to chemical pesticides because of their level of safety and minimal negative effect on environment.
Plant-associated bacteria are classified into beneficial, deleterious and neutral groups based on their effects on plant growth [5]. The beneficial organisms are referred as plant growth-promoting rhizobacteria (PGPR). Rhizosphere soil and vermicompost are good sources of PGPR. PGPR enhance plant growth directly by producing plant growth-promoting substances synthesized by the organism or by enhancing the uptake of nutrients or indirectly by exerting antagonistic activity against the fungal pathogens [5]. PGPR strains are also reported to enhance plant growth when they are inoculated into the seed [36]. PGPR are used as alternatives to chemical fertilizers and pesticides. Several PGPR strains, such as Bacillus, Pseudomonas, Rhizobium and actinomycetes, have been reported to control various soil-borne plant pathogenic fungi under greenhouse and field conditions [16, 27, 42].
Actinomycetes, a group of gram-positive bacteria with a higher G + C content (55–75%), are found commonly in rhizosphere soil. The term actinomycete was derived from the Greek words aktis (a ray) and mykes (fungus). Actinomycetes are known to produce antibiotics, that are effective against fungal plant pathogens, and possess plant growth-promoting traits [7, 11, 39]. The primary objective of the present investigation was to isolate and characterize actinomycetes, with broad-spectrum antagonistic activity against important fungal pathogens of chickpea, for their PGP potentials under greenhouse and field conditions.
Materials and Methods
Isolation of Actinomycetes
Ten grams of rhizosphere soils of chickpea were suspended in 90 ml of physiological saline (0.85% of NaCl) in a bottle and placed on an orbital shaker (at 100 rpm) at 28 ± 2 °C for 1 h. After shaking, the samples were serially diluted up to 107 dilutions with physiological saline, and the dilutions 104–106 were plated on actinomycetes isolation agar (AIA) by spread plate technique. The plates were incubated at 28 ± 2 °C for 4 days. The most prominent colonies (the ones that were found abundantly in the plate, produced pigments and inhibited the adjacent colonies) were isolated and maintained on AIA slants at 4 °C.
Screening of Actinomycetes for Antagonistic Properties
The actinomycete isolates were evaluated for antifungal activity against pathogens of chickpea, including Fusarium oxysporum f. sp. ciceri (FOC; causes Fusarium wilt), Rhizoctonia bataticola (causes dry root rot; three strains RB-6, RB-24 and RB-115) and Botrytis cinerea (causes BGM). The pathogens were acquired from Legumes Pathology Division, ICRISAT, Patancheru, India. The antagonistic activity of actinomycete isolates was evaluated by dual-culture assay as per the standard protocol [3, 11].The culture filtrates of the promising isolates were extracted by partitioning against ethyl acetate (EtOAc), and the resultant organic and aqueous fractions were evaluated against the three fungal pathogens. To achieve this, a fungal disc with a 6 mm diameter was bored and placed in the centre of the quarter-strength potato dextrose agar (PDA) plate supplemented with 10% of either organic or aqueous fractions. The control plates contained no samples. The plates were incubated at 28 ± 2 °C for 5 days, and fungal growth was recorded on a scale of 0–3 as follows: 0 = no inhibition; 1 = slight inhibition; 2 = moderate inhibition; and 3 = good inhibition.
Evaluation of Actinomycetes for Physiological Traits and Fungicide Tolerance
The physiological properties of the potential actinomycetes were studied which include pH, temperature and salinity tolerance. For this, the isolates were streaked on Bennet’s agar, adjusted to different pH (5, 7, 9 and 11) and saline concentrations (0–12% at the interval of 2%) and incubated at 28 °C for 5 days. For the temperature analysis, the Bennet’s agar plates were streaked with the actinomycetes and incubated at different temperatures (20, 30 and 40 °C) for 5 days, whereas for incubation at 50 °C, the isolates were inoculated in Bennet’s broth. The fungicide tolerance of the actinomycetes was evaluated at field application levels as per the standard protocols [13]. The actinomycetes were streaked on AIA plates supplemented with fungicides like Bavistin, Thiram, Benlate, Captan, and Ridomil and at field application levels of 2500, 3000, 4000, 3000 and 3000 ppm, respectively. The plates were incubated at 28 °C for 5 days. At the end of incubation, the growth of actinomycetes was recorded on a scale of 0–3 as follows: 0 = no growth; 1 = slight growth; 2 = moderate growth; and 3 = good growth.
In Vitro Evaluation for PGP and Biocontrol Traits
The actinomycetes were evaluated for their PGP and biocontrol traits, including production of siderophore [38], chitinase [18], cellulase [17], lipase, protease [6], hydrocyanic acid (HCN) [22], β-1,3-glucanase [11] and indole acetic acid (IAA) [33] using standard protocols. The rating scales for siderophore, chitinase, cellulase, lipase and protease production are given as follows: 0 = no halo zone; 1 = halo zone of <1 mm; 2 = halo zone of 2–3 mm; 3 = halo zone of 4–6 mm, 4 = halo zone of 7–9 mm; and 5 = halo zone of ≥10 mm. For HCN production, the following rating scale was used: 0 = no colour change; 1 = light reddish brown; 2 = medium reddish brown; and 3 = dark reddish brown.
Evaluation for PGP Traits Under Greenhouse Conditions
The seven promising isolates were evaluated for PGP traits on chickpea under greenhouse conditions. A total of eight treatments including seven isolates and one control (without actinomycetes) with three replications were maintained. A pot mixture comprising of sterilized black soil, sand and farm yard manure (3:2:1) was prepared in plastic pots (8″). Chickpea seeds (ICCV 2) were surface sterilized with 3% chlorax for 5 min, rinsed 8–10 times with sterilized water and incubated with the respective actinomycetes (107 cfu ml−1; grown in starch casein broth {SCB}) for 1 h before sowing. Three seeds were sown on each pot but thinned to one after germination. Booster doses of actinomycetes (5 ml per seedling, 107 cfu ml−1) were applied on 15, 30 and 45 days after sowing (DAS) by the soil drench method. Forty-five days after sowing, observations including the root length, root volume, leaf area, leaf weight, root weight, shoot weight and nodule weight were noted, whereas at final harvest, the shoot weight, root weight, pod numbers and pod weight were recorded.
Evaluation for PGP Traits Under Field Conditions
Field trials were performed in the 2012 Rabi (post-rainy) season at ICRISAT, Patancheru (17°30′N; 78°16′E; altitude = 549 m) in the Telangana State of India. The experimental field soil was composed of 53% clay, 21% silt and 25% sand with an organic carbon content of 0.4–0.6% and an alkaline pH of 7.5–8.2. The mineral composition of the top soil (0–15 cm) was found to contain 23.7 mg kg−1 available N, 8.9 mg kg−1 available P and 291 mg kg−1 available K. The soil depth of the experimental site used was at least 1.2 m, and this soil retained 210 mm of the plant available water. The plot sizes of 4 × 3 m ridges in a randomized complete block design (RCBD) were prepared, and three replications per treatment were maintained.
The selected isolates were grown in SCB for 5 days, soaked with chickpea seeds (variety ICCV-2) for 1 h and sown by hand. A booster dose of actinomycetes (108 CFU ml−1) was applied to soil at an interval of 15 days until flowering. The control plots contained no actinomycetes. Irrigation was performed on 0, 25 and 50 DAS. Weeding was performed as and when required. No incidence of insect–pest or phytopathogen attack was observed during the cropping period. At 60 DAS, parameters such as nodule number, leaf area, leaf weight, pod number, shoot weight and root weight were recorded. At final harvest, parameters including seed weight, seed number, stover yield, grain yield and total dry matter were recorded. Soil samples (from the 0–15-cm soil profile) were collected at flowering (60 DAS) and harvesting and analysed for organic carbon, available P and total N using the standardized protocols [28,29,30].
Molecular Identification
The selected actinomycetes were identified by 16S rDNA analysis. Pure cultures of actinomycetes were grown in SCB until log phase (4 days), and the genomic DNA was isolated [4]. Amplification of the 16S rDNA gene was performed using the universal primers 1492R (5′-TAC GGY TAC CTT GTTACG ACT T-3′) and 27F (5′-AGA GTT TGA TCM TGG CTC AG-3′) as per the standard conditions described [32]. The PCR product was sequenced at Macrogen Inc. (Seoul, Korea). The sequences obtained were compared with those from GenBank using the BLAST program [2] and aligned with the Clustal W software [44], and phylogenetic trees were inferred by neighbour-joining method [37]. Bootstrap analysis using the MEGA version 4 software was performed to estimate the statistical stability of the branches in the cluster with 1000 replicates [43].
Colonization Studies in Chickpea Root by Actinomycetes
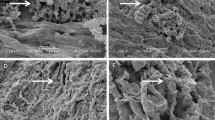
The colonization of the actinomycete isolates on the roots of chickpea was demonstrated through scanning electron microscopy (SEM) analysis. To achieve this, the chickpea seeds (ICCV 2) were surface sterilized with 3% chlorax for 5 min and washed several times with sterilized water. The seeds were then soaked in 70% ethanol in water for 5 min, rinsed with sterilized water and maintained for germination on a Petri dish containing blotter paper for 2 days under dark conditions. The tubes were prepared with coarse sand (50 g) and autoclaved twice at an interval of 24 h at 121 °C for 20 min. The germinated seeds were treated with selected actinomycete isolates (107 cfu ml−1) for 1 h and sowed in the sand tubes. One millilitre of cultures was added to each tube. The tubes were maintained under controlled conditions in a light chamber, where the temperature was maintained at 20 ± 2 °C, and the average illumination and photosynthetic photon flux at the surface of the plant tube were 9600 lx and 350 µE m−2 s −1, respectively, for 15 days. At the end of the incubation period, the plants were removed, and the roots were washed in phosphate buffer (pH 7). The root tips of the plants were cut, fixed in 2.5% glutaraldehyde in 0.1 M phosphate buffer (pH 7.2) for 24 h at 4 °C and post-fixed in 2% aqueous osmium tetroxide for 4 h. The root samples were dehydrated with a series of graded alcohols and dried to a critical point with a CPD unit. The processed samples were mounted over the stubs with double-sided carbon conductivity tape, and the samples were coated with a thin layer of gold using an automated sputter coater (Model:JEOL JFC-1600) for 3 min and scanned under SEM (Model: JOEL-JSM 5600) at the required magnifications using standard procedures at RUSKA Lab, College of Veterinary Science, SVVU, Rajendranagar, Hyderabad, India.
Statistical Analysis
The data were analysed through analysis of variance (ANOVA) with the SAS GLM (General Linear Model) procedure (SAS Institute 2002-08, SAS version 9.3) considering the isolates and replications as fixed variables in a randomized complete block design. Depth-wise ANOVA was performed for the traits root length, volume and dry mass. The isolate means were tested for significance and compared using Fisher’s protected least significant difference.
Results
A total of 35 actinomycetes, the most prominent isolates (the ones which were found abundantly, produced pigments and inhibited the adjacent colonies) in the AIA plate, were isolated from the rhizosphere soils of chickpea and further screened for their antagonistic potential against FOC, R. bataticola and B. ciceri by in vitro dual-culture and metabolite production assays. Of the 35, seven isolates (BCA-546, BCA-659, BCA-667, BCA-689, BCA-698, CAI-133 and CAI-8) were found to have broad-spectrum antifungal activity (Table 1).
All the seven isolates grew well at temperatures between 20 and 40 °C (with the exception of CAI-133, which was found to tolerate high-temperature range of 30–50 °C), pH between 7 and 11 and saline conditions of up to 8% (with the exception of BCA-659 and CAI-133, which were found to tolerate only up to 4% while BCA-689 tolerated up to 10%). The selected isolates were also found highly tolerant to Bavistin, moderately tolerant to Captan (except BCA-546, BCA-667, BCA-689 and CAI-133) and susceptible or slightly tolerant to Thiram, Benlate and Ridomil at field application levels (Table 2).
Under in vitro conditions, all the seven actinomycetes were found to produce siderophore, chitinase (except CAI-133), cellulase, lipase, protease (except CAI-133 and BCA-689), HCN (except CAI-133), IAA and β-1,3-glucanase (Table 3).
The seven actinomycetes were identified by 16s rDNA analysis. A neighbour-joining dendrogram was generated using the sequences of the seven actinomycete isolates (1400 bp) and representative sequences from the databases. The phylogenetic analysis of the 16S rDNA sequences of the seven actinomycetes matched with the genus Streptomyces but different species (Fig. 1). The sequences of the seven actinomycetes were submitted to GenBank, NCBI, and the following accession numbers were obtained: BCA-546 (KF770898), BCA-659 (KF770889), BCA-667 (KF770888), BCA-689 (KF770899), BCA-698 (KF770900), CAI-133 (KF770895) and CAI-8 (KF770890).
Under greenhouse conditions, at 45 DAS, all the seven actinomycetes exhibited enhancements in the root length (up to 97%), root volume (up to 115%), leaf area (up to 77%), leaf weight (up to 50%), root dry weight (up to 173%), shoot weight (up to 20%) and nodule number (up to 87%) and at final harvest, the shoot weight (up to 84%), root weight (up to 57%), pod number (up to 102%) and pod weight (up to 84%) over the un-inoculated control. Among the seven actinomycetes, BCA-689, followed by BCA-698 and BCA-667, showed the highest PGP traits (Table 4).
Under field conditions, at 60 DAS, the actinomycete-treated plots increased the nodule number (up to 114%), leaf area (up to 35%), leaf weight (up to 23%), pod number (up to 86%), shoot weight (up to 36%) and root weight (up to 145%) and at final harvest, the 1000 seed weight (up to 9%), seed number (up to 22%), stover yield (up to 86%), grain yield (up to 17%) and total dry matter (up to 51%) over the un-inoculated control plots. Under field conditions, among the seven actinomycetes tested, CAI-8 produced the highest PGP traits while all the other isolates were equally good (Table 5). The soil mineral nutrient contents in the actinomycete-treated plots enhanced total N (up to 27 and 24%), available P (up to 20 and 18%) and organic carbon (up to 5 and 18%) at flowering and final harvest, respectively (Table 6).
The colonization of actinomycete isolates on the roots of chickpea was demonstrated by SEM. When observed under SEM, extensive colonization was observed on the chickpea roots by all the seven isolates. Both mycelial growth and sporulation were observed without damage to the root cells (Fig. 2).
Discussion
In the present study, a total of 35 actinomycete isolates were evaluated for their antagonistic potential against important pathogens of chickpea including FOC, R. bataticola (three strains: RB-6, RB-24 and RB-115) and B. cinerea, which causes wilt, dry root rot and BGM diseases, respectively. Based on the antagonistic activity determined by dual-culture as well as metabolite assays, seven broad-spectrum isolates (BCA-546, BCA-659, BCA-667, BCA-689, BCA-698, CAI-8 and CAI-133) were selected for further studies. Actinomycetes have been reported as biocontrol agents against plant pathogens such as Aspergillus niger, Helminthosporium and Fusarium spp., which causes mould, blight and wilt in many crops [10], Xanthomonas axonopodis, which causes bacterial blight in pomegranate (Punica granatum L.) [34] and Ralstonia solanacearum, which causes bacterial wilt in tobacco [21].
The seven promising isolates were found to have tolerance to wide range of physiological conditions including pH 11, salinity at 10% and temperature at 40 °C, thus making them a better organism of choice for different environmental conditions. When the seven actinomycete isolates were evaluated for their PGP and biocontrol traits, most of them produce siderophore, chitinase, cellulase, lipase, protease, HCN, IAA and β-1,3-glucanase. Among the seven isolates, BCA-698, followed by BCA-667 and CAI-8, produced the maximum levels of IAA, siderophore and β-1,3-glucanase. Actinomycetes are reported to produce growth hormones and increase plant growth [20, 34, 41]. The mechanisms through which actinomycetes promote plant growth include the production of plant growth regulators [9, 26]. IAA is a phytohormone which has a profound influence on plant growth. The production of IAA and its role in plant growth promotion have been widely reported [12, 15]. Siderophores bind Fe3+ from the environment and make it available for plant growth and also compete with pathogens for iron availability. Actinomycetes that produced cellulase, chitinase and β-1, 3-glucanase are reported to have antifungal activity [19]. Chitin, a linear β-1,4-linked polymer of N-acetyl glucosamine, is an important constituent of the fungal cell wall [8]. Many chitinolytic bacteria have been reported to lyse fungal hyphae. Extracellular enzymes, such as chitinase and cellulase, have been reported to control fungal diseases [24]. β-1,3-glucanase degrades the 1,3-glucan layer of many pathogens, thus inhibiting pathogen invasion. HCN is one of the antifungal secondary metabolites produced by many actinomycetes. HCN production plays an important role in disease suppression [12, 46]. In the present investigation, the selected seven actinomycetes showed many of these PGP and biocontrol traits and hence can be exploited for yield enhancement and plant disease control.
In the 16S rDNA analysis, the selected seven actinomycete isolates were identified as Streptomyces but different species. Five of the seven isolates clustered in one clade, whereas the other two belonged to two different clades in phylogenetic analysis. Streptomyces sp. are widely reported to have plant growth-promotion and biological control traits. In the present investigation, when the selected isolates were evaluated for PGP traits under greenhouse and field conditions, all the Streptomyces strains exhibited enhanced PGP traits, including root length, root volume, root dry weight, leaf dry weight, leaf area, shoot weight, nodule number, pod weight, stover yield, grain yield and total dry matter over the un-inoculated control. Under greenhouse conditions, among the seven Streptomyces, BCA-689 followed by BCA-698 and BCA-667 showed the highest PGP traits whereas under field conditions, CAI-8 produced the highest PGP traits while all other isolates were equally good. Streptomyces have been reported widely to promote plant growth in chickpea, rice, tomato and melon [14, 31, 47], but reports of Streptomyces having both PGP and biocontrol traits are rare. The soil mineral nutrient contents in the actinomycete-treated plots significantly enhanced total N, available P and organic carbon contents at both flowering and harvest over the un-inoculated plots. Streptomyces spp. has also been reported to enhance soil mineral contents. Further, extensive colonization of these isolates on the roots of chickpea was also observed under SEM. Streptomyces spp. have been reported to colonize the roots and enhance plant growth [23, 35]. The mechanism by which the actinomycetes enhanced morphological and yield traits of rice could be attributed not only to their direct PGP activities such as siderophore and IAA and/or indirect PGP activities like chitinase, cellulase, lipase, protease, hydrocyanic acid and β-1,3-glucanase production capabilities, but also to their ability to survive under harsh environments.
Conclusion
The present study was successful in identifying seven effective isolates of Streptomyces spp., from the rhizosphere of chickpea, which can be a useful component for integrated disease management and integrated nutrition management. Though, all the seven Streptomyces spp. have been demonstrated for their PGP potential in chickpea CAI-8, BCA-689, BCA-698 and BCA-667 were found to have superiority over other isolates. Hence, these isolates can be exploited as PGP agents in chickpea.
References
Akhtar MS, Siddiqui ZA (2010) Effect of AM fungi on plant growth and root-rot diseases of chickpea. Am Eurasian J Agric Environ Sci 8:544–549
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Anjaiah V, Koedam N, Nowak-Thompson B, Loper JE, Hofte M, Tambong JT, Cornelis P (1998) Involvement of phenazines and anthranilate in the antagonism of Pseudomonas aeruginosa PNA1 and Tn5 derivative toward Fusarium spp. and Pythium spp. Mol Plant Microbe Int 11:847–854
Bazzicalupo M, Fani R (1995) The use of RAPD for generating specific DNA probes for microorganisms. In: Clap JP (ed) Methods in molecular biology, species diagnostic protocols: PCR and other nucleic acid methods. Humana Press Inc., Totowa, pp 112–124
Beneduzi A, Ambrosini A, Passaglia LMP (2012) Plant growth-promoting rhizobacteria (PGPR): their potential as antagonists and biocontrol agents. Genet Mol Biol 35:1044–1051
Bhattacharya A, Chandra S, Barik S (2009) Lipase and protease producing microbes from the environment of sugar beet field. Ind J Agric Biochem 22:26–30
Bressen W (2003) Biological control of maize seed pathogenic fungi by use of actinomycetes. Biocontrol 48:233–240
De Boer W, Gunnewiek PJAK, Lafeber P, Janse JH, Spit BE, Woldendorp JW (1998) Anti-fungal properties of chitinolytic dune soil bacteria. Soil Boil Biochem 30:193–203
El-Tarabily KA (2003) An endophytic chitinase-producing isolate of Actinoplanes missouriensis, with potential for biological control of root rot of lupine caused by Plectosporium tabacinum. Aust J Bot 51:257–266
Evangelista-Martínez Z (2014) Isolation and characterization of soil Streptomyces species as potential biological control agents against fungal plant Pathogens. World J Microbiol Biotechnol 30:1639–1647
Gopalakrishnan S, Keerthi KB, Pagidi H, Vidya MS, Deepthi K, Simi J, Srinivas V, Alekhya G, Rupela OP (2011) Biocontrol of charcoal-rot of sorghum by actinomycetes isolated from herbal vermicompost. Afr J Biotechnol 79:18142–18152
Gopalakrishnan S, Suresh P, Mamta S, Humayun P, Keerthi KB, Sandeep D, Vidya MS, Deepthi K, Rupela OP (2011) Evaluation of actinomycete isolates obtained from herbal vermicompost for the biological control of Fusarium wilt of chickpea. Crop Prot 30:1070–1078
Gopalakrishnan S, Upadhyaya HD, Humayun P, Srinivas V, Sreevidya M, Alekhya G, Vijayabharathi R, Bhimineni RK, Seema M, Abhishek R, Rupela OP (2012) Plant growth-promoting traits of biocontrol potential bacteria isolated from rice rhizosphere. Springer Plus 1:71
Gopalakrishnan S, Srinivas V, Shravya A, Prakash B, Vijayabharathi R, Bhimineni RK, Rupela OP (2013) Evaluation of Streptomyces spp. for their plant-growth-promotion traits in rice. Can J Microbiol 59:534–539
Goudjal Y, Toumatia O, Sabaou N, Barakate M, Mathieu F, Zitouni A (2013) Endophytic actinomycetes from spontaneous plants of Algerian Sahara: indole-3-acetic acid production and tomato plants growth promoting activity. World J Microbiol Biotechnol 29:1821–1829
Hemissi I, Yassine M, Abdi N, Bouraoui M, Sifi B (2011) Effects of some Rhizobium strains on chickpea growth and biological control of Rhizoctonia solani. Afr J Microbiol Res 5:4080–4090
Hendricks CW, Doyle JD, Hugley B (1995) A new solid medium for enumerating cellulose-utilizing bacteria in soil. Appl Environ Microbiol 61:2016–2019
Hsu SC, Lockwood JL (1975) Powdered chitin Agar as a selective medium for enumeration of actinomycetes in water and soil. Appl Microbiol 29:422–426
Juan Z, Xue Q, Niu G, Lei X, Shen G, Jun-zhi D (2013) Extracellular enzyme production and fungal mycelia degradation of antagonistic Streptomyces induced by fungal mycelia preparation of cucurbit plant pathogens. Ann Microbiol 63:809–812
Khamna S, Yokota A, Peberdy JF, Lumyong S (2010) Indole-3-acetic acid production by Streptomyces spp. Isolated from some Thai medicinal plant rhizosphere soils. Eur Asia J BioSci 4:23–32
Liu YX, Shi JX, Feng YG, Yang XM, Li X, Shen QR (2013) Tobacco bacterial wilt can be biologically controlled by the application of antagonistic strains in combination with organic fertilizer. Biol Fert Soils 49:447–464
Lorck H (1948) Production of hydrocyanic acid by bacteria. Plant Physiol 1:142–146
Miller JJ, Liljeroth E, Williamsen-De Klein MJEIM, Veen JAV (1990) The dynamics of actinomycetes and fluorescent pseudomonads in wheat rhizoplane and rhizosphere. Symbiosis 9:389–391
Nagpure A, Choudhary B, Kumar S, Gupta RK (2014) Isolation and characterization of chitinolytic Streptomyces spp. MT7 and its antagonism towards wood-rotting fungi. Ann Microbiol 64:531–541
Namvar A, Sharifi RS, Khandan T (2011) Growth analysis and yield of chickpea (Cicer arietinum L.) in relation to organic and inorganic nitrogen fertilization. Ekologija 57:97–108
Nassar AH, El-Tarabily KA, Sivasithamparam K (2003) Growth promotion of bean (Phaseolus vulgaris L.) by a polyamine-producing isolate of Streptomyces griseoluteus. Plant Growth Regul 40:97–106
Nelson LM (2004) Plant growth-promoting rhizobacteria (PGPR): prospects for new inoculants. Crop Manag Netw. doi:10.1094/Cm-2004-0301-05-RV
Nelson DW, Sommers LE (1982) Total organic carbon and organic matter. In: Page AL, Miller RH, Keeney DR (eds) Methods of soil analysis, Part 3, chemical and microbiological properties. SSSA, Madison, pp 539–579
Novozamsky I, Houba VJG, Van ECKR, VanVark W (1983) A novel digestion technique for multiple element analysis. Commun Soil Sci Plant Anal 14:239–249
Olsen SR, Sommers LE (1982) Phosphorus. In: Page AL (ed) Methods of soil analysis, Agron No 9, Part 2, ‘chemical and microbial properties, 2nd edn. American Society of Agronomy, Madison, pp 403–430
Palaniyandi SA, Damodharan K, Yang SH, Suh JW (2014) Streptomyces sp. strain PGPA39 alleviates salt stress and promotes growth of ‘Micro Tom’ tomato plants. J Appl Microbiol 117:766–773
Pandey P, Kang SC, Maheswari DK (2005) Isolation of endophytic plant growth-promoting Burkholderia spp. MSSP from root nodules of Mimosa pudica. Curr Sci 89:177–180
Patten C, Glick BR (2002) Role of Pseudomonas putida in indole acetic acid in development of host plant root system. Appl Environ Microbiol 68:3795–3801
Poovarasan S, Mohandas S, Paneerselvam P, Saritha B, Ajay KM (2013) Mycorrhizae colonizing actinomycetes promote plant growth and control bacterial blight disease of pomegranate (Punica granatum L. cv Bhagwa). Crop Prot 53:175–181
Ranjeet KT, Janice LS, Carina MJ, Don LC, Michelle HS, Lee AD, Franklin BJ, Morra MJ (2002) Novel plant-microbe rhizosphere interaction involving Streptomyces lydicus WYEC108 and the pea plant (Pisum sativum). Appl Environ Microbiol 68:2161–2171
Saharan BS, Nehra V (2011) Plant growth promoting rhizobacteria: a critical review. Life Sci Med Res 21:1–30
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol 4:406–425
Schwyn B, Neilands JB (1987) Universal chemical assay for the detection and determination of siderophores. Anal Biochem 160:47–56
Shahidi BGH, Fooladi MH, Mahdavi MJ, Shahghasi A (2004) Broad spectrum, a novel antibacterial from Streptomyces spp. Biotechnology 3:126–130
Sharma M, Mangla UN, Krishnamurthy M, Vedez V, Pande S (2010) Drought and dry root rot of chickpea. Paper presented in 5th international food legumes research conference (IFLRCV) and European conference on Grain Legumes (AEP II), pp 26–30
Shrivastava S, Souza SFD, Desai PD (2008) Production of indole-3-acetic acid by immobilized actinomycetes (Kitasatospora spp.) for soil application. Curr Sci 94:1595–1604
Siddiqui ZA (2006) PGPR: prospective biocontrol agents of plant pathogens. In: Siddiqui ZA (ed) PGPR: biocontrol and biofertilization. Springer, Dordrecht, pp 111–142
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Thompson JD, Gibsom TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The clustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Verma JPB, Janardan Y, Kavindra NT, Ashok Kumar B (2013) Effect of indigenous Mesorhizobium spp. and plant growth-promoting rhizobacteria on yields and nutrients uptake of chickpea (Cicer arietinum L.) under sustainable agriculture. Ecol Eng 51:282–286
Wei G, Kloepper JW, Sadik T (1991) Induction of systemic resistance of cucumber to Colletotrichum orbiculare by selected strains of plant growth-promoting rhizobacteria. Phytopathology 81:1508–1512
Zhao J, Xue QH, Shen GH, Xue L, Duan JL, Wang DS (2012) Evaluation of Streptomyces spp. for biocontrol of gummy stem blight (Didymella bryoniae) and growth promotion of Cucumis melo L. Biocontrol Sci Technol 22:23–37
Acknowledgements
We are thankful to DST-INSPIRE for the financial support provided to G. Alekhya for her Ph.D. fellowship. This work has been undertaken as part of the CGIAR Research Program on Grain Legumes. ICRISAT is a member of CGIAR consortium. We also thank the staff of the Biocontrol Unit of ICRISAT, including V Srinivas, M Sreevidya, A Sathya, R Vijayabharathi, B Prakash, PVS Prasad, P Manohar, B Nagappa, D Barath, A Jabbar and S Rohini, for their significant inputs to the laboratory and field studies.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Alekhya, G., Gopalakrishnan, S. Biological Control and Plant Growth-Promotion Traits of Streptomyces Species Under Greenhouse and Field Conditions in Chickpea. Agric Res 6, 410–420 (2017). https://doi.org/10.1007/s40003-017-0278-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40003-017-0278-2