Abstract
Background and purpose
Studies on the relationship between Phosphodiesterase 4 D (PDE4D) gene polymorphism with the risk of ischemic stroke (IS) have shown discordant results. The present meta-analysis was aimed to clarify the relationship between PDE4D gene polymorphism with the risk of IS by estimating pooled analysis of published epidemiological studies.
Methods
A comprehensive literature search for all the published articles was performed in various electronic databases, including PubMed, EMbase, Cochrane Library, Trip Database, Worldwide Science, CINAHL, and Google Scholar up to 22nd December 2021. Pooled Odds ratios (ORs) with 95% Confidence Intervals (CIs) under dominant, recessive, and allelic models were calculated. Subgroup analysis based on ethnicity (Caucasian vs. Asian) was performed to examine the reliability of these findings. Sensitivity analysis was also performed to detect the heterogeneity between studies. Finally, Begg’s funnel plot was used to assess the potential for publication bias.
Results
In our meta-analysis, we identified a total of 47 case–control studies with 20,644 ischemic stroke (IS) cases and 23,201 control subjects, including 17 studies of Caucasian descent and 30 studies of Asian descent. Our findings suggest that there was a significant relationship between SNP45 gene polymorphism and risk of IS (Recessive model: OR = 2.06, 95% CI 1.31–3.23), SNP83 overall (allelic model: OR = 1.22, 95% CI 1.04–1.42), Asian (allelic model: OR = 1.20, 95% CI 1.05–1.37), and SNP89 Asian (Dominant model: OR = 1.43, 95% CI 1.29–1.59, recessive model: OR = 1.42, 95% CI 1.28–1.58) respectively. However, no significant relationship was found between SNP32, SNP41, SNP26, SNP56, and SNP87 gene polymorphisms and risk of IS.
Conclusion
Findings of this meta-analysis conclude that SNP45, SNP83, and SNP89 polymorphism could be capable of increasing stroke susceptibility in Asians but not in the Caucasian population. Genotyping of SNP 45, 83, 89 polymorphisms may be used as a predictor for the occurrence of IS.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Stroke is a heterogeneous complex disorder and the second most common cause of death and long-term disability with higher incidence among adults and older people worldwide and is a significant public health problem [1]. Stroke can be divided into two subtypes: 20% of cases are hemorrhagic (intracerebral hemorrhage), and 80% are ischemic stroke (IS) [2, 3]. Characterization of a sudden decrease in blood flow to one or more areas within the central nervous system is considered ‘Ischemic stroke’. In contrast, hemorrhagic stroke occurs when a weakened blood vessel rupture.
Both genetic and lifestyle-related factors may increase the risk of stroke. Genetic factors involving single nucleotide polymorphisms (SNPs) can indicate susceptibility to specific stroke subtypes. Numerous mutations in microsatellite markers and SNPs of the STRK1 locus, chromosome 5q12, encoding phosphodiesterase (PDE) type 4D (PDE4D), were identified as independent risk factors for IS [3]. PDEs are intracellular enzymes that degrade the cyclic nucleotides adenosine (cAMP) and/or guanosine monophosphate (cGMP), thereby modulating cellular signaling via the cAMP/cGMP pathways [4]. In IS, suppression of PDE4D protects the blood–brain barrier and reduces both inflammation and thrombosis. Inflammation plays a significant role in the development of atherosclerosis, and PDE4 has been significantly expressed in inflammatory cells during cerebral ischemia [5]. Variants and polymorphisms in the PDE4D gene have been investigated in different IS population to ascertain any associated risks [6]. In the present study, we retrospectively reviewed studies focusing on the relationship of PDE4D gene polymorphism with increasing or decreasing risk of IS in patients published to date.
Methods
Literature search
The current systematic review was reported according to the Preferred Reporting Items for Systematic Reviews and Meta-analysis (PRISMA) statement guidelines, and a meta-analysis was conducted with the extracted data [7]. Relevant publications (until 31st October 2021) were identified by searching various electronic databases, including PubMed, EMbase, Cochrane Library, Trip Database, Worldwide Science, CINAHL, and Google Scholar. The following key terms were used: ‘Phosphodiesterase 4 D’ OR ‘PDE4D’ AND ‘polymorphism’ OR ‘variant’ OR ‘mutation’ OR ‘genotype’ AND ‘stroke’ OR ‘ischemic stroke’ OR ‘Cerebral Infarction’ OR ‘Brain Infarction’ OR ‘Cerebrovascular disease’ OR ‘Cerebrovascular disorder’ in combination with ‘SNP 45’ OR ‘SNP 56’ OR ‘SNP 87’ OR ‘SNP 89’ OR ‘SNP 32’ OR ‘SNP 41’ OR ‘SNP 83’ OR SNP ‘26’ OR ‘rs966221’ OR ‘rs40512’ OR ‘rs12188959’ OR ‘rs702553’ OR ‘rs2910829’ OR ‘rs1396476’. Additionally, the reference list of the retrieved studies, review articles, and previous meta-analyses, were manually searched for collecting more relevant studies often missed while performing the electronic search. Two authors (MN and PS) independently conducted the search for various studies, and any discrepancy regarding the inclusion or exclusion of individual studies was resolved by consulting the corresponding author (PK).
Eligibility criteria
Studies were included if they met the following inclusion criteria: (1) case–control studies investigating the relationship of PDE4D gene polymorphism with the risk of IS; (2) clinically confirmed diagnosis of IS using CT or MRI scan; (3) patients aged < 18 years for pediatric and > 18 years for adult population; (4) numbers available for patient and control groups for PDE4D SNP 45, 32, 41, 26, 56, 83, 87, 89 genotype or data provided from which the numbers could be calculated. No restriction of language was applied.
Data extraction
Two investigators (PK and PS) independently extracted the data. Following data were extracted from each study: first author’s name, published year, ethnicity, country, number of cases and controls, matching criteria, sample source, duration of inclusion, mean age, genotyping method and frequency distribution of PDE4D SNP 45, 32, 41, 26, 56, 83, 87, 89 genotypes. Hardy–Weinberg Equilibrium (HWE) was calculated for allelic frequency distribution. Ethnicities were categorized as either Asian or Caucasian, and the population was classified into adult and paediatric groups.
Quality assessment
Newcastle–Ottawa Scale (NOS) [8] was used for assessing the quality of the included studies based on three components: selection, comparability and ascertainment of the outcome. Scores ranged from 01 to 09. Two authors (PS and MN) independently assessed the quality of the included studies. Discrepancies over quality scores were resolved by discussion among all the authors and subsequent consensus was reached.
Statistical analysis
Odds ratios (ORs) with 95% Confidence Intervals (CIs) were calculated to investigate the relationship between PDE4D (SNP 45, 32, 41, 26, 56, 83, 87, 89) gene polymorphism and risk of IS using fixed (Mantel–Haenszel method) or random effects (Dersimonian and Laird method) models [9, 10]. Heterogeneity between studies was compared using Cochran’s Q statistic and I2 metric [11, 12]. I2 metric was used to describe the degree of heterogeneity between included studies, where 0–25% indicated no observed heterogeneity and larger values showed increasing heterogeneity, with 25–50% regarded as low, 50–75% as moderate and 75–100% as high. If I2 < 50%, a fixed-effects model was used, and if I2 > 50%, a random-effects model was used. In addition, heterogeneity between studies was adjusted by subgroup analysis, HWE status, and meta-regression by quality score of the included studies. Meta-regression was conducted using the Restriction maximum likelihood (REML) estimate with Knapp-Hartung modification for determining the between-study variance [13].
One-way sensitivity analyses were performed to assess the stability of the results. A single study in the meta-analysis was deleted each time to reflect the influence of the individual dataset on the pooled OR. Funnel plots and Egger’s linear regression test were used to diagnose the potential publication bias [14, 15]. Presence of selection bias in control participants was evaluated by calculating HWE, and genotypic frequencies of the control subjects were compared using the chi-square test. Stratified analyses based on ethnicity (Asian vs. Caucasian) were performed. The reliability and accuracy of the results were validated by two investigators (PK and PS), who independently analyzed the data using the software. All statistical analyses were performed using STATA 13.0 and Review Manager 5.3 software. The p values were two-sided, and a p value < 0.05 was statistically significant.
Results
Literature search
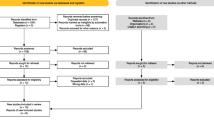
The initial search yielded 72 records from PubMed, EMbase, Scopus, Web of Science, CENTRAL (Cochrane Central Register of Controlled Trials), and Google scholar databases. Of them, 22 were excluded after the review of title/abstract, leaving 50 potential studies for full-text information review. Finally, 47 studies met the inclusion criteria and were included in this study (Fig. 1).
Characteristics of eligible studies
The main characteristics of included studies are presented in Table 1. The publication years of the studies included in our analysis ranged from 2003 to 2019. The sample size in each study ranged from 91 to 2890 in IS cases and 44 to 4412 in controls subjects for all the included studies in our meta-analysis. Forty-seven case–control studies (18 studies for SNP45, 12 for SNP56, 29 for SNP83, six for SNP89, five for SNP26, seven for SNP41, four studies for SNP32, and 27 studies for SNP87) were included in our meta-analysis. Studies were carried out in two major ethnic populations; 30 studies were in the Asian while 17 studies were in the Caucasian population. All studies in this meta-analysis had controls in HWE. The quality scores of all included studies were moderately high. Out of 47 studies, 27 studies had hospital-based and 20 studies had a population-based source of controls. Table 1 summarizes the characteristics and methodological quality of all the included studies. In our meta-analysis, a total of 47 case–control studies involving 20,644 IS cases and 23,201 controls were included.
Relationship between PDE4D SNP 45 gene polymorphism and ischemic stroke risk
No significant relationship was observed between PDE4D SNP45 gene polymorphism and risk of IS, under overall 18 studies, dominant model (OR = 1.00, 95% CI 0.95–1.04), recessive model (OR = 1.11, 95% CI 0.83–1.47), and allelic model (OR = 1.02, 95% CI 0.90–1.16). Upon conducting subgroup analysis on the basis of ethnicity of study population, significant association was observed in Asian population in case of recessive model (OR = 2.06, 95% CI 1.31–3.23); but not in dominant model (OR = 0.98, 95% CI 0.87–1.09) and allelic model (OR = 1.32, 95% CI 0.81–2.17) respectively [Figs. 2A, 3A, 4A)] [Table 2]. In Caucasian population, no significant association was observed under dominant model (OR = 1.00, 95% CI 0.95–1.05), recessive model (OR = 0.98, 95% CI 0.75–1.29) and allelic model (OR = 0.97, 95% CI 0.88–1.07) respectively [Figs. 2A, 3A, 4A] [Table 2].
Relationship between PDE4D SNP 56 gene polymorphism and ischemic stroke risk
Our results revealed that there were no significant relationship between PDE4D SNP56 gene polymorphism and risk of IS, under dominant model (OR = 1.01, 95% CI 0.96–1.07), recessive model (OR = 1.04, 95% CI 0.91–1.18), and allelic model (OR = 1.04, 95% CI 0.90–1.20). Upon subgroup analysis of the data on ethnicity of the study population, no significant association was observed in the Asian population under dominant model (OR = 1.06, 95% CI 0.94–1.20), recessive model (OR = 1.17, 95% CI 0.87–1.57) and allelic model (OR-1.16, 95% CI 0.87–1.55) respectively [Figs. 2B, 3B, 4B)] [Table 2]. Additionally, we did not find any correlation of SNP56 polymorphism with an increased or decreased risk of IS in all the three genetic models in Caucasian population, as can be observed under dominant model (OR = 0.99, 95% CI 0.93–1.07), recessive model (OR = 1.00, 95% CI 0.90–1.11) and allelic model (OR = 0.97, 95% CI 0.84–1.12) respectively [Figs. 2B, 3B, 4B)] [Table 2].
Relationship between PDE4D SNP 83 gene polymorphism and ischemic stroke risk
For SNP83, upon analysing 29 studies, results showed no significant relationship between PDE4D SNP83 gene polymorphism and risk of IS, under dominant model (OR = 0.98, 95% CI 0.94–1.03), and recessive model (OR = 1.11, 95% CI 0.97–1.27); but a significant association was observed under allelic model (OR = 1.22, 95% CI 1.04–1.42). Upon performing subtype analysis on the basis of ethnicity of the study population, no significant association was observed in the Asian population in dominant model (OR = 0.97, 95% CI 0.91–1.03), and recessive model (OR = 1.19, 95% CI 0.97–1.45); but a significant association in allelic model (OR-1.20, 95% CI 1.05–1.37) was observed [Figs. 2D, 3D, 4D] [Table 2]. In the Caucasian population, no significant association was observed under dominant model (OR = 1.01, 95% CI 0.94–1.08), recessive model (OR = 0.98, 95% CI 0.89–1.09) and allelic model (OR = 1.26, 95% CI 0.80–1.98) respectively [Figs. 2D, 3D, 4D] [Table 2].
Relationship between PDE4D SNP 26 gene polymorphism and ischemic stroke risk
For SNP26, we failed to find a significant genetic association between PDE4D SNP26 gene and risk of IS, under overall dominant model (OR = 0.99, 95% CI 0.91–1.08), recessive model (OR = 0.99, 95% CI 0.89–1.11), and allelic model (OR = 0.98, 95% CI 0.89–1.08) [Figs. 2F, 3F, 4F] [Table 2].
Relationship between PDE4D SNP 89 gene polymorphism and ischemic stroke risk
We did not find any association of the SNP89 gene polymorphism with increased or decreased risk of stroke under dominant model (OR = 1.06, 95% CI 0.88–1.28), recessive model (OR = 1.02, 95% CI 0.85–1.24), and allelic model (OR = 0.99, 95% CI 085–1.15) respectively. After the data were stratified according to ethnicity of the study population, our results showed significant association in the Asian population under dominant model (OR = 1.43, 95% CI 1.29–1.59), and recessive model (OR = 1.42, 95% CI 1.28–1.58); but no significant association was observed in allelic model (OR–0.93, 95% CI 0.74–1.16) [Figs. 2E, 3E, 4E] [Table 2]. In the Caucasian population, no significant association was observed under dominant model (OR = 0.97, 95% CI 0.90–1.05), recessive model (OR = 0.97, 95% CI 0.88–1.05) and allelic model (OR = 1.00, 95% CI 0.83–1.21) respectively [Figs. 2E, 3E, 4E] [Table 2].
Relationship between PDE4D SNP 32 gene polymorphism and ischemic stroke risk
No significant relationship was observed between PDE4D SNP32 gene polymorphism and risk of IS, under overall dominant model (OR = 0.89, 95% CI 0.71–1.12), recessive model (OR = 1.08, 95% CI 0.82–1.44), and allelic model (OR = 1.25, 95% CI 0.96–1.61). Upon conducting subgroup analysis on the basis of ethnicity of study population, no significant association was observed in Asian population under dominant model (OR = 0.87, 95% CI 0.65–1.17), recessive model (OR = 1.11, 95% CI 0.73–1.68) and allelic model (OR-1.33, 95% CI 0.94–1.90) respectively [Figs. 2H, 3H, 4H] [Table 2]. Also in the Caucasian population, no significant association was observed under dominant model (OR = 0.98, 95% CI 0.76–1.26), recessive model (OR = 1.08, 95% CI 0.78–1.50) and allelic model (OR = 1.04, 95% CI 0.81–1.33) respectively [Figs. 2H, 3H, 4H] [Table 2].
Relationship between PDE4D SNP 41 gene polymorphism and ischemic stroke risk
The pooled effect among all studies estimates that there were no significant association between PDE4D SNP41 gene polymorphism and risk of IS, under overall dominant model (OR = 1.08, 95% CI 0.98–1.19), recessive model (OR = 0.91, 95% CI = 0.81–1.02), and allelic model (OR = 0.89, 95% CI 0.76–1.05) respectively. Upon conducting subgroup analysis on the basis of ethnicity of the study population, no significant association was observed in the Asian population under dominant (OR = 1.15, 95% CI 0.98–1.35) and allelic models (OR = 0.89, 95% CI 0.70–1.12) respectively. However, a protective association was observed under the recessive model (OR = 0.80, 95% CI 0.66–0.97) [Figs. 2G, 3G, 4G] [Table 2]. Also in Caucasian population no significant association was observed under dominant model (OR = 1.07, 95% CI 0.89–1.30), recessive model (OR = 0.98, 95% CI 0.84–1.13) and allelic model (OR = 0.89, 95% CI 0.70–1.14) respectively [Figs. 2G, 3G, 4G] [Table 2].
Relationship between PDE4D SNP 87 gene polymorphism and ischemic stroke risk
We conducted a meta-analysis for 27 eligible studies. In the overall population, no significant association was observed between SNP87 and IS under dominant model (OR = 0.98, 95% CI 0.95–1.02), recessive model (OR = 0.99, 95% CI 0.94–1.03), and allelic model (OR = 1.04, 95% CI 0.97–1.12) respectively. Eighteen independent analyses of Asian and nine independent analyses of Caucasian populations were conducted. Subgroup analysis on the basis of ethnicity of study population observed no significant association in the Asian population under dominant model (OR = 0.98, 95% CI 0.93–1.02), recessive model (OR = 0.99, 95% CI 0.94–1.05) and allelic model (OR—1.06, 95% CI 0.95–1.19) respectively [Figs. 2C, 3C, 4C] [Table 2]. Also in the Caucasian population, no significant association was observed under dominant model (OR = 0.99, 95% CI 0.94–1.05), recessive model (OR = 0.98, 95% CI 0.90–1.06) and allelic model (OR = 1.02, 95% CI 0.94–1.10) respectively [Figs. 2C, 3C, 4C] [Table 2].
Publication bias
For SNPs 45, 32, 41, 26, 56, 83, 87, 89, publication bias arising from the literature was qualitatively estimated by funnel plots and quantitatively examined by Begg’s and Egger’s test. It was observed that all the plots were roughly symmetrical, indicating no publication bias was present as shown in the supplementary file as figures: S1-A, S2-A, S3-A, S4-A, S5-A, S6-A, S7-A and S8-A. In addition, Begg’s and Egger’s test did not have any significant p values for the different effect sizes (p value > 0.05).
Sensitivity analyses
Moreover, sensitivity analyses were carried out to assess the influence of each of the SNPs (SNP 45, 32, 41, 26, 56, 83, 87, 89) on the overall study by sequential omission of each eligible study. By removing individual studies, no statistical variation of pooled OR was seen. Our result indicated that no study influenced the quality of the pooled ORs and the current meta-analysis was reliable and robust [Figures: S1-B, S2-B, S3-B, S4-B, S5-B, S6-B, and S7-B].
Meta-regression analysis
Meta-regression analysis based on the quality score for the relationship between PDE4D SNP45 gene polymorphism and the risk of IS did not confirm any deviation from the findings of the meta-analysis (p = 0.86) [Figures: S1-C, S2-C, S3-C, S4-C, S5-C, S6-C, S7-C, and S8-B].
Discussion
The current systematic review and meta-analysis involving 47 studies consisted of 20,644 IS cases and 23,201 controls. The association between eight PDE4D SNPs (45, 32, 41, 26, 56, 83, 87, 89) and the risk of IS was analyzed. For SNP83, we included 29 eligible studies, comprising 10,639 cases and 14,234 controls. SNP83 was associated with a statistically increased risk of IS in overall analysis (OR = 1.22, 95% CI 1.04–1.42) under the allelic model. Moreover, we found a significant association in the subgroup analysis of the Asian population in the allelic model of the polymorphism (OR—1.20, 95% CI 1.05–1.37). In a previously published meta-analysis by Wang et al., 2017 [61], a total of 26 studies were included and a significant association was observed between the SNP83 genetic polymorphism and risk of IS in the Asian population under dominant (OR = 1.19, 95% CI 1.02–1.38) and allelic (OR = 1.25, 95% CI 1.06–1.48) models. Moreover, Yan et al. [62] observed a significant association in the overall population under dominant (OR = 1.15, 95% CI 1.02–1.30), recessive (OR = 1.21, 95% CI 1.02–1.42), and allelic (OR = 1.19, 95% CI 1.06–1.33) models. On subgroup analysis, a significant association was found in the Asian population under dominant (OR = 1.20, 95% CI 1.13–1.44), recessive (OR = 1.48, 95% CI 1.22–1.79), and allelic (OR = 1.35, 95% CI 1.16–1.57) models, respectively. There was no significant association of SNP83 of PDE4D in the Asian subgroup analysis in our study with the risk of IS under the dominant and recessive genetic models. In the studies by Wu et al. [63] and Xue et al. [64], a significant association was seen in the overall analysis (OR = 1.45, 95% CI 1.19–1.76 and OR = 1.42, 95% CI 1.44–1.77, respectively). However, Yoon et al. [65] only reported the significance of SNP83 association with the risk of IS in the Asian subgroup populations under all three genetic models. Our study did not find any overall significance of SNP83 with the risk of IS. Moreover, the Caucasian populations did not have any significant association of SNP83 in PDE4D and IS risk.
For SNP45, we observed that the PDE4D gene was not significantly associated with IS in the overall analysis, which is in accordance with the findings of Yoon et al. [65]. However, on conducting subgroup analysis stratified by ethnicity, a significant association between SNP45 and risk of IS was found in the Asian population, only under the recessive model (OR = 2.06, 95% CI 1.31–3.23). A meta-analysis by Zhang et al. [66] involving 8731 cases and 10,756 controls showed no significant association of SNP56 in PDE4D with the risk of IS, which is consistent with the findings of our study. Yoon et al. [65] also observed no significant association in their meta-analysis. The analysis of SNP56 consisting of 13 published studies with 6064 cases and 9612 controls, failed to find an association between SNP56 gene polymorphism and risk of IS. The inclusion of SNP87 with 27 studies comprised of 12,651 cases and 16,133 controls also demonstrated that there was no relation between SNP87 and IS. Liang et al. [67] did not find any significance for SNP87 polymorphism in PDE4D and the risk of IS after incorporating data from 18 studies with 8363 cases and 12,223 controls. Moreover, Xu et al. [64] also reported no observable associations between SNP87 PDE4D polymorphism and risk of IS. The present meta-analysis on SNP87 also did not show any association with increased risk of IS in the overall population and the ethnicity subgroups. For SNPs 26 and 89, no correlation was observed with increased or decreased risk of IS among Asian or European populations [65]. We also did not observe any significant association of SNP26 under any of the genetic models of PDE4D with IS risk for Asian and Caucasian ethnicities. However, SNP89 of PDE4D has a significant association with the risk of IS in the Asian population under dominant and recessive models in our meta-analysis, providing a correlation of SNP89 in patients of Asian descent. We also reported the analysis of two additional SNPs, 32 and 41, which were not included in previous meta-analyses. Although SNP 32 was not associated with any significant risk of IS, SNP41 of PDE4D showed significant protective association with IS in the Asian subgroup under the recessive model genetic correlation. Although, the sample sizes for these two SNPs were relatively lower, further studies are warranted to establish their association with IS pathophysiology. The detailed effect sizes of the different meta-analyses previously published on different SNPs of PDE4D have been depicted in Table 3.
We included eight SNPs in our meta-analysis which accounts for the largest meta-analysis on PDE4D done to date for analyzing the association of the risk of IS occurrence. Unlike previous studies, we did not find any significant association between the eight SNPs of PDE4D with the risk of IS in the overall population and the Caucasian subgroup. The significance of risk association of PDE4D polymorphism with IS was observed in the Asian population under specific genetic models in SNPs 45, 83, and 89. Moreover, SNP45 showed a protective association against IS in the Asian population. Polymorphism in PDE4D is diverse, and its relation to the associated risk of cerebral ischemic stroke has been quite intriguing. Despite the heterogeneity in sample size, the overall effect size of the studies showed a significant association in the results without any deviation. The risk conferred by the SNPs of the gene is non-significant in the overall and the Caucasian population. Although the Asian population is predisposed towards IS risk associated with specific polymorphisms of PDE4D, several elements need to be conferred upon, which could have added possible confounding effects to the resultant effect size. Interest in PDE4D stems from its significant involvement in the pathophysiology of IS governing inflammation and structural damage to the neural architecture. SNPs 39 and 45 are found to be involved in small vessel infarction in an isolated Caucasian population group, suggesting the allelic significance of PDE4D polymorphisms in IS physiology [21]. Moreover, targeted PDE4D inhibitors have also depicted favorable outcomes through neuroplastic improvements and anti-inflammatory responses in stroke recovering patients [68].
Despite the significance of a large-scale meta-analysis, our study had certain limitations to be considered for interpreting the results. First, there was significant heterogeneity in the effect size of the outcomes. The studies had variable sample sizes, which affected the individual strength of the studies on overall results. Since most of the studies were of case–control design, the sample sizes were usually < 1000. However, sensitivity analyses and meta-regression did not depict any significant deviation from the observed effect size, validating our results. Therefore, large-scale prospective cohort-based and case–control studies should be conducted to validate these polymorphisms. Second, most of the included studies were from the Asian population, and the observed overall effect size could have been affected by their association. However, the number of Caucasian studies included in the meta-analysis was the highest to date, and the effect size observed provides significant credibility. Third, the effect of confounders such as age, sex, and IS subtypes was not analyzed in the study, which could have impacted the desired outcome and overall heterogeneity. Lastly, unpublished or ongoing studies were not incorporated in the analysis, possibly introducing selection bias in the observed effect size.
Conclusion
The association between PDE4D gene polymorphism and the risk of IS is debatable. The present meta-analysis showed some possible associations of PDE4D with the risk of IS in Asian populations while other studies did not. Further large-scale genome-wide association studies are warranted to ascertain the association of PDE4D gene polymorphism and IS risk.
Data availability
All data used in this study are included in the article and its supplementary materials.
References
Boehme AK, Esenwa C, Elkind MSV (2017) Stroke risk factors, genetics, and prevention. Circ Res 120:472–495. https://doi.org/10.1161/CIRCRESAHA.116.308398
Adams HP, Bendixen BH, Kappelle LJ, Biller J, Love BB, Gordon DL, Marsh EE (1993) Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial. TOAST. Trial of Org 10172 in Acute Stroke Treatment. Stroke 24:35–41. https://doi.org/10.1161/01.STR.24.1.35
Gretarsdottir S, Thorleifsson G, Reynisdottir ST, Manolescu A, Jonsdottir S, Jonsdottir T, Gudmundsdottir T, Bjarnadottir SM, Einarsson OB, Gudjonsdottir HM, Hawkins M, Gudmundsson G, Gudmundsdottir H, Andrason H, Gudmundsdottir AS, Sigurdardottir M, Chou TT, Nahmias J, Goss S, Sveinbjörnsdottir S, Valdimarsson EM, Jakobsson F, Agnarsson U, Gudnason V, Thorgeirsson G, Fingerle J, Gurney M, Gudbjartsson D, Frigge ML, Kong A, Stefansson K, Gulcher JR (2003) The gene encoding phosphodiesterase 4D confers risk of ischemic stroke. Nat Genet 35:131–138. https://doi.org/10.1038/ng1245
Omori K, Kotera J (2007) Overview of PDEs and their regulation. Circ Res 100:309–327. https://doi.org/10.1161/01.RES.0000256354.95791.f1
Pincelli C, Schafer PH, French LE, Augustin M, Krueger JG (2018) Mechanisms underlying the clinical effects of apremilast for psoriasis. J Drugs Dermatol JDD 17:835–840
Das S, Roy S, Munshi A (2016) Association between PDE4D gene and ischemic stroke: recent advancements. Int J Neurosci 126:577–583. https://doi.org/10.3109/00207454.2015.1051621
Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, Shamseer L, Tetzlaff JM, Akl EA, Brennan SE, Chou R, Glanville J, Grimshaw JM, Hróbjartsson A, Lalu MM, Li T, Loder EW, Mayo-Wilson E, McDonald S, McGuinness LA, Stewart LA, Thomas J, Tricco AC, Welch VA, Whiting P, Moher D (2021) The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. https://doi.org/10.1136/bmj.n71
Stang A (2010) Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 25:603–605. https://doi.org/10.1007/s10654-010-9491-z
DerSimonian R (1996) Meta-analysis in the design and monitoring of clinical trials. Stat Med 15:1237–1248 (Discussion 1249–1252)
Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22:719–748
Higgins JPT, Thompson SG (2002) Quantifying heterogeneity in a meta-analysis. Stat Med 21:1539–1558. https://doi.org/10.1002/sim.1186
Higgins JPT, Thompson SG, Deeks JJ, Altman DG (2003) Measuring inconsistency in meta-analyses. BMJ 327:557–560. https://doi.org/10.1136/bmj.327.7414.557
Hartung J, Knapp G (2001) A refined method for the meta-analysis of controlled clinical trials with binary outcome. Stat Med 20:3875–3889. https://doi.org/10.1002/sim.1009
Begg CB, Mazumdar M (1994) Operating characteristics of a rank correlation test for publication bias. Biometrics 50:1088–1101
Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315:629–634. https://doi.org/10.1136/bmj.315.7109.629
Lin H-F, Liao Y-C, Liou C-W, Liu C-K, Juo S-HH (2007) The phosphodiesterase 4D gene for early onset ischemic stroke among normotensive patients. J Thromb Haemost 5:436–438. https://doi.org/10.1111/j.1538-7836.2007.02350.x
Banerjee I, Gupta V, Ahmed T, Faizaan M, Agarwal P, Ganesh S (2008) Inflammatory system gene polymorphism and the risk of stroke: a case–control study in an Indian population. Brain Res Bull 75:158–165. https://doi.org/10.1016/j.brainresbull.2007.08.007
Munshi A, Babu MS, Kaul S, Shafi G, Anila AN, Alladi S, Jyothy A (2009) Phosphodiesterase 4D (PDE4D) gene variants and the risk of ischemic stroke in a South Indian population. J Neurol Sci 285:142–145. https://doi.org/10.1016/j.jns.2009.06.024
Fidani L, Clarimon J, Goulas A, Hatzitolios AI, Evans W, Tsirogianni E, Hardy J, Kotsis A (2007) Association of phosphodiesterase 4D gene G0 haplotype and ischaemic stroke in a Greek population. Eur J Neurol 14:745–749. https://doi.org/10.1111/j.1468-1331.2007.01767.x
Kostulas K, Gretarsdottir S, Kostulas V, Manolescu A, Helgadottir A, Thorleifsson G, Gudmundsson LJ, Thorsteinsdottir U, Gulcher JR, Stefansson K, Hillert J (2007) PDE4D and ALOX5AP genetic variants and risk for ischemic cerebrovascular disease in Sweden. J Neurol Sci 263:113–117. https://doi.org/10.1016/j.jns.2007.06.042
van Rijn MJE, Slooter AJC, Schut AFC, Isaacs A, Aulchenko YS, Snijders PJLM, Kappelle LJ, van Swieten JC, Oostra BA, van Duijn CM (2005) Familial aggregation, the PDE4D gene, and ischemic stroke in a genetically isolated population. Neurology 65:1203–1209. https://doi.org/10.1212/01.wnl.0000178744.42953.b7
Staton JM, Sayer MS, Hankey GJ, Attia J, Thakkinstian A, Yi Q, Cole VJ, Baker R, Eikelboom JW (2006) Association between phosphodiesterase 4D gene and ischaemic stroke. J Neurol Neurosurg Psychiatry 77:1067–1069. https://doi.org/10.1136/jnnp.2006.092106
Zee RYL, Brophy VH, Cheng S, Hegener HH, Erlich HA, Ridker PM (2006) Polymorphisms of the phosphodiesterase 4D, cAMP-specific (PDE4D) gene and risk of ischemic stroke: a prospective, nested case-control evaluation. Stroke 37:2012–2017. https://doi.org/10.1161/01.STR.0000230608.56048.38
He Y, Yang DZ, Yu H, Li MY, Feng QC, Zheng H (2013) Genetic variants of phosphodiesterase 4D gene are associated with an enhanced risk for ischemic stroke in young Chinese population. Neurol India 61:21. https://doi.org/10.4103/0028-3886.108131
He Y, Bai J-Y, Song B, Tan S, Chang Y-S, Li T, Shi C-C, Zhang H, Feng Q-C, Qi H, Song G-Y, Zheng H, Xu Y-M (2012) Sex-dependent association of phosphodiesterase 4D gene polymorphisms with ischemic stroke in Henan Han population. Chin Med J (Engl) 125:2255–2259
Kalita J, Somarajan BI, Kumar B, Kumar S, Mittal B, Misra UK (2011) Phosphodiesterase 4 D gene polymorphism in relation to intracranial and extracranial atherosclerosis in ischemic stroke. Dis Markers 31:191–197. https://doi.org/10.3233/DMA-2011-0810
Li N, He Z, Xu J, Liu F, Deng S, Zhang H (2010) Association of PDE4D and IL-1 gene polymorphism with ischemic stroke in a Han Chinese population. Brain Res Bull 81:38–42. https://doi.org/10.1016/j.brainresbull.2009.09.009
Sun Y, Huang Y, Chen X, Liu Y, Lu X, Shi Y, Tang W, Yang J, Chen W, Zhao X, Gao L, Li S, Feng G, He L (2009) Association between the PDE4D gene and ischaemic stroke in the Chinese Han population. Clin Sci 117:265–272. https://doi.org/10.1042/CS20080471
Xue H, Wang H, Song X, Li W, Sun K, Zhang W, Wang X, Wang Y, Hui R (2009) Phosphodiesterase 4D gene polymorphism is associated with ischaemic and haemorrhagic stroke. Clin Sci 116:335–340. https://doi.org/10.1042/CS20080162
Munshi A, Roy S, Thangaraj K, Kaul S, Babu MS, Jyothy A (2012) Association of SNP41, SNP56 and a novel SNP in PDE4D gene with stroke and its subtypes. Gene 506:31–35. https://doi.org/10.1016/j.gene.2012.06.079
Saleheen D, Bukhari S, Haider SR, Nazir A, Khanum S, Shafqat S, Anis MK, Frossard P (2005) Association of Phosphodiesterase 4D Gene with ischemic stroke in a pakistani population. Stroke 36:2275–2277. https://doi.org/10.1161/01.STR.0000182242.59466.ee
Ma J, Sun Q, Zhang X, Du H (2014) Correlation between the single nucleotide polymorphisms of the human phosphodiesterase 4D gene and the risk of cerebral infarction in the Uygur and Han ethnic groups of Xinjiang, China. Exp Ther Med 7:155–160. https://doi.org/10.3892/etm.2013.1370
Hsieh M-S, Yu S-C, Chung W-T, Hsueh Y-M, Chen F-C, Chiu W-T, Lee H-M (2009) Phosphodiesterase 4D (PDE4D) gene variants and risk of ischemic stroke in the taiwanese population. Lab Med 40:87–90. https://doi.org/10.1309/LM4X5PCRN4AECXBB
Shao M, Yi X, Chi L, Lin J, Zhou Q, Huang R (2015) Ischemic stroke risk in a southeastern Chinese population: insights from 5-lipoxygenase activating protein and phosphodiesterase 4D single-nucleotide polymorphisms. J Formos Med Assoc 114:422–429. https://doi.org/10.1016/j.jfma.2013.12.004
Skvortsova VI, Limborskaya SA, Shetova IM, Slominskii PA, Shamalov NA, Bondarenko EA, Timofeev DYu (2012) Association between Polymorphisms in the Phosphodiesterase 4D (PDE4D) Gene and the Development of Cerebral Stroke in Patients in the Moscow Population. Neurosci Behav Physiol 42:838–841. https://doi.org/10.1007/s11055-012-9645-4
Meschia JF, Brott TG, Brown RD, Crook R, Worrall BB, Kissela B, Brown WM, Rich SS, Case LD, Evans EW, Hague S, Singleton A, Hardy J (2005) Phosphodiesterase 4D and 5-Lipoxygenase Activating Protein in Ischemic Stroke. Ann Neurol 58:351–361. https://doi.org/10.1002/ana.20585
Woo D, Kaushal R, Kissela B, Sekar P, Wolujewicz M, Pal P, Alwell K, Haverbusch M, Ewing I, Miller R, Kleindorfer D, Flaherty M, Chakraborty R, Deka R, Broderick J (2006) Association of Phosphodiesterase 4D with ischemic stroke: a population-based case-control study. Stroke 37:371–376. https://doi.org/10.1161/01.STR.0000198843.72824.0a
Song Q, Cole JW, O’Connell JR, Stine OC, Gallagher M, Giles WH, Mitchell BD, Wozniak MA, Stern BJ, Sorkin JD, McArdle PF, Naj AC, Xu Q, Gibbons GH, Kittner SJ (2006) Phosphodiesterase 4D polymorphisms and the risk of cerebral infarction in a biracial population: the Stroke Prevention in Young Women Study. Hum Mol Genet 15:2468–2478. https://doi.org/10.1093/hmg/ddl169
Kuhlenbäumer G, Berger K, Huge A, Lange E, Kessler C, John U, Funke H, Nabavi DG, Stögbauer F, Ringelstein EB, Stoll M (2006) Evaluation of single nucleotide polymorphisms in the phosphodiesterase 4D gene (PDE4D) and their association with ischaemic stroke in a large German cohort. J Neurol Neurosurg Psychiatry 77:521–524. https://doi.org/10.1136/jnnp.2005.073577
Tomonaga M, Michiaki K, Koji Y, Toshiharu N, Kyota A, Bailing L, Jun H, Yasufumi D, Takanari K, Setsuro I, Mitsuo I, Yutaka K, Yusuke N (2009) Lack of Association Between Variations of PDE4D and Ischemic Stroke in the Japanese Population. Stroke 40:1245–1251. https://doi.org/10.1161/STROKEAHA.108.527408
Wang HM, Chen XL, Ye HH, Bi Y, Pan DB, Xu LY et al (2012) Association ot phosphodiesterase 4D gene with Atherothrombosis ischemic Strok. J Med Res 41:134–137
Cheng H, Jin Q, Li L, Ding X, Song X, Zeng Y (2011) Association of ALOX5AP and PDE4D with the risk of lacunar infarct in people from Jiangsu Province, China. China Nerve Regeneration Research 6:935–940.
Zhang HL, Wang SR, Li SM (2009) Study on association of the single nucleotide polymorphism of phosphodiesterase 4D with stroke. J Chin Microcirc 13:624–627
Xu S, Zhang Y, Lin X (2008) Relationship between phosphodiesterase 4D gene polymorphism and ischemic cerebral vascular disease. J Clin Neurol 21:249–252
Zhao J, Wang X, Xu J, Li N, Shang X, He Z, Yang J (2012) Association of inflammatory response gene polymorphism with atherothrombotic stroke in Northern Han Chinese. Acta Biochim Biophys Sin 44:1023–1030. https://doi.org/10.1093/abbs/gms088
Luo M et al (2014) Relation between phosphodiesterase 4D gene polymorphism and ischemic stroke in Guangdong Han population. Chinese journal of geriatric heart brain and vessel. Diseases 16:503–506
Yue X, Lixia L, Yan H, Zhang P, Gui Y, Song J (2019) Association between PDE4D polymorphism and ischemic stroke in young population, Saudi. J Biol Sci 26:1023–1026. https://doi.org/10.1016/j.sjbs.2019.04.007
Zhang L, Ding R, Kuang P, Wang L, Deng H, Xiong Q, Jiang H (2019) Interaction between CONNEXIN37 and PDE4D gene polymorphisms with susceptibility to ischemic stroke in Chinese population. Exp Biol Med 244:1642–1647. https://doi.org/10.1177/1535370219885079
Shi JP, Chen WD, Zhou JQ, Xue MM, Xue F, Li HZ, Xu ZP (2015) Investigation of single nucleotide polymorphisms in phosphodiesterase 4D gene in Mongol and Han patients with ischemic stroke in Inner Mongolia. Genet Mol Res 14:10281–10287. https://doi.org/10.4238/2015.August.28.13
Bevan S, Porteous L, Sitzer M, Markus HS (2005) Phosphodiesterase 4D gene, ischemic stroke, and asymptomatic carotid atherosclerosis. Stroke 36:949–953. https://doi.org/10.1161/01.STR.0000162713.06519.41
Brophy VH, Ro SK, Rhees BK, Li-Yung L, Lee JM, Nanette U, Gordon BL, Jia Li, Suzanne C, Browner Warren S, Erlich HA (2006) Association of phosphodiesterase 4D polymorphisms with ischemic stroke in a US population stratified by hypertension status. Stroke 37:1385–1390. https://doi.org/10.1161/01.STR.0000221788.10723.66
Elin L, Andreas G, Mueller JC, Tõnis O, Erich W, Gerhard H, Thomas M, Martin D (2005) ALOX5AP Gene and the PDE4D Gene in a Central European population of stroke patients. Stroke 36:731–736. https://doi.org/10.1161/01.STR.0000157587.59821.87
Nilsson-Ardnor S, Wiklund P-G, Lindgren P, Nilsson AK, Janunger T, Escher SA, Hallbeck B, Stegmayr B, Asplund K, Holmberg D (2005) Linkage of ischemic stroke to the PDE4D region on 5q in a Swedish population. Stroke 36:1666–1671. https://doi.org/10.1161/01.STR.0000174188.04716.8d
Lövkvist H, Smith JG, Luthman H, Höglund P, Norrving B, Kristoffersson U, Jönsson A-C, Lindgren AG (2008) Ischaemic stroke in hypertensive patients is associated with variations in the PDE4D genome region. Eur J Hum Genet 16:1117–1125. https://doi.org/10.1038/ejhg.2008.62
Nakayama T, Asai S, Sato N, Soma M (2006) Genotype and haplotype association study of the STRK1 region on 5q12 among Japanese: a case-control study. Stroke 37:69–76. https://doi.org/10.1161/01.STR.0000194961.17292.33
Kumar A, Misra S, Kumar P, Sagar R, Gulati A, Prasad K (2017) Relationship of phosphodiesterase 4D (PDE4D) gene polymorphisms with risk of ischemic stroke: a hospital based case-control study. Neurol Res 39:689–694. https://doi.org/10.1080/01616412.2017.1333975
Wang X, Sun Z, Zhang Y, Tian X, Li Q, Luo J (2017) Impact of the PDE4D gene polymorphism and additional SNP–SNP and gene–smoking interaction on ischemic stroke risk in Chinese Han population. Neurol Res 39:351–356. https://doi.org/10.1080/01616412.2017.1289309
Kim M-K, Kim J-T, Choi S-M, Lee S-H, Park M-S, Cho K-H (2009) Phosphodiesterase 4D gene and risk of noncardiogenic ischemic stroke in a Korean population. J Korean Med Sci 24:307–310. https://doi.org/10.3346/jkms.2009.24.2.307
Milton AG, Aykanat VM, Hamilton-Bruce MA, Nezic M, Jannes J, Koblar SA (2011) Association of the phosphodiesterase 4D (PDE4D) gene and cardioembolic stroke in an Australian Cohort. Int J Stroke. https://doi.org/10.1111/j.1747-4949.2011.00616.x
Zhang XN, Du HB, Wang J et al (2012) Investigation on the single-nucleotide polymorphism of phosphodiesterase 4D gene in Uygur and Han patients with ischemic stroke in Xinjiang district. J Clin Neurol
Wang P, Yang F, Liu CX, Wu YM, Gu C, Zhu HJ (2018) Association between PDE4D rs966221 polymorphism and risk of ischemic stroke: a systematic review and meta-analysis. Metab Brain Dis 33:637–645. https://doi.org/10.1007/s11011-017-0158-2
Yan Y, Luo X, Zhang J, Su L, Liang W, Huang G, Wu G, Huang G, Gu L (2014) Association between phosphodiesterase 4D polymorphism SNP83 and ischemic stroke. J Neurol Sci 338:3–11. https://doi.org/10.1016/j.jns.2013.12.012
Wu W-L, Feng X-W, Qiu C-F, Lin J, Bao X-J (2017) A meta-analysis of PDE-gene polymorphism and cerebral infarction risk. Exp Ther Med 13:2905–2911. https://doi.org/10.3892/etm.2017.4318
Xu X, Li X, Li J, Ou R, Sheng W (2010) Meta-analysis of association between variation in the PDE4D gene and ischemic cerebral infarction risk in Asian populations. Neurogenetics 11:327–333. https://doi.org/10.1007/s10048-010-0235-8
Yoon D, Park SK, Kang D, Park T, Park JW (2011) Meta-analysis of homogeneous subgroups reveals association between PDE4D gene variants and ischemic stroke. Neuroepidemiology 36:213–222. https://doi.org/10.1159/000327915
Zhang X, Wan Q, Zhu D (2016) No association between single-nucleotide polymorphism 56 (SNP56) in phosphodiesterase 4D (PDE4D) gene and susceptibility to ischemic stroke: a meta-analysis of 15 studies. Med Sci Monit 22:3820–3827. https://doi.org/10.12659/MSM.896904
Liang W, Zhang D, Mang J, He J, Liu H, Shao Y, Han F, Xu Z (2015) Association between phosphodiesterase 4D (PDE4D) SNP 87 and ischemic stroke: a meta-analysis. Int J Clin Exp Med 8:1715–1725
Ponsaerts L, Alders L, Schepers M, de Oliveira RMW, Prickaerts J, Vanmierlo T, Bronckaers A (2021) Neuroinflammation in ischemic stroke: inhibition of cAMP-specific phosphodiesterases (PDEs) to the rescue. Biomedicines 9:703. https://doi.org/10.3390/biomedicines9070703
Acknowledgements
None.
Funding
This research did not receive any grants from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
PK and PS were involved in study selection and data extraction for the included study; MN and SM contributed to writing the manuscript to its final version. PK contributed to the concept, designing, statistical analysis, and writing the manuscript. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
No potential conflict of interest.
Ethical approval
Ethical approval was not required for this manuscript as it was a systematic review and meta-analysis done by using existing published data and tusing existing published data. In addition, the research was not directly conducteddid not involve in any human subjects.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Nath, M., Swarnkar, P., Misra, S. et al. Phosphodiesterase 4 D (PDE4D) gene polymorphisms and risk of ischemic stroke: A systematic review and meta-analysis. Acta Neurol Belg 123, 2085–2110 (2023). https://doi.org/10.1007/s13760-023-02218-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13760-023-02218-w