Abstract
Epimorphic regeneration in vertebrates involves the restoration of lost tissue or organs through the formation of a regeneration blastema and occurs through a complex interaction of a number of molecular signaling pathways. Of the many effectors of successful tail regeneration in the lizard Hemidactylus flaviviridis, one crucial pathway is the cyclooxygenase-2 (COX-2) mediated PGE2 signaling pathway. The current study was aimed at understanding whether COX-2 signaling plays any role in the expression of Wnt/β-Catenin signaling components during regenerative outgrowth in H. flaviviridis. Etoricoxib—selective inhibitor of the inducible isoform of COX-2—was administered to lizards orally. We tested the expression of β-Catenin during wound epidermis and blastema stages in the regenerating tail and found a reduction in its expression in response to drug treatment. Further, it was observed that the expression of canonical Wnt ligands was greatly altered due to COX-2 inhibition. Our results provide evidence of a cross-talk between the COX-2 induced PGE2 pathway and Wnt/β-Catenin signaling in the regenerating lizard tail. An understanding of the interaction among various signaling pathways will help elucidate the mechanism underlying epimorphosis in lizards, the only amniotes capable of appendage regeneration.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Regeneration in vertebrates is a phenomenon that has been vastly studied since the mid 18th century and yet our understanding about the process in its entirety seems limited. Among the commonly studied vertebrate models, the Zebrafish and Salamanders are remarkable for their capacity to regenerate a wide variety of tissues. Beyond these, many lizards are also capable of appendage regeneration, albeit limited to the tail. Appendage regeneration in all the above mentioned animals proceeds through the formation of a regeneration blastema, a population of progenitor cells responsible for regenerative outgrowth [1]. Blastema formation requires and follows the formation of a wound epidermis, which is a thick multilayer covering of the amputation wound by migrating epidermal cells [2, 3].
The current study makes use of northern house gecko Hemidactylus flaviviridis, which has been a subject of research for numerous parameters related to tail regeneration [4,5,6,7,8]. A complete wound epidermis in this lizard is formed at 4–5 days post amputation (dpa) and blastema at 7–8 dpa at 37 °C. Each of these stages results from drastic changes in the molecular environment that take effect immediately following amputation. Among the variety of regulatory factors induced during regeneration is cyclooxygenase-2 (COX-2), an inducible isoform of cyclooxygenase enzyme, which catalyses the formation of prostaglandin E2 (PGE2) from arachidonic acid [9]. COX-2 mediated PGE2 is classically known to play crucial roles in pain, fever and inflammation [10]. More recently, its role has been acknowledged in the progression of cancers [11,12,13].
Reports of COX-2 having any role in initiation and progression of appendage regeneration are limited [7, 14,15,16]. Our lab has previously shown that inhibition of COX-2 activity, by the use of NSAIDs specific for the isoform, result in a retardation of wound epidermis formation and in its ability to give rise to a blastema [7]. We hypothesize that the PGE2 pathway is functioning upstream of some developmentally important pathways, thereby ensuring at an early stage that an amputation wound takes its course towards regenerative repair. In the present study, we make use of Etoricoxib, a selective COX-2 inhibitor, to investigate whether COX-2 mediated PGE2 pathway interacts with the Wnt/β-Catenin pathway, which is attributed with numerous functions related to cell proliferation, specification and differentiation.
2 Material and methods
2.1 Animal housing and maintenance
Northern House Geckos Hemidactylus flaviviridis of both the sexes, weighing about 10–12 g were captured from the wild and maintained in wooden cages. All animals were screened for parasitic infections and the healthy ones were used for the experiment. Cockroach nymphs were fed once a day and purified water was provided ad libitum. A temperature range of 36 ± 2 °C was maintained with 12:12 hour light to dark cycles. All experimental protocols were approved by the Institutional Animal Ethics Committee (IAEC) constituted as per the guidelines of Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA), India (IACUC no. 984/07/2014-2).
2.2 Drug administration and experimental design
Animals were randomized and divided into two experimental groups, namely control and treatment, each group having 20 individuals. Treatment group animals were orally administered Etoricoxib (gift by Sun Pharma Advanced Research Centre, Vadodara, India) at 25mg/kg body weight once daily, based upon a dose-range study previously carried out in the lab. The drug was given as a suspension in purified water. Control animals were orally given the equivalent volume of purified water. Lizard tails were amputated a day after commencement of drug dosing by inducing autotomy after the third segment from the base of the tail. Regenerating tail tissue was collected for analysis at two major stages, viz., Wound Epithelium and Blastema, from six animals per group at each stage.
2.3 Liquid chromatography–mass spectrometry
Tail tissue was collected after amputation and a 10% homogenate was prepared in Chloroform:Methanol (2:1 ratio), according to the method of Folch et al. [17]. After centrifugation at 10,000g for 10 min, the supernatant was collected and vaporized in a rotary vaporizer at 50 °C under high vacuum. The solid residues were washed twice with Acetonitrile, which was subsequently vaporized under the same conditions. Finally, the solid residue was dissolved in Acetonitrile, and filtered through a Nitrocellulose membrane filter of pore size 0.2µm. This solution was used further for LC/MS analysis. Separation was carried out on an Eksigent Ekspert UltraLC 100 machine coupled with an ABSciex 3200 Q Trap machine with Analyst software (version 1.6.2). The conditions for LC/MS were similar to those used by Bräutigam et al. [18]. A Reprosil 100 C18 RP column was used. Mobile phase was acetonitrile-water (90:10). Flow rate was 0.8 ml/min, with an injection volume of 20 µl.
2.4 COX-2 activity assay
10% homogenates of tail tissue were prepared in cold Tris-EDTA from six samples pooled in each group. A microplate format COX activity assay kit (Cayman Chemical Co., USA) was used and results were reported in nmol/min/ml homogenate.
2.5 Western blot
Regenerating tail tissue at Wound Epithelium and Blastema stages was collected from six animals of each experimental group, pooled and homogenised in Tris-SDS lysis buffer with protease inhibitor (Sigma Aldrich, USA). 10% homogenates were assayed for total protein content by Bradford method [19]. Equal amount of total protein was loaded and separated by SDS-PAGE on 10% gels. Protein was transferred onto nitrocellulose membrane by semi-dry transfer at 100 mA for 30 min. The membrane was probed separately with Anti-β-Catenin and Anti-β-Actin (IgG antibodies raised in Mouse). The ALP system was used for staining the protein of interest.
2.6 Real time RT-PCR
Total RNA was isolated from regenerating tail tissue of six animals of each group using TRIzol reagent (Applied Biosystems, USA). 1 μg total RNA was reverse transcribed to cDNA using a one-step cDNA synthesis kit (Applied Biosystems, USA). Primer sequences from 5′ to 3′ used for the subsequent real-time PCR were as follows: wnt1 fwd: AAGTCGGGAAGGAGAGGTGA; wnt1 rev: GAGCCATCTGAAACTGCCCT; wnt2b fwd: CCTGTTGGCTGGCTATGTCT; wnt2b rev: TGTCAGCCACCATAAAGCCA; wnt3a fwd: TCCTTTGTGCCAGCATACCA; wnt3a rev: TGGATGCCGATCTTTACCCC; wnt4 fwd: CGCAAGGTGGGATCTACCAA; wnt4 rev: TTCTGTTACACTGGCGTCCC; wnt6 fwd: TTGGTCATGGACCCCAACAG; wnt6 rev: CCTCGCTGACGATTTCTGGT; wnt7a fwd: CTGCGGAAGGGGATACAACA; wnt7a rev: TAACGTAGCAGCACCACAGG; wnt7b fwd: TAGCCAAGGCAACCTAAGCC; wnt7b rev: CCACGCCGTACTTGACATCT; wnt8a fwd: TAACAACGAGGCGGGAAGAC; wnt8a rev: GCAACTTCCAGACACCCCAT; wnt10a fwd: CGAGGAGGCATTTCGACTCA; wnt10a rev: ACCGTGAACCATCCCTTTCC; wnt16 fwd: CTAAACAGTGACCAGTGCCG; wnt16 rev: GTCAGGGGTCAAGGACAGAAC; 18S rRNA fwd: GGCCGTTCTTAGTTGGTGGA; 18S rRNA rev: TCAATCTCGGGTGGCTGAAC. The suitability of 18S rRNA as an internal loading control was first checked. For this, the cDNA (which was prepared from equal amounts of total RNA) was itself quantified using the Qubit 3.0 fluorimeter assay (Life Technologies. USA) and was found to be not significantly different among samples. A qPCR of 18S rRNA was then performed and the Cq values were correlated with the total cDNA concentrations. It was found to correlate, with no variation among samples (data not shown). Quantitative real-time PCR was performed on a Lightcycler96 (Roche Diagnostics, Switzerland) with the following program: 3 min at 95 °C, 45 cycles (each cycle of 10 s at 95 °C, 10 s at 60 °C and 10 s at 72 °C). Melt curve analysis was used to confirm specific product formation. Data was represented as mean Cq values normalized with 18S rRNA levels and data was analysed by one-way ANOVA using Prism v5.03 (GraphPad Software Inc., USA). Fold change in expression was calculated using the 2−ΔΔCq method of Livak and Schmittgen [20].
3 Results
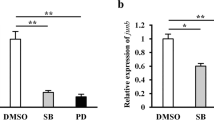
To achieve inhibition of cyclooxygenase-2 activity in vivo, we orally administered Etoricoxib to H. flaviviridis. Before the effect of COX-2 inhibition on Wnt/β-Catenin signaling could be tested, it was necessary to validate this inhibition. For this, we collected tail tissue after 24 hours of a single oral dose of Etoricoxib and prepared an extract in Acetonitrile (LC/MS grade). The sample was subjected to LC/MS and Etoricoxib was quantified in the tail tissue. A linearity graph was obtained using Etoricoxib standards of concentrations 1, 50, 100, 200 and 300 ng/ml (Fig. 1). Further, samples were run wherein control sample produced no peak while treated group sample produced a peak corresponding to 32.8 ng/ml. This corresponds to 32.8 ng Etoricoxib in 100 mg of tail tissue. Next, to test the inhibition of COX-2 in tail tissue, an activity assay was performed. COX-2 activity in the wound epidermis and blastema stages was found to be significantly lower in the treated group animals (p ≤ 0.001 and p ≤ 0.01 respectively), as compared to controls (Fig. 2). Moreover, the activity assay revealed that COX-2 is most active in during the wound epidermis stage (p ≤ 0.001 when compared to resting stage), followed by the blastema stage (p ≤ 0.01 when compared to resting stage).
The present study was taken up to understand the effect of COX-2 inhibition on Wnt/β-Catenin signaling. When Wnt signals are present, intracellularly β-Catenin is stabilized and allowed to translocate into the nucleus to induce the transcription of target genes. With this, it regulates various developmental processes including cell proliferation and differentitation. In the absence of the Wnt signal, however, β-Catenin is targeted for proteasomal degradation. We carried out a western blot to check the expression of β-Catenin in response to Etoricoxib treatment. It was found that drug treatment lead to a reduced level of β-Catenin in the tail tissue at both wound epidermis and blastema stages (Fig. 3).
Reduced β-Catenin signals prompted us to check the status of the Wnt ligands in wound epidermis and blastema tissue to understand whether COX-2 signaling is directly interacting with β-Catenin [21] or it is causing a change in the expression of Wnt ligands. Moreover, it was so far not known precisely which Wnt ligands are expressed during the specific stages of regeneration in lizards. Therefore, we screened for twelve Wnt ligands, classically known to induce the ‘canonical’ (β-Catenin dependent) pathway, for their expression in the two stages of tail regeneration. Of these, we found expression of transcripts of 10 ligands, viz., wnt1, wnt2b, wnt3a, wnt4, wnt6, wnt7a, wnt7b, wnt8a, wnt10a and wnt16 in both wound epidermis and blastema stages. Further, real-time PCR results showed that wnt2b, wnt4 and wnt7a had heightened expression (p ≤ 0.001, p ≤ 0.05 and p ≤ 0.001 respectively) in wound epidermis stage as compared to the resting tail and the blastema stages, whereas wnt1 and wnt6 showed significantly greater (p ≤ 0.001) expression in the blastema stage (Table 1; Fig. 4).
For the current study, we analysed the expression of the abovementioned 10 Wnt ligands in response to COX-2 inhibition during the two stages, at transcript level using real time PCR. We found skewed expression of a number of these in both the stages. Among the two genes found majorly during wound epidermis stage, wnt4 showed an increase in Cq value pointing towards heavily decreased expression (p ≤ 0.001) compared to control (Table 2; Fig. 5). wnt7a, on the other hand, was found to be upregulated (p ≤ 0.05) as compared to control. During the blastema stage, wnt1 and wnt16 showed sharp decrease (p ≤ 0.001 as compared to controls) in expression in response to Etoricoxib treatment, as reflected by the high Cq values (Table 3; Fig. 6). Over and above these, a statistically significant increase in expression was found for wnt2b, wnt3a, wnt7a, wnt7b, wnt8a and wnt10a in the blastema stage.
4 Discussion
Inflammation is an inevitable process immediately following an injury. In both regenerating and non-regenerating systems, an inflammatory response is essential to proper healing of the wound. It is well known that scar-free wound healing in regenerating vertebrate limbs involves immune molecules [22]. One such inducible mediator of inflammation is cyclooxygenase-2, the importance of which in regeneration has been highlighted in few earlier studies [7, 14,15,16]. Suresh et al. [23] demonstrated the impeding effects of COX-2 inhibition on angiogenesis, myogenesis and cell synthetic activities in the wound epidermal and blastemal cells of H. flaviviridis. However, the mechanism with which COX-2 mediated PGE2 influences regenerative healing and outgrowth is still at large.
Of the molecular signaling pathways orchestrating the variety of processes which result in tail restoration, the Wnt/β-Catenin pathway is vastly important for its role in cell migration, wound repair and proliferation in a wide variety of developing systems [21, 24, 25]. It has been identified as a regulator of growth and patterning during appendage regeneration in amphibians [26,27,28]. Importantly, it has been demonstrated in a diverse set of biological systems that PGE2 modulates Wnt signaling by altering the expression of various pathway components [21, 29,30,31]. Therefore, it was thought pertinent to test whether PGE2 signals through Wnt/β-Catenin to bring about successful regeneration of tail in H. flaviviridis. Our study involved the inhibition of COX-2 using its pharmacological inhibitor Etoricoxib and noting its influence on the expression of selected Wnt/β-Catenin pathway genes.
We orally administered Etoricoxib once daily to lizards up until the end of the experiment. Since there is no earlier record of oral treatment of animals with Etoricoxib, we first checked for its assimilation in the tail tissue by LC/MS and further assayed for COX-2 activity. We found significant inhibition of COX-2 activity in the tail after Etoricoxib treatment. Following commencement of drug treatment, the lizard tails were autotomised from a pre-determined point. Tissue sampling was done at two major stages of regeneration – wound epidermis and blastema, considering the importance of these stages and their implication in successful tail replacement.
Analysis of β-Catenin expression showed a definite reduction in both wound epidermis and blastema stages in response to Etoricoxib. This points, at least partially, towards the lack of Wnt signal thereby allowing its proteasomal degradation. As a transcriptional coactivator, β-Catenin functions to regulate a battery of genes associated with cell proliferation, cell polarity and cell fate determination [32]. In order to ascertain whether altered expression of the Wnt ligands was responsible for the reduced β-Catenin expression, we performed a relative quantification of the transcripts of a set of Wnt ligands, classically attributed with the function of β-Catenin stabilization. A scan through literature, however, reveals that there has so far not been a screen identifying the Wnt ligands expressed during lizard tail regeneration and we are the first to list these. RT-PCR results show the presence of wnt1, wnt2b, wnt3a, wnt4, wnt6, wnt7a, wnt7b, wnt8a, wnt10a and wnt16 at both wound epidermis and blastema stages. At this stage, we may attribute the function of wound epidermis and blastema formation to the Wnt ligands showing heightened expression in these respective stages, although its ascertainment would need further detailed studies to be taken up.
Relative quantification of transcripts by real time PCR shows that among the Wnt ligand transcripts, wnt4 and wnt7a had significantly higher expression during the wound epidermis stage than the in the resting tail and blastemal stage. wnt4 gene expression in treated animals demonstrated a significant decrease, relative to control, during the wound epidermis stage. This may explain the incomplete healing which is seen in Etoricoxib treated animals since Wnt4 has been previously shown to be important for timely wound healing and is upregulated in response to trauma [24]. During the blastema stage, wnt1 and wnt6 were expressed at a higher level than in the resting tail and the preceding wound epidermis stage. In samples treated with Etoricoxib, there was significantly reduced expression of wnt1 and wnt16 as compared to control. Wnt1 may be important in the late blastema owing to its property of inducing satellite cell proliferation [25]. It has to be acknowledged here that wnt2b, wnt3a, wnt7a, wnt7b, wnt 8a and wnt10a were all significantly higher in expression in treated samples versus control. While the work of Ramos-Solano et al. [33] supports the notion that Wnt7a may signal to decrease cell proliferation in some systems, one finds conflicting results from other studies [34, 35]. In order to draw a crisp conclusion about the role of individual Wnt ligands in tail regeneration, a lot more must be done in addition to the current work, since β-Catenin lies at the convergence point of multiple pathways and is not regulated by the Wnt ligands alone.
Our results here have nevertheless provided us with evidence that there is a cross-talk between the COX-2 induced PGE2 signal and the Wnt/β-Catenin pathway in the regeneration process and that together, they are essential for successful achievement of the wound epidermis and blastema stages. We hypothesize that this interaction could be through the Fibroblast Growth Factor (FGF) signaling pathway. This stems from a personal observation that the attenuation of COX-2-induced PGE2 leads to reduced expression of FGF signaling components in both the mentioned stages of lizard tail regeneration (unpublished data). Additionally, the Wnt/β-Catenin pathway is known to be activated in the presence of the FGF signal [3, 36]. Thus deviant levels of Wnt ligands and β-Catenin in response to COX-2 inhibition by Etoricoxib could be a result of reduced FGF activation, leading to delayed and imperfect wound closure, reduced blastemal cell proliferation and hence retardation of the regenerative response. An understanding of the signaling pathways interacting with PGE2 signaling will give us valuable information about many developmental processes, more so since COX-2 is also considered as a target for cancer therapeutics.
References
Wallace H. Vertebrate limb regeneration. Chichester: Wiley; 1981.
Call MK, Tsonis PA. Vertebrate limb regeneration. Adv Biochem Eng Biotechnol. 2005;93:67–81.
Stoick-Cooper C, Weidinger G, Riehle KJ, Hubbert C, Major MB, Fausto N, Moon RT. Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development. 2007;134:479–89.
Kumar A, Pilo B. Influence of catecholamine and acetylcholine neurotransmitters on tail regeneration in gekkonid lizard Hemidactylus flaviviridis. Ind J Exp Biol. 1994;32:767–71.
Pilo B, Suresh B. Effect of EGF on progress of tail regeneration in the gekkonid lizard Hemidactylus flaviviridis. J Anim Morph Physiol. 1994;41:61–6.
Pilo B, Kumar A. Effect of chemical adrenalectomy and corticosterone administration on tail regeneration in the gekkonid lizard Hemidactylus flaviviridis. Ind J Exp Biol. 1995;33:917–20.
Sharma P, Suresh B. Influence of COX-2-induced PGE on the initiation and progression of tail regeneration in Northern House Gecko, Hemidactylus Flaviviridis. Folia Biol (Praha). 2008;54:193–200.
Yadav M, Buch P, Desai I, Suresh B. Exogenous administration of EGF augments nucleic acid biosynthesis and cell proliferation in the regenerating tail of wall lizard. Eur J Exp Biol. 2014;4:113–23.
Smith WL. Prostanoid biosynthesis and mechanisms of action. Am J Physiol. 1992;263:F181–91.
Sugimoto Y, Narumiya S. Prostaglandin E receptors. J Biol Chem. 2007;282:11613–7.
Greenhough A, Smartt HJM, Moore AE, Roberts HR, Williams AC, Paraskeva C, Kaidi A. The COX-2/PGE 2 pathway: key roles in the hallmarks of cancer and adaptation to the tumour microenvironment. Carcinogenesis. 2009;30:377–86.
Saini M, Sanyal S. Evaluation of chemopreventive response of two Cycloxygenase-2 inhibitors, Etoricoxib and Diclofenac in rat colon cancer using FTIR and NMR spectroscopic techniques. Nutr Hosp. 2010;25:577–85.
Li W, Yue W, Zhang L, Zhao X, Ma L, Yang X, Zhang C, Wang Y, Gu M. COX-2 silencing inhibits proliferation in A549 cell. Chin Ger J Clin Oncol. 2011;10:P423–7.
Finetti F, Solito R, Morbidelli L, Giachetti A, Ziche M, Donnini S. Prostaglandin E2 regulates angiogenesis via activation of fibroblast growth factor receptor-1. J Biol Chem. 2008;283:2139–46.
Veliça P, Bunce CM. Prostaglandins in muscle regeneration. J Muscle Res Cell Motil. 2008;29:163–7.
Anusree P, Saradamba A, Tailor N, Desai I, Suresh B. Caudal fin regeneration is regulated by Cox-2 induced PGE2 in Teleost fish Poecilia latipinna. J Cell Tissue Res. 2011;11:2795–801.
Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509.
Brautigam L, Nefflen JU, Geisslinger G. Determination of Etoricoxib in human plasma by liquid chromatography–tandem mass spectrometry with electrospray ionisation. J Chromatogr B. 2003;788:309–15.
Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods. 2001;25:402–8.
Wong CT, Ahmad E, Li H, Crawford DA. Prostaglandin E2 alters Wnt-dependent migration and proliferation in neuroectodermal stem cells: implications for autism spectrum disorders. Cell Commun Signal. 2014;12:19.
Godwin J, Kuraitis D, Rosenthal N. Extracellular matrix considerations for scar-free repair and regeneration: insights from regenerative diversity among vertebrates. Int J Biochem Cell Biol. 2014;56:47–55.
Suresh B, Sharma P, Desai I. Evidence of the involvement of Prostaglandin E2 in different cellular activities during epimorphic regeneration. J Cell Tissue Res. 2009;9:1883–90.
Labus MB, Stirk CM, Thompson WD, Melvin WT. Expression of Wnt genes in early wound healing. Wound Repair Regen. 1998;6:58–64.
Otto A, Schmidt C, Luke G, Allen S, Valasek P, Muntoni F, Lawrence-Watt D, Patel K. Canonical Wnt signalling induces satellite-cell proliferation during adult skeletal muscle regeneration. J Cell Sci. 2008;121:2939–50. doi:10.1242/jcs.026534.
Caubit X, Nicolas S, Le Parco Y. Possible roles for Wnt genes in growth and axial patterning during regeneration of the tail in urodele amphibians. Dev Dyn. 1997;210:1–10.
Caubit X, Nicolas S, Shi D, Le Parco Y. Reactivation and graded axial expression pattern of Wnt-10a gene during early regeneration stages of adult tail in amphibian urodele Pleurodeles waltl. Dev Dyn. 1997;208:139–48.
Yokoyama H, Ogino H, Stoick-Cooper CL, Grainger RM, Moon RT. Wnt/β-catenin signaling has an essential role in the initiation of limb regeneration. Dev Biol. 2007;306:170–8.
Shao J, Jung C, Liu C, Sheng H. Prostaglandin E2 stimulates the β-catenin/T cell factor-dependent transcription in colon cancer. J Biol Chem. 2005;280:26565–72.
North TE, Babu IR, Veddera LM, Lord AM, Wishnok JS, Tannenbaum SR, Zon LI, Goessling W. PGE2-regulated Wnt signaling and N-acetylcysteine are synergistically hepatoprotective in zebrafish acetaminophen injury. Proc Natl Acad Sci. 2010;107:17315–20.
Liu X, Kirschenbaum A, Weinstein BM, Zaidi M, Yao S, Levine AC. Prostaglandin E2 modulates components of the Wnt signaling system in bone and prostate cancer cells. Biochem Biophys Res Commun. 2010;394:715–20.
MacDonald BT, Tamai K, He X. Wnt/β-catenin signailng: components. Mech Dis Dev Biol. 2010;17:9–26.
Ramos-Solano M, Meza-Canales ID, Torres-Reyes LA, Alvarez-Zavala M, Alvarado-Ruíza L, Rincon-Orozcod B, Garcia-Chagollan M, Ochoa-Hernández AB, Ortiz-Lazareno PC, Rösld F, Gariglio P, Jave-Suárez LF, Aguilar-Lemarroy A. Expression of WNT genes in cervical cancer-derived cells: implication of WNT7A in cell proliferation and migration. Exp Cell Res. 2015;335:39–50.
Lyu J, Joo C. Wnt-7a up-regulates matrix metalloproteinase-12 expression and promotes cell proliferation in corneal epithelial cells during wound healing. J Biol Chem. 2005;280:21653–60.
Qu Q, Sun G, Li W, Yang S, Ye P, Zhao C, Yu RT, Gage FH, Evans RM, Shi Y. Orphan nuclear receptor TLX activates Wnt/β-catenin signalling to stimulate neural stem cell proliferation and self-renewal. Nat Cell Biol. 2010;12:1–20.
Stulberg MJ, Lin A, Zhao H, Holley SA. Crosstalk between Fgf and Wnt signaling in the zebrafish tailbud. Dev Biol. 2012;369:298–307.
Acknowledgements
The authors are thankful to DST-SERB, Govt. of India for financial assistance in the form of Project No. SB/SO/AS-008; CSIR, New Delhi for research fellowship to PB; DBT-MSUB-ILSPARE, The M. S. University of Baroda for providing the LC/MS facility; Adityarao Khnavilkar for his assistance in LC/MS.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest whatsoever.
Ethical statement
All experimental protocols were approved by the Institutional Animal Ethics Committee (IAEC) constituted as per the guidelines of Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA), India (IACUC no. 984/07/2014-2).
Rights and permissions
About this article
Cite this article
Buch, P.R., Sarkate, P., Uggini, G.K. et al. Inhibition of Cyclooxygenase-2 Alters Wnt/β-Catenin Signaling in the Regenerating Tail of Lizard Hemidactylus flaviviridis . Tissue Eng Regen Med 14, 171–178 (2017). https://doi.org/10.1007/s13770-017-0037-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13770-017-0037-2