Abstract
Treatment of pyridine-2,6-dicarboxylic acid and 2-aminopyrimidine with Co(NO3)2∙6H2O under hydrothermal conditions led to a new Co (II) complex [Co(amp)(pydc)(H2O)2]∙H2O (1), which was characterized by infrared spectroscopy, elemental analysis as well as X-ray diffraction studies. The DNA-binding behavior of the complex has been studied by UV–Vis absorption and fluorescence spectroscopic titration, viscosity measurements, thermal denaturation and circular dichroism (CD). The experimental results indicated that Co (II) complex was bound to DNA by an intercalative mode. The intrinsic binding constant of Co (II) complex with DNA was (3.80 ± 0.02) × 104 M−1. The biological effects of the Co (II) complex were also studied by MTT assay in MCF-7 and HT-29 cancer cell lines. Treatment of MCF-7 and HT-29 cells with Co (II) complex resulted in a concentration-dependent cell growth inhibition.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
N-Heterocyclic compounds have been used for the development of pharmaceutically and therapeutically important molecules. In the family of N-heterocyclic compounds, pyrimidines, pyridine and their derivatives are important classes of compounds in medicinal chemistry [1, 2]. Pyrimidines, being an integral part of DNA and RNA, exhibit diverse pharmacological properties as effective bactericides against viruses including polio herpes viruses, diuretic, antitumor, anti HIV, and cardiovascular [3, 4]. The transition metal complexes of N-heterocyclic ligands are very important due to their capacity for binding and cleaving DNA under physiological conditions [5, 6]. Accordingly, the design of new transition metal-based anticancer drugs, which exhibit increased selectivity and various non-covalent DNA-binding interaction modes, e.g., intercalation, minor groove binding, major groove and outside electrostatic binding, are of considerable interest [7].
Cobalt is accepted as an indispensable metal widely dispensed in biological systems. For some elements, the genetic code involves proteins, which recognize specific transition metal complexes. For example, only one specific cobalt complex, the coenzyme vitamin B12, appears to be essential [8]. Some cobalt (II) complexes, such as [Co(tfa)(happ)], are known to function as specific probes for DNA bulges due to their ability to cleave DNA specifically. Therefore, the interaction of cobalt complexes with DNA has engrossed much attention [9]. In this regard, mixed-ligand transition metal complexes were found to be especially useful because of their potential to bind DNA via a multitude of interactions and cleave the duplex by virtue of their intrinsic chemical, electrochemical, and photochemical reactivity [10–12].
In continuation of our research on the synthesis and characterization of transition metal complexes with pyridine-2,6-dicarboxylic acid (pydcH2) in the presence of some amino compounds [13–15], in the present work, we attempted to design proposed anticancer Co (II) complexes with pyridine-2,6-dicarboxylic acid and 2-aminopyrimidine in three stages: (1) synthesis and characterization of the complex by spectroscopic and X-ray diffraction studies; (2) study of the ability of the complex to interact with DNA and investigation of its interaction mechanism using UV–Vis and fluorescence spectroscopy, thermal denaturation, circular dichroism (CD) spectra and dynamic viscosity measurements; (3) study of the cytotoxicity effects of the Co (II) complex on MCF-7 human breast cancer and HT-29 colon cancer cell line.
Experimental
Materials and instrumentation
The UV–Vis spectra were recorded on a Shimadzu 2550 UV–Vis spectrophotometer using a 1-cm path length cell. IR spectra were recorded on a Shimadzu IR-470 spectrophotometer. Elemental analyses were performed using a Costech ECS 4010 CHNS analyzer. The thermal denaturation spectra were recorded on a Varian BioCary-100 UV–Vis spectrophotometer using a 1-cm path length cell. Circular dichroism measurements were carried out on a Jasco-810 spectropolarimeter at room temperature with a rectangular quartz cell of 1 cm path. Viscosity experiments were conducted on a Brookfield Circulating Bath Ultra DV III viscometer maintained at 25.0 ± 0.1 °C in a circulating water bath.
All chemicals were obtained from commercial sources. Double-strand calf thymus DNA (CT-DNA) from Sigma was used as received. The stock solution of DNA (pH 7.4) was prepared by dissolving the appropriate amount of DNA in 5 mmol L−1 Tris–HCl [Tris (hydroxymethyl)-aminomethane] containing 50 mmol L−1 NaCl and adjusted to (pH 7.4) with hydrochloric acid and stored at 4 °C. The ratio of the absorbance at 260 and 280 nm (A 260/A 280) was checked to be 1.89, indicating that the DNA was sufficiently free from protein [16]. The DNA concentration per nucleotide was determined by ultraviolet absorption at 260 nm (ε = 6600 L mol−1 cm−1) [17].
Preparation of [Co(amp)(pydc)(H2O)2].H2O (1)
Pyridine-2,6-dicarboxylic acid (0.167 g, 1 mmol) and NaOH (0.04 g, 1 mmol) were dissolved in distilled water (15 mL) and stirred for 30 min at room temperature. A solution of 2-aminopyrimidine (0.09 g, 1 mmol) in methanol (5 mL) was added and the reaction mixture was stirred for 10 min at room temperature. The aqueous solution of Co(NO3)2·6H2O (0.291 g, 1 mmol) was added to the above solution. The reaction mixture was then placed in a Parr Teflon-lined stainless steel reactor sealed and heated at 130 °C for 8 h. The reaction mixture was gradually cooled to room temperature and then kept at 4 °C until blue crystals of complex suitable for X-ray diffraction were obtained.
IR (KBr) (υ, cm−1): 3500–3225 (b), 1655 (s), 1385 (s). UV–Vis (aqueous solution) (λ, nm): 218, 268, 518. Anal. Calc. for C11H14CoN4O7 (M = 373.19): C, 35.37, H, 3.75, N, 15.00 %. Found C, 34.49, H, 3.28, N, 14.68 %.
Single crystal structure determination
The selected crystals of 1 were covered with per fluorinated oil and mounted on the tip of a glass capillary under a flow of cold gaseous nitrogen. The orientation matrix and unit cell dimensions were determined from 8000 (Stoe IPDS II (1), (graphite-monochromated Mo-K α radiation in all cases; λ = 71.073 pm). In addition, absorption corrections were applied for 1 (numerical). The structures were solved by the direct methods (SHELXS-97 [18]) and refined against F 2 by full-matrix least squares using the SHELXL-97 program [19]. ORTEP-3 [20], software was used to prepare the material for publication: WinGX [21]. Further details can be obtained free of charge on application to the Cambridge Crystallographic Data Centre, 12 Union Road, Cambridge CB2 1EZ, UK [Fax: (internet) +44 (0)1223 336033; E-mail: deposit@ccdc.cam.ac.uk] quoting the depository number CCDC 772300 for 1.
Cell viability assay with MTT reduction
The MCF-7 breast and HT-29 colon cell lines were obtained from Pasture Institute of Iran. The cells were adherently grown in RPMI-1640 supplemented with 10 % fetal bovine serum, 100 units mL−1 of penicillin and 100 mg mL−1 of streptomycin and were maintained at 37 °C in a humidified incubator with 5 % CO2. Cell viabilities were determined using the MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide] assay [22]. Cells were seeded at 5 × 103 cells per well onto 96-well plates. After overnight culture, the cells were treated with various concentrations of specific compounds and incubated for 48 h individually. Afterwards, 20 µL of MTT (5 mg mL−1) were added to each well and the cells were incubated for another 4 h at 37 °C. The supernatants were then carefully aspirated and 200 µL of dimethyl sulfoxide was added to each well at room temperature over a period of 20 min. The absorbance values were read using a micro-plate reader at 570 nm. Data were collected from several experiments and the percentage of cell growth inhibition was determined by comparison with the control cells.
Statistical analysis
Non-parametric one-way analysis of variance (ANOVA) was performed with the Dunnett’s test, using GraphPad Prism software. Each experiment was carried out in triplicate and repeated 3–4 times independently. P < 0.05 was taken as the level of significance and such differences are indicated in the figures by an asterisk.
Results and discussion
Crystal structure of complex
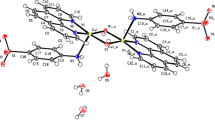
According to the crystallographic data (Table 1), the compound consists of [Co(amp)(pydc)(H2O)2] complex and uncoordinated water molecules (Fig. 1). Co (II) atom is coordinated by an O,N,O′-tridentate dipicolinate (bound via the pyridine and two carboxylate oxygen atoms and forming a five-membered chelate ring), heterocyclic nitrogen atom of 2-aminopyrimidine and two oxygen atoms from two coordinated water molecules. The geometry of the resulting CoN2O4 coordination can be described as a distorted octahedral. The bond angles around Co (II) ion involving trans pairs of donor atoms are in the range of 151.89 (4)°–177.00 (5)° for 1 and deviate from linearity. For the cis pairs of donor atoms, this range is 76.04 (5)°–106.93 (5)° (Table 2). These values indicate large distortion from the ideal octahedral geometry due to the binding of dipicolinate ligand to Co (II) ion in tridentate-O,N,O′ fashion in the complex. The bond distances Co–N and Co–O (Table 2) are in accordance with the values from the literature [23, 24]. The extensive O–H···O, N–H···O hydrogen bonding interactions between complex and uncoordinated water molecule play an important role in stabilizing the crystal packing.
Infrared spectra
The IR spectrum of the complex exhibits broad and strong bands in the 3500–3225 cm−1 range, which can be assigned to O–H and N–H stretching vibrations of the O–H···O and N–H···O hydrogen bonded groups. In addition, the aromatic C–H stretching vibrations of the aromatic rings should also occur in the range of 3100–3000 cm−1 [15, 25]. The carboxylate ions may coordinate to a metal atom in one of the unidentate, bidentate or bridging modes [26]. The characteristic strong bands of carboxylate group appear at 1655 cm−1 (for asymmetric stretching) and 1385 cm−1 (for symmetric stretching). The difference (Δυ = 270 cm−1) between υ asym (COO–) and υ sym (COO–) bands indicate the presence of carboxylate group coordinated to cobalt (II) ion in unidentate mode [27].
Electronic spectra
Complex 1 displays strong bands at about 218 nm (ε = 8030.5 mol−1 dm3 cm−1) and 368 nm (ε = 21,100 mol−1 dm3 cm−1), which are due to the inter-p p transitions of ligands. The spectrum also shows broad absorption band at 518 nm (ε = 810 mol−1 dm3 cm−1) which is assigned to the d d transition.
DNA-binding studies
Electronic absorption titrations
Electronic absorption spectroscopy is universally employed to examine the binding characteristics of transition metal complexes with DNA [28]. Intercalative binding of complexes with DNA generally results in hypochromism and bathochromism due to the intercalative mode involving a strong stacking interaction between an aromatic chromophore and the base pairs of DNA [29, 30]. The strength of this electronic interaction is expected to decrease as the cube of the distance between the aromatic chromophore and the DNA bases [16].
The experiments were performed by maintaining a constant concentration of the complex while varying the nucleic acid (CT-DNA) concentration. Since the metal to ligand charge transfer (MLCT) band is so weak, the intra-ligand (IL) band around 262 nm is monitored as a function of added DNA (Fig. 2). Since CT-DNA is an absorbing species in the absorption range of the complex, the titrations were carried out by adding the same amounts of CT-DNA solution to both references and measuring cells to eliminate the absorbance of DNA itself. Addition of increasing amounts of CT-DNA significantly perturbs the spectrum pattern of the complex showing hypochromism as well as bathochromism. The hypochromism reached 35.4 %, with bathochromism at about 2 nm.
The binding constant (K b) obtained from fittings for data matrix in MATLAB according to equilibrium (1) is obtained as 3.80 (±0.02) × 104 M−1. The equilibrium constants of the model were calculated by using Newton–Gauss Levenberg/Marquardt (NGL/M) method for nonlinear least-squares fitting. NGL/M is fast, robust and additionally delivers basic statistical analysis of the resulting parameters [31, 32].
where M is the titrant (CT-DNA), L is the complex molecule, and ML is the binary complex, whose resultant constant is K b. Comparing the intrinsic binding constant of the Co (II) complex with those of DNA-intercalative Co (II) complexes, such as [Co(GA)2(phen)] (3.8 × 104 M−1) [33] [Co(bpy)2(BHBMe)]ClO4 (1.23 × 104 M−1), and [Co(bpy)2(BHBNO2)]ClO4 (2.06 × 104 M−1) [34], the K b of the Co (II) complex is larger, but obviously smaller than the typical intercalators such as [Co(dpq)3]3+ (dpq = dipyrido[3,2-f:20,30-h]-quinoxaline, 8.02 × 105 M−1) [35] and [Co(phen)2(dppz)]3+ (9.0 × 105 M−1) [36], suggesting the moderate-intensity association of the cobalt complex with DNA. This might be caused by the relatively small planar structure of the intercalative ligand (pyridine) in the complex as compared with the other large intercalative ligands.
Fluorescence titration
The cobalt complex exhibited emission bands at 396 nm when excited at 348 nm. In the absence of CT-DNA, moderate luminescence is emitted in 5 mM Tris–HCl and 50 mM NaCl buffers at ambient temperature, which is the characteristic of the MLCT[Co(dπ) → ligand(dπ*]. Titration of complex with CT-DNA led to a remarkable enhancement of the intensity of the emission for Co (II) complex, as illustrated in Fig. 3.
This enhancement implies that cobalt complex can interact with DNA and can be efficiently protected, since the hydrophobic environment inside the DNA helix reduces the accessibility of solvent water molecules to the complex and the complex mobility is restricted at the binding site, leading to a decrease in the vibrational modes of relaxation. In addition, energy transfer from DNA to metal complexes could also induce fluorescence enhancement [37].
Thermal denaturation studies
The melting of DNA is an important tool for studying the interaction of transition metal ion with nucleic acid. The interaction of transition metal complexes with a double helix is known to increase the helix melting temperature (T m) [38]. It is well known that when the temperature of the solution increases, the double-stranded DNA dissociates into single strands, generating a hyperchromic effect in the absorption spectra of DNA bases (λ max = 260). This transition of double-stranded DNA to single-stranded DNA is termed as the melting temperature (T m) of DNA. According to the literatures [39, 40], the thermal behavior of DNA in the presence of complex can provide some information on the interaction of the complexes with DNA. The effect of the Co (II) complex on the melting temperature (T m) of DNA in buffer is shown in Fig. 4. The results reveal that the T m of free DNA duplex was 78.05 ± 0.02 °C,which increased by 3.70 ± 0.01 °C in the presence of complex at a molar ratio of [Co]/[DNA] = 0.5. Common DNA metallo-intercalators cause ΔT m values of between 4 and 14 °C depending on their intrinsic DNA-binding constant [41, 42]. Hence, the increased ΔT m with the titled cobalt complex suggests an intercalation also inferred from spectral titrations.
Circular dichroism spectra studies
Circular dichroism (CD) spectrum is useful in diagnosing changes in DNA morphology during drug–DNA interactions as the CD signals are quite sensitive to the mode of DNA interactions with small molecules and transition metal complexes [43]. The chirality of DNA double helix originates from the DNA geometry, resulting from the coupling of bases, phosphates backbone and chiral sugar units will exhibit CD spectra [44].
CT-DNA is the B-form DNA, a right-handed double helix with the base pairs stacked in the center and the base plane perpendicular to the helical axis causing two well-known grooves; the major and minor grooves. The CD spectrum of DNA consists of a positive band at 275 nm due to bases tacking and a negative band at 245 nm due to helicity, which is also characteristic of DNA in a right-handed B-form [41].
CD spectral variations of DNA in the UV region were recorded by the addition of Co (II) complex, as shown in Fig. 5. It is generally accepted that the intercalative interaction of classical intercalation enhances the intensities of both the bands due to the strengthening of the bases tacking and stabilizing the right-handed B conformation of DNA, while simple groove binding and electrostatic interaction of small molecules show less or no perturbation on the bases tacking and helicity bands [45]; whereas, interaction of the titled Co (II) complex with DNA in a molar ratio of 0.5 causes intensity increase in the positive band and intensity decrease in the negative bands. These changes are indicative of a typical intercalation involving π-stacking and reflected by the enhancement in the positive bands and the partial unwinding of DNA helix, as reflected by the intensity decrease in the negative bands, hence stabilization of the right-handed B-form of DNA [28].
Viscometric studies
To further clarify the binding of the cobalt (II) complex with CT-DNA, viscosity measurements were carried out by varying the concentration of the added complex. Optical photo-physical probes provide necessary, but not sufficient, clues to support a binding mode. Hydrodynamic measurements, such as determination of viscosity, which is exquisitely sensitive to the change of length of DNA (i.e., viscosity and sedimentation), may be the most effective means for studying the binding mode of complex to DNA in solution in the absence of X-ray crystallographic or NMR structural data [46].
Intercalation of the transition metal complex to DNA demands that the DNA helix lengthen as base pairs separated to accommodate the binding complex, hence leading to a significant increase in DNA viscosity. In contrast, groove-face or electrostatic interactions and non-classical intercalation typically cause a bend (or kink) in DNA helix, reducing its effective length and thereby its viscosity [46]. In addition, some complexes such as [Ru(bpy)3]2, which interact with DNA by an electrostatic binding mode, have no influence on DNA viscosity [47, 48]. Figure 6 shows the effect of Co (II) complex on the viscosity of CT-DNA at 25 °C. Data are presented as (η/η 0)1/3 versus the ratio of the concentration of the compound and DNA, where η is the viscosity of DNA in the presence of the compound and η 0 is that of DNA alone [49]. It was observed that increasing the Co (II) complex concentration led to an increase in DNA viscosity. The complex with planer ligands such as pyridine, thiadiazole rings could partially insert into the DNA base pair and form partial stacking of the complexes with the base pairs . The interaction of DNA with cobalt complex results in the stabilization of the double helix because of hydrogen bonding interaction between oxygen, nitrogen atoms of complex and DNA base pairs. The results obtained from the viscosity experiments validate those obtained from the spectroscopic studies. Based on all the spectroscopic studies together with the viscosity measurements, we find that the Co (II) complex can bind to DNA via an intercalation mode.
MTT viability assay
To evaluate the effect of the Co (II) complex on the viability of the cancer cell lines, MTT assay was carried out. The MTT assay measures the activity of mitochondrial dehydrogenase enzyme based on its ability to cleave tetrazolium ring to produce formazan. Thus, the assay can be used as an index of cell viability. As shown in Fig. 7, Co (II) complex significantly inhibited the viability of both MCF-7 and HT-29 cells in a dose-dependent manner. In the MCF-7 cells, significant inhibitory effect was observed at 10 µM, which reached a maximum at 1000 µM (from 93.5 ± 2.5 % for 10 µM to 34 ± 3 % for 1000 µM vs. control 100 %, respectively). In the HT-29 cells, significant inhibitory effect was observed at 100 µM, which reached a maximum at 1000 µM (from 92.4 ± 2 % for 100 µM to 55.3 ± 4 % for 1000 µM vs. control 100 %, respectively). The effective doses of Co (II) complex, which inhibited 50 % growth (IC50) of MCF-7 and HT-29 cells after 48 h of treatment were 208 and 402 µM, respectively. The results showed that the Co (II) complex is a more potent cancer compound in MCF-7 cell lines.
Cytotoxicity effect of Co (II) complex in MCF-7 and HT-29 cancer cell lines. Cells were treated with different concentrations of Co (II) complex for 48 h, and cytotoxicity was assessed by MTT assay. * P < 0.05 and ** P < 0.001 are significant. Statistical analysis was performed by ANOVA. Each value is presented as mean ± SD of three experiments. Each experiment was conducted in triplicate
Conclusion
In this paper, preparation and characterization of cobalt (II) complex having two ligands, pyridine-2,6-dicarboxylic acid and 2-aminopyrimidine were reported. The molecular structure of complex was determined by single crystal X-ray diffraction. The cytotoxicity effects and binding affinity of the complex with native calf thymus DNA in physiological buffer was investigated by UV–Vis and fluorescence spectroscopic, thermal denaturation, circular dichroism (CD) spectra and dynamic viscosity parameters were calculated as well. The experimental results such as hypochromism (34 %) and bathochromic shift (2 nm) in spectra from electronic absorption titration, increasing in viscosity with increasing [complex]/[DNA] ratio, change of the DNA melting temperature, T m of about 1.70 ± 0.01 °C, show that cobalt (II) complex as a small molecule has intercalative binding with DNA. These results are supported by competitive binding experiment.
References
T. Phaechamud, J. Mahadlek, J. Charoenteeraboon, S. Choopun, Indian J. Pharm. Sci. 74, 498–504 (2012)
S. Biswal, U. Sahoo, S. Sethy, H.K.S. Kumar, M. Banerjee, Asian J. Pharm. Clin. Res. 15, 1–6 (2011)
C. Oliver Kappe, Tetrahedron 49, 6937–6963 (1993)
M. Bhuiyan, K.M. Rahman, M. Hossain, M. Rahim, M. Hossain, Croat. Chem. Acta 78, 633 (2005)
K. Akdi, R.A. Vilaplana, S. Kamah, F. Gonzalez-Vilchez, J. Inorg. Biochem. 99, 1360–1368 (2005)
K.E. Erkkila, D.T. Odom, J.K. Barton, Chem. Rev. 99, 2777–2796 (1999)
A.M. Pyle, J.P. Rehmann, R. Meshoyrer, C.V. Kumar, N.J. Turro, J.K. Barton, J. Am. Chem. Soc. 111, 3051–3058 (1989)
N.K. Karpowich, H.H. Huang, P.C. Smith, J.F. Hunt, J. Biol. Chem. 278, 8429–8434 (2003)
C.-C. Cheng, Y.-N. Kuo, K.-S. Chuang, C.F. Luo, W.J. Wang, Angew. Chem. Int. Ed. 38, 1255–1257 (1999)
D.S. Sigman, A. Mazumder, D.M. Perrin, Chem. Rev. 93, 2295–2316 (1993)
N.J. Turro, J.K. Barton, D.A. Tomalia, Acc. Chem. Res. 24, 332–340 (1991)
T. Ghosh, B.G. Maiya, A. Samanta, A.D. Shukla, D.A. Jose, D.K. Kumar, A. Das, J. Biol. Inorg. Chem. 10, 496–508 (2005)
M. Tabatabaee, Chem. Cent. J. 6, 1–8 (2012)
M. Tabatabaee, F. Abbasi, B.-M. Kukovec, N. Nasirizadeh, J. Coord. Chem. 64, 1718–1728 (2011)
M. Tabatabaee, B.-M. Kukovec, M. Kazeroonizadeh, Polyhedron 30, 1114–1119 (2011)
C.Z. Huang, Y.F. Li, P. Feng, Talanta 55, 321–328 (2001)
Z.M. Lighvan, A. Abedi, M. Bordbar, Polyhedron 42, 153–160 (2012)
G.M. Sheldrick, SHELXS-97, Program for Crystal Structure Determination (University of Göttingen, Germany, 1997)
G.M. Sheldrick, SHELXTL, Siemens Analytical X-Ray Instruments Inc. (Madison, WI, USA, 1990)
L.J. Farrugia, J. Appl. Crystallogr. 30, 565 (1997)
F. Fallahian, F. Karami-Tehrani, S. Salami, M. Aghaei, FEBS J. 278, 3360–3369 (2011)
M. Tabatabaee, B.-M. Kukovec, V. Razavimahmoudabadi, Z. Naturforsch. B Chem. Sci. 66b, 813–818 (2011)
M. Tabatabaee, V. Razavimahmoudabadi, B.-M. Kukovec, M. Ghassemzadeh, B. Neumüller, J. Inorg. Organomet. Polym. Mat. 21, 450–457 (2011)
M. Tabatabaee, M. Tahriri, M. Tahrir, Y. Ozawa, B. Neumüller, H. Fujioka, K. Toriumi, Polyhedron 33, 336–340 (2012)
G.B. Deacon, R.J. Phillips, Coord. Chem. Rev. 33, 227–250 (1980)
L.-J. Zhang, J.-Q. Xu, Z. Shi, W. Xu, T.-G. Wang, Dalton Trans. 6, 1148–1152 (2003)
H.-F. Wang, R. Shen, N. Tang, Eur. J. Med. Chem. 44, 4509–4515 (2009)
J.-G. Liu, Q.-L. Zhang, X.-F. Shi, L.-N. Ji, Inorg. Chem. 40, 5045–5050 (2001)
S.A. Tysoe, R.J. Morgan, A.D. Baker, T.C. Strekas, J. Phys. Chem. 97, 1707–1711 (1993)
M. Maeder, Y.M. Neuhold, Practical Data Analysis in Chemistry (Elsevier, Oxford, UK, 2007)
M. Maeder, A.D. Zuberbuehler, Anal. Chem. 62, 2220–2224 (1990)
H.B. Lin, Q.X. Wang, C.M. Zhang, W.Q. Li, Chin. Chem. Lett. 22, 969–972 (2011)
S. Srinivasan, J. Annaraj, P.R. Athappan, J. Inorg. Biochem. 99, 876–882 (2005)
Y. Sun, Q.-X. Zhou, J.-R. Chen, Y.-J. Hou, W.-H. Lei, X.-S. Wang, B.-W. Zhang, J. Inorg. Biochem. 103, 1658–1665 (2009)
S. Arounaguiri, B.G. Maiya, Inorg. Chem. 35, 4267–4270 (1996)
B.C. Baguley, B.M. Le, Biochemistry 23, 937–943 (1984)
B. Jiang, X.-N. Xiong, C.-G. Yang, Biorg. Med. Chem. Lett. 11, 475–477 (2001)
M. Cusumano, A. Giannetto, J. Inorg. Biochem. 65, 137–144 (1997)
B. Peng, X. Chen, K.-J. Du, B.L. Yu, H. Chao, L.-N. Ji, Spectrochim. Acta Part A 74, 896–901 (2009)
V.I. Ivanov, L.E. Minchenkova, A.K. Schyolkina, A.I. Poletayev, Biopolymers 12, 89–110 (1973)
P. Lincoln, E. Tuite, B. Nordén, J. Am. Chem. Soc. 119, 1454–1455 (1997)
A.M. Polyanichko, V.V. Andrushchenko, E.V. Chikhirzhina, V.I. Vorobèv, H. Wieser, Nucleic Acids Res. 32, 989–996 (2004)
A. Rodger, B. Nordén, Circular Dichroism and Linear Dichroism (Oxford University Press, Oxford, 1997)
W. Saenger, Principles of Nucleic Acid Structure (Springer, New york, 1984)
S. Satyanarayana, J.C. Dabrowiak, J.B. Chaires, Biochemistry 31, 9319–9324 (1992)
F.-H. Li, G.-H. Zhao, H.-X. Wu, H. Lin, X.-X. Wu, S.-R. Zhu, H.-K. Lin, J. Inorg. Biochem. 100, 36–43 (2006)
J.-G. Liu, B.-H. Ye, H. Li, Q.-X. Zhen, L.-N. Ji, Y.-H. Fu, J. Inorg. Biochem. 76, 265–271 (1999)
M. Eriksson, M. Leijon, C. Hiort, B. Norden, A. Graeslund, Biochemistry 33, 5031–5040 (1994)
N. Wang, Q.-Y. Lin, J. Feng, Y.-L. Zhao, Y.-J. Wang, S.-K. Li, Inorg. Chim. Acta 363, 3399–3406 (2010)
Acknowledgments
We gratefully acknowledge the support from Qom University, Payam Noor University and Yazd branch Islamic Azad University.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bordbar, M., Tabatabaee, M., Alizadeh-Nouqi, M. et al. Synthesis, characterization, cytotoxic activity and DNA-binding studies of cobalt (II) mixed-ligand complex containing pyridine-2,6-dicarboxylate ion and 2-aminopyrimidine. J IRAN CHEM SOC 13, 1125–1132 (2016). https://doi.org/10.1007/s13738-016-0826-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13738-016-0826-x