Abstract
The lipA gene, encoding a solvent-tolerant extracellular lipase from Proteus sp. SW1, was displayed on the cell surface of Escherichia coli by fusing it to an antigen 43 anchoring motif. The display of LipA on the Escherichia coli cell surface was directly confirmed by immunofluorescence microscopy and flow cytometry. After 6 days of incubation in media containing 1 % used cooking oil, an Escherichia coli strain expressing surface displayed lipase was able to degrade 27 % of the oil. The biosurfactant, pseudopyronine B, was purified from culture supernatants of Pseudomonas sp. SL31. Its critical micelle concentration was determined to be 1400 mg/l, and the surfactant was stable within a temperature range from 0 to 120 °C and a pH range of 3–11. Pseudopyronine B-containing crude media extracts efficiently removed up to 51 % of the cadmium from contaminated water. We demonstrated the oil degradation ability of the mixed culture of four bacterial strains, namely the recombinant Escherichia coli expressing cell surface displayed lipase (pKKJlipA), His-tagged lipase (pETlipA), extracellular lipase-producing Proteus sp. SW1, and pseudopyronine B-producing Pseudomonas sp. SL31 by culturing in LB media containing 1 % oil. The consortium degraded 29 % of oil in one day and reached 84 % after 7 days.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Lipids are an important component in domestic waste that causes severe environmental pollution. They can form oil films on water surfaces, preventing the diffusion of oxygen from air into water and leading to the death of many forms of aquatic life. Microbial products, particularly lipases and surfactants, have been investigated for use in the breakdown of lipids in wastewater (Mongkolthanaruk and Dharmsthiti 2002). Lipases are triacylglycerol hydrolases (E.C.3.1.1.3) that catalyze the hydrolysis of triacylglycerol into di- or monoglycerides, free fatty acids, and glycerol, at the lipid–water interface (Gupta et al. 2015). They are ubiquitous in nature and are found in plants, animals, and microorganisms. Microbial lipases are usually secreted into the media (extracellular), although a few cell-bound and intracellular enzymes have also been reported (Gurung et al. 2013). Extracellular enzymes are preferable since they are easy to purify and are advantageous in industrial applications (Gurung et al. 2013). The utility of these enzymes has spurred demand for new lipases that are better suited for specific applications, for example lipase expressed on the cell surface which, in principle, attacks bulky substrates more efficiently. Previously, our research group reported the characterization of a novel extracellular lipase from Proteus sp. SW1 that is highly organic solvent tolerant, and investigated its suitability for biodiesel production (Whangsuk et al. 2013). Further improvements of this lipase, such as expressing it on the microbial cell surface, would be desirable to increase its utility.
The ability of cells to display proteins on their surface is essential for a wide range of biological processes, such as adhesion, colonization, biofilm formation, cell–cell recognition, and signal transduction (Becker et al. 2005). In recent years, a variety of expression systems have been developed for the display of heterologous peptides on cell surfaces. The advantage of displaying enzymes at the cell surface of microorganisms is that neither substrate nor product needs to be membrane permeable, and both could be protected from any unwanted attack by intracellular enzymes (Jose et al. 2012). Many different types of surface proteins from gram-negative bacteria have been used as vehicles for the display of foreign proteins on cell surfaces. These include antigen 43 (Kjaergaard et al. 2002), EstA (Becker et al. 2005), and OprF (Lee et al. 2005).
Antigen 43 (Ag43) is a surface protein of Escherichia coli (E. coli) that is a member of the autotransporter family, for which all information required for transport to the outer membrane and secretion through the cell envelope is contained in the protein itself (Henderson and Owen 1999). Ag43 is encoded by flu, and it is synthesized as a 1039 amino acid precursor. The precursor is then processed into α- and β-subunits of 499 and 488 amino acid residues, respectively, that comprise the mature Ag43 dimer (Hasman et al. 1999; Henderson and Owen 1999). The β-subunit integrates into the outer membrane where it forms a pore-like β-barrel structure (Henderson and Owen 1999). The α-subunit reaches the cell exterior assisted by the β-subunit and remains attached to the cell surface, presumably through non-covalent interaction with the β-subunit (Henderson and Owen 1999). While there is information on wild-type microorganisms that degrade oil, specific information concerning the use of recombinant surface displayed enzymes for oil biodegradation and their use as a consortium is unavailable.
Biosurfactants are amphiphilic compounds that contain hydrophilic and hydrophobic moieties and are derived from living organisms. They are widely used in the industrial applications such as food, cosmetic, and pharmaceutical industries (Desai and Banat 1997). They can be excreted and accumulated between two fluid phases where they reduce the surface and interfacial tensions at the fluid surface and fluid-phase interface, respectively (Jorfi et al. 2013). They are emulsifiers that enhance lipase activity by (a) increasing the available substrate surface area (dispersion) and (b) improving the affinity of lipase-producing microbes for the substrate (Chang et al. 2004; Cammarota and Freire 2006). Rhamnolipids produced by Pseudomonas aeruginosa, which is the best-studied microbial surfactant, showed the ability to accelerate the biodegradation of total petroleum hydrocarbons in liquid medium from 32 to 61 % at 10 days of incubation when expressed as part of a microbial consortium (Abalos et al. 2004). Moreover, a consortium of five bacteria showed an excellent oily sludge degradation capacity, reducing 90.7 % of the aliphatic fraction and 51.8 % of the aromatic fraction. Biosurfactant activity could also be detected in the culture media, achieving a 39.4 % reduction in the surface tension of the culture medium and an emulsifying activity of 55.1 % (Cerqueira et al. 2011). Recently, the production of pseudopyronine B (PPB) biosurfactant (initially designated α-pyrone I) was reported in the marine bacterium Pseudomonas sp. F92S91. This biosurfactant inhibited the growth of a number of pathogenic microorganisms (Singh et al. 2003). While the oil biodegradation properties of rhamnolipids are well documented, no investigation of PPB properties besides antimicrobial activity has yet been reported.
In this study, we report the isolation and characterization of the pseudopyronine B (PPB)-producing bacterium, Pseudomonas sp. SL31 and its potential for the bioremediation of oil in co-culture with lipase-producing bacterial strains, i.e., the extracellular lipase-producing bacterium, Proteus sp. SW1, recombinant E. coli expressing lipase on its surface using antigen 43 as anchoring motif, and a lipase-overexpressing recombinant E. coli strain.
This research project was performed at the Chulabhorn Research Institute, Bangkok, Thailand.
Materials and methods
Bacteria and media
Escherichia coli cells, DE3 (Novagen, USA) (Robichon et al. 2011), and DH5α were grown at 37 °C in Luria–Bertani (LB) medium (0.5 % yeast extract, 1 % tryptone, 0.5 % NaCl) with 100 μg/ml ampicillin to select recombinant cells. Proteus sp. SW1 and Pseudomonas sp. SL31 were grown in LB medium at 28 °C.
Construction of pKKJlipA
The lipase lipA gene was amplified from the chromosome of a Proteus sp. SW1 (Whangsuk et al. 2013) with primer BT3629 (GATTAACTTAAAGATCTCACCACCACCACCACCACCCAACTACATATCCAATT) and BT3527 (CAGGAGATCTAAGCTTTTT), which contains a BglII site (underlined) (Whangsuk et al. 2013). The primer BT3629 contained the 6 histidine codons fused in frame between amino acids 147 and 148 in the α-subunit, thus enabling detection of lipase on the cell surface using antibodies to either the His-tag motif or native lipase. An 890-bp PCR product was digested with BglII and inserted into the BglII site of pKKJ135, which was kindly provided by Professor Per Klemm, Denmark (Kjaergaard et al. 2002).
Immunofluorescence microscopy
Cells were prepared for immunofluorescence microscopy by washing with phosphate-buffered saline (PBS) solution and fixing with 2 % formaldehyde for 10 min. Fixed cells were washed three times with PBS and resuspended in PBS solution supplemented with 2 % bovine serum albumin and incubated for 18 h at 4 °C. Cells were then incubated in PBS solution containing 3 % (w/w) bovine serum albumin and rabbit anti-LipA antibody for 2 h on ice. After washing three times with PBS solution, the cell–antibody complex was incubated for 1 h at 37 °C with goat anti-rabbit IgG conjugated with Texas red (Jackson immune research, USA). Prior to microscopic observation, cells were washed three times with PBS solution to remove unbound goat anti-rabbit IgG conjugated with Texas red. Cells expressing lipase on their surface were detected by fluorescence microscopy.
Flow cytometry or fluorescence-activated cell sorting (FACS)
For immunofluorescence staining, cells were harvested and washed three times with PBS. The washed cells were resuspended in 1 ml PBS containing 1 % BSA and rabbit anti-LipA antibody and incubated for 1 h. After washing three times with PBS, the cells were incubated with Texas red-conjugated goat anti-rabbit IgG antibody on ice for 1 h. The Texas red-labeled cells were examined using a flow cytometer (BD, FACS Canto, USA).
Oil degradation
Escherichia coli strain DH5α::pKKJlipA displaying lipase was cultured in LB medium while shaking at 180 rpm for 18 h at 37 °C. The overnight culture (1 %) was subcultured in LB broth, grown to an OD600 of 1.0, induced with 1 mM arabinose for 2 h at 28 °C, added with 1 % used soya bean oil, and shaken (180 rpm) for 7 days at 28 °C. During cultivation, samples were taken for lipid degradation analysis.
In the case of bacterial pure culture and consortia, E. coli (DE3) harboring pETlipA (Whangsuk et al. 2013), E. coli (DH5α) harboring pKKJlipA, Proteus sp. SW1 (Whangsuk et al. 2013), and Pseudomonas sp. SL31 were separately grown in LB broth overnight. The overnight culture (1 %) was subcultured in LB broth, grown to an OD600 of 1.0, and induced with either 200 µM IPTG for E. coli DE3::pETlipA or 1 mM arabinose for E. coli DH5α::pKKJlipA, for 2 h at 28 °C. After induction, the lipase activities of both cultures and also Proteus sp. SW1 culture were determined by monitoring the cleavage of ρ-nitrophenyllaurate (pNL) substrate as previously described (Whangsuk et al. 2013). The biosurfactant activity of Pseudomonas sp. SL31 culture was determined by the oil displacement test.
In the case of lipid degradation experiments using pure cultures, the 20 ml culture was mixed with oil (1 % v/v) and shaken (180 rpm) for 7 days at 28 °C. In the case of consortia, 5 ml of each culture was mixed with oil (1 % v/v) and shaken (180 rpm) for 7 days. The addition of crude extract of PPB to a consortium was performed by mixing 3 mg of PPB in methanol to 20 ml culture. Lipid degradation was analyzed using a chloroform–methanol extraction method described previously (Koma et al. 2003). Briefly, a chloroform–methanol mixture (20 ml; 3:1, v/v) was added to a 20-ml culture and shaken. The chloroform layer was transferred to a new tube and completely dried in a rotary evaporator. The amount (weight) of extracted lipid was then measured.
Pseudomonas sp. SL31 Identification
16S ribosomal RNA gene sequencing primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and BT1525R (5′-AAGGAGGTGATCCAGCC-3′), complementary to conserved regions (Lane, 1991), were used to PCR amplify the 16S rRNA gene resulting in a 1.5-kb fragment.
PCR detection of rhamnolipid biosynthesis gene
The gene encoding rhamnosyltransferase I (rhlB), which is a part of the rhamnolipid biosynthesis pathway, was PCR amplified to determine the presence of rhlB in the chromosome of Pseudomonas sp. SL31 using previously determined forward (5′-GCCCACGACCAGTTCGAC-3′) and reverse (5′CATCCCCCTCCCTATGAC-3′) primers (Bodour et al. 2003). The expected 226-bp PCR product was considered indicative of the presence of rhlB in the template DNA (Bodour et al. 2003).
The oil displacement test
The oil displacement test was performed as described by Rodrigues et al. (2006). Briefly, 50 ml of distilled water was added to a 15-cm petri dish. Crude oil (20 µl) was dropped onto the surface of the water, followed by the addition of 10 µl of cell culture supernatant. After 30 s, the visualized clear zone was observed under visible light.
The drop-collapse test
Bacterial culture supernatants (100 µl) were added to wells in a 96-well microplate. Crude oil (5 µl) was added to the surface of the culture supernatant. Biosurfactant production was indicated by a uniform (i.e., flat) dispersion of the oil across the surface (Płaza et al. 2006).
Purification and structure elucidation of the biosurfactant produced by Pseudomonas sp. SL31
Pseudomonas sp. SL31 was grown overnight in LB at 28 °C. Cells were pelleted by centrifugation (6000 rpm) at 4 °C for 20 min, and the supernatant was collected for biosurfactant extraction. The supernatant was extracted twice with hexane and completely dried in rotary evaporator. The crude extract was subjected to semi-preparative HPLC (Water Delta 600, Massachusetts, USA) on a Hichrom reverse-phase column (21.2 × 150 mm) (the Markham Center, Berkshire, England). Binary solutions of milli-Q water and methanol were used as the mobile phase with a flow rate of 15 ml/min. Molecules were eluted using a continuous gradient from 3 % (v/v) methanol to 100 % (v/v) over 13 min with a final wash of 3 % (v/v) methanol. The structure of purified compounds was elucidated by mass spectrometry and nuclear magnetic resonance (NMR) (Bruker AVANCE 300 MHz).
Surface tension measurement
The surface tension of aqueous solutions at different surfactant concentrations was measured using a tensiometer at room temperature (McInerney et al. 1990). The surface tension measurement was taken at 25 ± 1 °C after soaking the platinum ring in the solution in order to attain equilibrium conditions. The measurement was repeated three times and the average value calculated. The critical micelle concentration (CMC) was then determined from the break point of the surface tension versus the log of bulk concentration curve. The instrument was calibrated by measuring the surface tension of pure water before each set of experiments.
Stability testing
Samples of crude biosurfactant and synthetic surfactant were prepared at the CMC for thermal and pH stability measurement (Pornsunthorntawee et al. 2008). The surfactant solutions were incubated in a water bath at different temperatures and different time intervals before cooling to room temperature. The pH stability was investigated by adjusting the solutions to different pH values. For both thermal and pH stability testing, the surface tension was measured and used to indicate the stability.
Cadmium (Cd) removal by PPB
Cd removal by surfactant was assayed according to a previously described report (Kim and Vipulanandan 2006). The surfactant solution with cadmium was filtered through a 0.2-µm filter to separate the cadmium-containing micelles from the water-soluble cadmium ions. SDS was used as a surfactant control and distilled water (DW) was used as a negative control. The filter was then acid-washed using concentrated nitric acid (HNO3) to recover the cadmium in the micelle phase. Cd removal efficiency (R w) in wastewater was defined as:
where C e is the amount of cadmium in the filtrate and C m is the amount of cadmium retained in the filter (micelle phase) (Kim and Vipulanandan 2006). Cd concentrations in the filtrate and in the filter were measured by atomic absorption spectrometry (Perkin-Elmer, USA).
Results and discussion
Construction of an antigen 43–lipA gene fusion
As a first step in generating an E. coli strain DH5α that displays LipA on its surface, the entire coding sequence of Proteus sp. SW1 lipA containing 6 histidines at the N terminus of LipA was inserted in frame between codons 147 and 148 of E. coli flu gene in plasmid pKKJ135 with a fusion product of 4010 bp encoding 1323 amino acid protein with a molecular mass of approximately 144 kDa (Fig. 1). In this construct, expression of the flu–lipA gene is under the control of the arabinose-inducible araBAD promoter.
A previous report had shown that in-frame fusion of heterologous sequences between codons 147 and 148 in the α-subunit of Ag43 did not affect the production, surface localization, or autoaggregation of Ag43 (Gupta et al. 2015). This was confirmed in cells producing either wild-type Flu or the LipA–Flu fusion protein by looking for the appearance of frizzy colony morphology and the autoaggregation of cells; two obvious phenotypes of Ag43 expression (Hasman et al. 1999). E. coli containing either pKKJlipA or pKKJ135 displayed a distinct frizzy colony morphotype when grown on arabinose-containing solid medium (data not shown). Also, when cells carrying either pKKJlipA or pKKJ135 were left standing after growth in liquid medium in the presence of arabinose, they were observed to autoaggregate (data not shown). These results are similar to those of cells expressing the well-characterized epitope peptides, CTP3 and Chlam12, which were used as heterologous passengers and introduced into Ag43. The CTP3 epitope, comprising amino acids 50–64 of the CTB chain, is known to form a conformational loop on the surface of CTB (Hasman et al. 1999). Our results indicated that introduction of the heterologous lipase did not interfere with the functionality of Ag43.
Expression of surface displayed lipase LipA protein
The recombinant fusion plasmid pKKJlipA was introduced into E. coli strain DH5α. The transformant colonies produced a hydrolytic clear zone on LA media containing 1 mM arabinose and 1 % tributyrin. E. coli (DH5α) harboring pKKJ135 plasmid did not, suggesting the successful display of active lipase on the E. coli DH5α::pKKJlipA cell surface (data not shown).
The display of lipase on the E. coli DH5α::pKKJlipA cell surface was also directly confirmed by flow cytometry and immunofluorescence microscopy. We used a specific rabbit anti-LipA antibody to confirm the surface exposure of lipase via flow cytometry. The antibodies can only bind to surface-exposed structures because they are too large to cross the outer membrane. Both arabinose-induced E. coli DH5α::pKKJlipA and E. coli DH5α::pKKJ135 cells were marked with fluorescently labeled anti-LipA antibodies and analyzed by flow cytometry (Fig. 2a). The sample (E. coli DH5α::pKKJlipA) containing the lipase-expressing cells showed a ten fold increase in mean fluorescence intensity values (Fig. 2a, intensity value 121 vs. 12) compared to the vector control (E. coli DH5α::pKKJ135), which showed no increased fluorescence signal.
a Monitoring surface displayed lipase by flow cytometry: an arrow indicates the fluorescence signal observed by E. coli harboring pKKJlipA compared to plasmid vector. b Immunofluorescence micrographs of E. coli harboring pKKJlipA (1, 2) and plasmid vector (3, 4) (1 and 3: phase contrast images, 2 and B4 fluorescence images)
To further confirm the display of lipase on the cell surface, E. coli DH5α::pKKJlipA, expressing the Ag43–lipA fusion, and the vector control, E. coli DH5α::pKKJ135, were incubated with anti-lipase antibody followed by labeling with a secondary antibody conjugated to Texas red. Labeled cells were examined by immunofluorescence microscopy. E. coli DH5α::pKKJlipA clearly fluoresced [Fig. 2b(1, 2)], indicating that lipase was expressed on the surface of E. coli. By contrast, E. coli DH5α::pKKJ135 cells, which carry plasmid vector alone, did not fluoresce (Figs. 2b, 3, 4). These results indicate that the Ag43–LipA fusion was successfully displayed on the cell surface of E. coli DH5α::pKKJlipA and that the LipA domain was surface exposed.
Oil degradation by cells displaying an Ag43–LipA fusion protein on their surface
After confirming that lipase was displayed on the cell surface, we examined whether lipase-expressing cells can be used to treat lipid-containing water. E. coli DH5α::pKKJlipA was tested for its ability to degrade lipid by incubating cells in the LB medium containing 1 mM arabinose and 1 % used soya cooking oil for 7 days. It was found that 10 % of the oil was degraded after 24 h and reached the highest level of 27 % at day 6 (Fig. 3). To further characterize lipid degradation by E. coli DH5α::pKKJlipA, the changes in the composition of residual lipids was analyzed by thin layer chromatography (data not shown). After 3 days of incubation, the triglycerides were degraded efficiently for up to 6 days of incubation, while free fatty acids together with diglycerides accumulated.
Isolation of a PPB-producing Pseudomonas sp. SL31
Lipid biodegradation is generally enhanced by emulsification of the lipid substrates with biosurfactant, which increases the interaction between microbial enzymes and lipids (Desai and Banat 1997). The ability of surfactants to emulsify hydrocarbon–water mixtures has been demonstrated to increase hydrocarbon degradation significantly and is thus potentially useful for oil spill remediation (Saeki et al. 2009). Because they can easily be incorporated directly into the biological process, thus eliminating the need for additional processes and resulting in lesser operational costs, therefore, a high potential biosurfactant-producing bacterial strain is required.
Two sensitive and rapid methods, drop collapse and oil displacement, were employed to screen bacterial colonies for the production of biosurfactant (Płaza et al. 2006; Youssef et al. 2004). A drop of a culture supernatant is placed on an oil-coated surface. The drops of culture supernatant containing biosurfactant collapse while non-surfactant-containing drops remain stable (Desai and Banat 1997). Oil displacement tests were performed in which a sample of culture supernatant was gently placed at the center of an oil film overlaid on the surface of distilled water. Culture supernatants containing biosurfactants produce an area of clear halo visualized. Of the many types of surfactants produced by microorganisms, rhamnolipid is the most well studied (Sandrin et al. 2000). Since the goal was to isolate novel bacterial surfactants, PCR was performed on chromosomal DNA samples from surfactant-producing isolates using primers specific to rhlB, which encodes rhamnosyltransferase I, a key enzyme in the rhamnolipid biosynthesis pathway (Bodour et al. 2003). Isolates that were positive for rhlB were excluded.
One surfactant-producing rhlB − colony (SL31) was selected for further characterization. Strain SL31 exhibited the highest activity for both oil displacement test and drop collapsing. It is a gram negative, short rod that forms small, convex, yellow-white, smooth, and mucoid colonies on LB agar plates when cultivated for 24 h, at 28 °C. The 16SrRNA gene sequence of the SL31 isolate was compared to previously published sequences in GenBank and was found to share a high degree of sequence identity with the 16SrRNAs of many Pseudomonas species. Strain SL31 was therefore designated as Pseudomonas sp. SL31.
The culture supernatant produced by bacterial strain SL31 was found to be active in oil film displacement test (Fig. 4a). We found that culture supernatants of strain Pseudomonas sp. SL31 showed the largest oil displacement area. This strain did not contain rhlB as determined by PCR (Fig. 9S, supplementary data), suggesting that strain SL31 does not possess rhamnolipid biosynthesis genes.
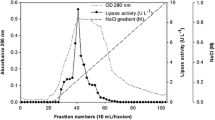
Purification and identification of PPB
To purify the active biosurfactant from strain SL31, culture supernatants were extracted twice with hexane and further purified by analytical reverse-phase HPLC resulting in two well-separated major peaks and two small peaks at different retention times (data not shown). The separation of the individual compounds in the fractions containing the peaks was achieved by reverse-phase semi-preparative HPLC. Based on the fact that one well-separated fraction obtained from semi-preparative HPLC exhibited oil displacement activity, it was subjected to structure elucidation by NMR and IR spectroscopy, as well as mass spectrometry (Fig. 1S–8S, supplementary data). The elucidated structure was found to be identical to the previously isolated compound, pseudopyronine B (PPB) (Giddens et al. 2008) (Fig. 4b), which has been demonstrated to possess antimicrobial and antimalarial activities (Giddens et al. 2008; Singh et al. 2003).
PPB is known to be produced by the marine Pseudomonas sp. F92S91 where it was initially designated α-pyrone I (Singh et al. 2003). PPB has also been isolated from cultures of Pseudomonas fluorescens (Chu et al. 2002). PPB of Pseudomonas sp. F92S91 is known to inhibit the growth of a number of pathogenic microorganisms (Singh et al. 2003). Of the bacteria tested, Pseudomonas sp. F92S91 was most active against gram-positive pathogens where its primary mode of action appeared to be the disruption of membrane function. No inhibitory activity was detected against either gram-negative bacteria or yeast; however, growth of the malaria parasite, Plasmodium falciparum, was inhibited (Singh et al. 2003). It has been further shown, in vitro, that PPB inhibits the function of three Plasmodium falciparum enzymes involved in fatty acid biosynthesis (PfFabG; the β-ketoacyl-ACP-reductase, PfFabI; the enoyl-ACP-reductase and PfFabZ; and the β-hydroxyacyl-ACP-dehydratase) (Giddens et al. 2008). PPB was found to inhibit both PfFabG and PfFabI of P. falciparum, fatty acid biosynthesis enzymes, thus providing evidence that the fatty acid biosynthesis pathway may be the cellular target of this compound (Giddens et al. 2008).
Characterization of the biosurfactant properties of PPB
While the antimicrobial properties of PPB are well documented, no examination of its biosurfactant properties has yet been reported. Therefore, the effect of PPB-containing crude hexane extracts of Pseudomonas sp. SL31 culture media on the surface tension of water was measured. The surface tension was measured using a DuNuoy ring detachment apparatus as described previously using a tensiometer at room temperature (McInerney et al. 1990), and the results were compared to those of a positive control solution of SDS. A plot of the surface tension versus initial surfactant concentration for the crude extract and SDS is shown in Fig. 5. The critical micelle concentration (CMC) is found as the point at which the two lines intersect, the baseline of minimal surface tension and the slope, where surface tension shows a linear decline. In the case of PPB, the surface tension rapidly decreased as the concentration increased, and a minimum surface tension of 41.6 mN/m was found at an initial crude extract concentration >1400 mg/l. From the break point of surface tension versus its log of the concentration curve, the CMC of the PPB is approximately 1400 mg/l. SDS is able to reduce the surface tension of water to 28.6 mN/m. The corresponding CMC value is approximately 1400 mg/l. The surface tension of distilled water is noted to be 72 mN/m, and addition of biosurfactant can lower it from 72 to 30 mN/m (Desai and Banat 1997). Incremental addition of biosurfactant to water reduces its surface tension to a critical level above which amphiphilic molecules readily form supramolecular structures like micelles, bilayers, and vesicles. This point is known as the critical micelle concentration (CMC). CMC is therefore defined as the ability of a biosurfactant to form micelles within an aqueous phase and is commonly used to measure the efficiency of a biosurfactant (Desai and Banat 1997).
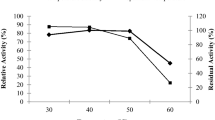
When used in wastewater treatment, a biosurfactant that can remain stable over a broad pH and temperature range is needed. In this study, solutions of either Pseudomonas sp. SL31 PPB-containing crude extract or SDS (as a surfactant control) were prepared at the CMC for use in thermal and pH stability tests. This entailed the measurement of the surface tension of each solution over a range of temperature and pH. The surfactant activity of PPB-containing extract was relatively stable within a temperature range from 0 to 120 °C and a pH range of 3–11 (Fig. 6). The surface tension of the crude extract decreased from 50.00 to 39.67 mN/m after heat treatment. In the case of SDS, the surface tension decreased slightly from 33.00 to 28.67 mN/m, indicating that PPB is stable under extreme conditions of temperature and pH.
Application of PPB in cadmium (Cd) removal from contaminated water
The application of biosurfactants in the remediation of heavy metal-contaminated soil/water has recently been investigated (Kim and Vipulanandan 2006; Rufino et al. 2012). The US Environmental Protection Agency (US EPA) lists 5 of the top 20 hazardous substances as metals including arsenic (As), lead (Pb), mercury (Hg), cadmium (Cd), and chromium (Cr) (Maier et al. 2001). Cd, one of the most toxic metals, was selected as a model in this study. PPB crude extract efficiently removed the highest amount of Cd from contaminated water samples achieving a 50.84 % Cd removal efficiency when 1 CMC was used. The removal efficiencies of DW and SDS were 4.31 and 3.45 %, respectively (Fig. 7). Therefore, PPB removed the highest amount of Cd from contaminated water compared to SDS and water, indicating that micelles formed by PPB were effective in trapping Cd in the solution. The possible mechanisms by which surfactants remove heavy metals include ion exchange, precipitation–dissolution and counterion association. Metals are removed by forming complexes with surfactants on the soil surface followed by detachment from the soil into solution due to a lowering of the interfacial tension and hence association with surfactant micelles (Juwarkar et al. 2007). However, the use of SDS and water to remove Cd from the soil had little effect compared to EDTA due to the fact that they lack chelating capacity (Chang et al. 2005). The anionic surfactant SDS has previously been shown to have only a small effect (<6 %) on cadmium removal at pH 5–11 as a consequence of its very low cadmium chelating ability (Chang et al. 2005). By contrast, similar studies using an anionic monorhamnolipid demonstrated that complexation of Cd by the biosurfactant was rapid and stable (Tan et al. 1994). It was reported that 92 % of Cd(II) (0.72 mM) was complexed by 7.3 mM rhamnolipid, while the maximum complexation capacity was 0.2 mol Cd(II)/mole rhamnolipid (Tan et al. 1994). The complexation structure between Cd(II) and PPB can be proposed based on the structural similarity between PPB and d-glucono-1,5-lactone. Previously, the interaction of d-glucono-1,5-lactone with Zn(II), Cd(II), and Hg(II) ions in both a solid and aqueous solution has been studied using NMR, FTIR, and X-ray powder diffraction measurements (Tajmir-Riahi 1989). Cd(II) ion was found to bind to two d-gluconate anions via the O-1, O-2, and O-3 of each sugar anion, giving a six-coordinate Cd(II) ion (Tajmir-Riahi 1989). The coordination stereochemistry of Cd(II) is unusually varied, and it is often found in a large number of complexes with coordination numbers ranging from 2 to 8 (Andersen 1984). The exact complexation structure between Cd(II) and PPB remains to be determined.
The ability of a number of surfactants, including biosurfactant, anionic, and nonionic surfactants, was investigated for their ability to remove Pb from contaminated water (Kim and Vipulanandan 2006). Of the surfactants used, an unidentified biosurfactant from Flavobacterium sp. had the best Pb removal efficiency of 75 % at an initial Pb concentration of 100 mg/l in water at pH > 12, while the removal efficiencies of SDS and Triton X-100 were 26 and 6 %, respectively (Kim and Vipulanandan 2006). A lipopeptide biosurfactant produced by Candida lipolytica UCP 0988 has been shown to be effective in the removal of heavy metals and reduced the surface tension of the production medium from 50 to 25 mN/m, with a yield of 8 g/l, after 72 h of fermentation (Rufino et al. 2012). The yeast lipoprotein also removed 50 % of Cd from contaminated soil (Rufino et al. 2012) which is similar to our PPB Cd removal efficiency of 51 %.
Oil degradation of bacterial pure culture and consortium
Large amounts of lipid-containing wastewater generated by factories, restaurants, and other human activities have become an important environmental problem. Lipid can cover the surface of water, thus reducing the oxygen concentration leading to the death of fish and other aquatic organisms. As described previously, lipases break down lipids into fatty acids and glycerol, so they could be used to treat lipid-containing wastewater before its release to the environment. To overcome the need for high amounts of purified enzyme, a bacterium expressing surface displayed lipase was constructed and used together with other beneficial bacteria in a consortium to efficiently degrade lipid in contaminated wastewater.
In order to ensure that all bacterial members produced lipase, we checked whether the PPB-producing strain, Pseudomonas sp. SL31, also produced lipase. This was indeed the case. Its colonies displayed lipolytic clear zones on tributyrin plates and the crude cell lysates contained specific lipase activity of 0.07 U/mg when assayed using ρ-nitrophenyllaurate (pNL) substrate. Next, we determined the oil degradation ability of monocultures of each of the bacterial strains used, namely recombinant E. coli expressing the cell surface displayed lipase-Ag43 fusion protein (pKKJlipA), E. coli expressing intracellular His-tagged lipase (pETlipA), the extracellular lipase-producing Proteus sp. SW1 (Whangsuk et al. 2013), and the PPB-producing strain, Pseudomonas sp. SL31, by culturing in LB media containing 1 % oil for 7 days. As shown in Fig. 8a, E. coli expressing His-tagged lipase (pETlipA) and Proteus sp. SW1 efficiently degraded oil (70 and 68 %, respectively), whereas Pseudomonas sp. SL31 moderately digested oil (58 %). The lowest efficiency (23 %) is the recombinant E. coli expressing cell surface display lipase (pKKJlipA) suggesting the low expression of lipase on cell surface membrane.
Oil degradation by various pure (a) and mixed cultures (b, c). a The monoculture of recombinant E. coli expressing cell surface displayed lipase (pKKJlipA) and His-tagged lipase (pETlipA). The extracellular lipase-producing Proteus sp. (SW1) and biosurfactant PPB-producing Pseudomonas sp. (SL31). b Consortium comprising all four microorganisms as in (a). The results shown are the average of three independent assays. c Consortium comprising all four microorganisms along with exogenously added PPB. Error bars show SD
To improve the oil degradation efficiency further, all of the bacterial strains were combined into a single mixed culture. Each pure bacterial culture was grown to OD600 1, induced with arabinose (in the case of recombinant E. coli strains) and biological activity checked by chromogenic assay using ρ-nitrophenyllaurate as a substrate for lipase activity and oil displacement test for PPB activity to ensure the expression of lipase and PPB before they were combined (1:1:1:1, 5 ml each) and continued shaking in the presence of oil for 7 days. The consortium degraded 29 % of used cooking oil (1 %) after a day and reached 84 % after 7 days (Fig. 8b). The result showed that the ability of the consortium culture to degrade oil was superior to that of either of the single bacterial strains (pETlipA, SW1, SL31, and pKKJlipA; 84 vs. 70, 68, 58, and 23 %, respectively) (Fig. 8a, b). In order to demonstrate that the addition of PPB-producing bacteria (SL31) to cultures of lipase-producing strains is more practical than exogenously adding purified PPB, the oil degradation rate of the PPB- and lipase-producing bacteria was compared with that of the same lipase-producing bacteria in which PPB crude extract was added. The consortium containing the PPB-producing strain was more efficient at degrading oil compared to the lipase-producing consortium to which PPB was added (84 vs. 76 %, Fig. 8c). Therefore, a consortium of PPB- and lipase-producing bacteria is more cost efficient and practical. The mixed culture demonstrated the better performance in oil degradation when compared to the single cultures of each bacterial strain. This result is in good agreement with many previous reports in which consortia out performed single strains in enzymatic degradation of a number of different pollutants (Dharmsthiti and Kuhasuntisuk 1998; Mukherjee and Bordoloi 2011; Phugare et al. 2011). A mixed culture of P. aeruginosa and Providencia sp. was shown to biodegrade dyes much better than single bacterial strains. It was hypothesized that the efficiency of the consortium depended upon versatile enzyme activities since each strain possessed two of four different dye decolorizing enzymes. This gave the mixed culture a biodegradative advantage compared to the single bacterial strains (Phugare et al. 2011). Similarly, a consortium of B. subtilis and P. aeruginosa demonstrated a 76 % petroleum oil removal from the contaminated soil after 180 days (Mukherjee and Bordoloi 2011). Mongkolthanaruk and Dharmsthiti (2002) evaluated a mixed culture composed of P. aeruginosa, Acinetobacter calcoaceticus (both lipase-producing bacteria), and Bacillus sp. (an amylase- and protease-producing bacterium) for the ability to lower the biochemical oxygen demand (BOD) value and lipid content of lipid-rich wastewater. The BOD and the lipid content were reduced from 3600 and 21,000 mg/l, respectively, at day 0, to <20 mg/l within 12 days under aerobic conditions (Mongkolthanaruk and Dharmsthiti 2002). Our PPB surfactant can dissolve hydrocarbons by forming micelles in aqueous solution so the direct interactions between cells and micelles can occur. A number of species of bacteria are able to degrade liquid hydrocarbons after adhering to the surfaces of droplets (Stelmack et al. 1999). This direct contact between a bacterial cell and a target hydrocarbon can significantly increase the rate of diffusion into the cell and change the adhesion of the hydrocarbon to the cell surface, thereby enhancing growth and increasing the apparent rate of dissolution of the hydrocarbon.
Conclusion
The lipA gene, encoding a solvent-tolerant extracellular lipase from Proteus sp. SW1, was successfully expressed on the cell surface of E. coli employing antigen 43 as an anchoring system. An E. coli strain expressing surface displayed lipase was able to degrade 27 % of the oil after 6 days of incubation. Pseudomonas sp. SL31 was isolated and found to produce PPB, the critical micelle concentration of which was determined to be 1400 mg/l. PPB was stable within a temperature range from 0 to 120 °C and a pH range of 3–11. PPB-containing crude extracts efficiently removed cadmium (up to 50.84 %) from contaminated water samples. The consortium was composed of: E. coli expressing a cell surface displayed lipase–antigen 43 fusion protein along with an intracellular His-tagged lipA, Proteus sp. SW1, and Pseudomonas sp. SL31. The consortium showed 84 % oil removal after 7 days in media containing 1 % oil.
References
Abalos A et al (2004) Enhanced biodegradation of casablanca crude oil by a microbial consortium in presence of a rhamnolipid produced by Pseudomonas aeruginosa AT10. Biodegradation 15(4):249–260
Gurung N et al (2013) A broader view: microbial enzymes and their relevance in industries, medicine, and beyond. BioMed Res Int 2013:1–18
Andersen O (1984) Chelation of cadmium. Environ Health Perspect 54:249–266
Becker S et al (2005) A generic system for the Escherichia coli cell-surface display of lipolytic enzymes. FEBS Lett 579(5):1177–1182
Bodour AA, Drees KP, Maier RM (2003) Distribution of biosurfactant-producing bacteria in undisturbed and contaminated arid southwestern soils. Appl Environ Microbiol 69(6):3280–3287
Cammarota MC, Freire DMG (2006) A review on hydrolytic enzymes in the treatment of wastewater with high oil and grease content. Bioresour Technol 97(17):2195–2210
Cerqueira VS et al (2011) Biodegradation potential of oily sludge by pure and mixed bacterial cultures. Bioresour Technol 102:11003–11010
Chang JS et al (2004) Enhancement of phenanthrene solubilization and biodegradation by trehalose lipid biosurfactants. Environ Toxicol Chem 23(12):2816–2822
Chang SH et al (2005) Remediation of metal-contaminated soil by an integrated soil washing-electrolysis process. Soil Sediment Contam 14(6):559–569
Chu M et al (2002) Structure of sch 419560, a novel α-pyrone antibiotic produced by Pseudomonas fluorescens. J Antibiot 55(2):215–218
Desai JD, Banat IM (1997) Microbial production of surfactants and their commercial potential. Microbiol Mol Bio Rev 61(1):47–64
Dharmsthiti S, Kuhasuntisuk B (1998) Lipase from Pseudomonas aeruginosa LP602: biochemical properties and application for wastewater treatment. J Ind Microbiol Biotechnol 21(1–2):75–80
Giddens AC et al (2008) Natural product inhibitors of fatty acid biosynthesis: synthesis of the marine microbial metabolites pseudopyronines A and B and evaluation of their anti-infective activities. Tetrahedron Lett 64(7):1242–1249
Gupta R et al (2015) Molecular and functional diversity of yeast and fungal lipases: their role in biotechnology and cellular physiology. Prog Lipid Res 57:40–54
Hasman H, Chakraborty T, Klemm P (1999) Antigen-43-mediated autoaggregation of Escherichia coli is blocked by fimbriation. J Bacteriol 181(16):4834–4841
Henderson IR, Owen P (1999) The major phase-variable outer membrane protein of Escherichia coli structurally resembles the immunoglobulin A1 protease class of exported protein and is regulated by a novel mechanism involving dam and OxyR. J Bacteriol 181(7):2132–2141
Jorfi S et al (2013) Application of biosurfactants produced by Pseudomonas aeruginosa SP4 for bioremediation of soils contaminated by pyrene. Soil Sediment Contam 22(8):890–911
Jose J, Maas RM, Teese MG (2012) Autodisplay of enzymes—molecular basis and perspectives. J Biotechnol 161(2):92–103
Juwarkar AA et al (2007) Biosurfactant technology for remediation of cadmium and lead contaminated soils. Chemosphere 68(10):1996–2002
Kim J, Vipulanandan C (2006) Removal of lead from contaminated water and clay soil using a biosurfactant. J Environ Eng 132:777–786
Kjaergaard K et al (2002) Antigen 43-mediated autotransporter display, a versatile bacterial cell surface presentation system. J Bacteriol 184(15):4197–4204
Koma D et al (2003) Degradation of car engine base oil by Rhodococcus sp. NDKK48 and Gordonia sp. NDKY76A. Biosci Biotechnol Biochem 67(7):1590–1593
Lee SH, Lee SY, Park BC (2005) Cell surface display of lipase in Pseudomonas putida KT2442 using OprF as an anchoring motif and its biocatalytic applications. Appl Environ Microbiol 71(12):8581–8586
Maier RM et al (2001) Remediation of metal-contaminated soil and sludge using biosurfactant technology. Int J Occup Med Environ Health 14(3):241–248
McInerney MJ, Javaheri M, Nagle DP (1990) Properties of the biosurfactant produced by Bacillus licheniformis strain JF-2. J Ind Microbiol 5(2–3):95–101
Mongkolthanaruk W, Dharmsthiti S (2002) Biodegradation of lipid-rich wastewater by a mixed bacterial consortium. Int Biodeterior Biodegrad 50(2):101–105
Mukherjee AK, Bordoloi NK (2011) Bioremediation and reclamation of soil contaminated with petroleum oil hydrocarbons by exogenously seeded bacterial consortium: a pilot-scale study. Environ Sci Technol 18(3):471–478
Phugare SS et al (2011) Textile dye degradation by bacterial consortium and subsequent toxicological analysis of dye and dye metabolites using cytotoxicity, genotoxicity and oxidative stress studies. J Hazard Mater 186(1):713–723
Płaza GA, Zjawiony I, Banat IM (2006) Use of different methods for detection of thermophilic biosurfactant-producing bacteria from hydrocarbon-contaminated and bioremediated soils. J Pet Sci Technol 50(1):71–77
Pornsunthorntawee O et al (2008) Structural and physicochemical characterization of crude biosurfactant produced by Pseudomonas aeruginosa SP4 isolated from petroleum-contaminated soil. Bioresour Technol 99(6):1589–1595
Robichon C et al (2011) Engineering Escherichia coli BL21 (DE3) derivative strains to minimize E. coli protein contamination after purification by immobilized metal affinity chromatography. Appl Environ Microbiol 77(13):4634–4646
Rodrigues LR et al (2006) Physicochemical and functional characterization of a biosurfactant produced by Lactococcus lactis 53. Colloids Surf B 49(1):79–86
Rufino RD et al (2012) Application of the biosurfactant produced by Candida lipolytica in the remediation of heavy metals. Chem Eng Trans 27:61–66
Saeki H et al (2009) Oil spill remediation by using the remediation agent JE1058BS that contains a biosurfactant produced by Gordonia sp. strain JE-1058. Bioresour Technol 100(2):572–577
Sandrin TR, Chech AM, Maier RM (2000) A rhamnolipid biosurfactant reduces cadmium toxicity during naphthalene biodegradation. Appl Environ Microbiol 66(10):4585–4588
Singh MP et al (2003) Novel alpha-pyrones produced by a marine Pseudomonas sp. F92S91: taxonomy and biological activities. J Antibiot 56:1033–1044
Stelmack PL, Gray MR, Pickard MA (1999) Bacterial adhesion to soil contaminants in the presence of surfactants. Appl Environ Microbiol 65(1):163–168
Tajmir-Riahi HA (1989) Carbohydrate metal ion complexes. Interaction of d-glucono-1,5-lactone with Zn(II), Cd(II), and Hg(I1) ions in the solid and aqueous solution, studied by 13C-NMR, FT-IR, and X-ray powder diffraction measurements. Can J Chem 67:651–654
Tan H et al (1994) Complexation of cadmium by a rhamnolipid biosurfactant. Environ Sci Technol 28(13):2402–2406
Whangsuk W et al (2013) Gene cloning and characterization of a novel highly organic solvent tolerant lipase from Proteus sp. SW1 and its application for biodiesel production. Mol Biotechnol 53(1):55–62
Youssef NH et al (2004) Comparison of methods to detect biosurfactant production by diverse microorganisms. J Microbiol Methods 56:339–347
Acknowledgments
This research work was supported in part by a grant from the Chulabhorn Research Institute and Center of Excellence on Environmental Health and Toxicology, Thailand. Special thanks go to Piyajit Watcharasit for Texas red-conjugated anti-rabbit IgG antibody, Nuchanart Rangkadilok and Supachai Ritruechai for preparative HPLC, Amnart Khongmanee for his kind assistance with fluorescence microscopes, James Dubbs for critical reading and discussion, Pakwilai Chouichit for technical assistance, and also to Piyapol Munpiyamit for photograph preparation. We appreciate the funding by the Royal Golden Jubilee Ph.D. Program (PHD/0144/2554) to Sirinthra Thiengmag.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Thiengmag, S., Chuencharoen, S., Thasana, N. et al. Bacterial consortium expressing surface displayed, intra- and extracellular lipases and pseudopyronine B for the degradation of oil. Int. J. Environ. Sci. Technol. 13, 2067–2078 (2016). https://doi.org/10.1007/s13762-016-1034-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13762-016-1034-z