Abstract
Two complexes, [Co(pyzdc) ∙ (H2O)3]n (I) and [Cd2(pyzdc)2 · (H2O)3]n (II) (H2pyzdc = pyrazine-2,3-dicarboxylic acid) have been synthesized and characterized by IR, single crystal X-ray diffraction (CIF files CCDC nos. 1830635 (I) and 1830636 (II)) and powder X-ray diffraction measurement. The crystal structure of I was refined in the monoclinic space group P21/c. The central atom is coordinated to a nitrogen atom, two oxygen atoms from H2pyzdc ligand and other three oxygen atom from coordinated water molecules. Complex II was refined in the monoclinic space group P21/c, consisting of two cadmium atoms, two repeated H2pyzdc ligand and three coordinated water. The interaction with DNA was carried out by absorption titration experiment, competition experiments, gel electrophoresis assay and molecular docking. The results demonstrated that the two complexes have the ability to cleave DNA and binding to DNA through intercalation mode. And the binding affinity was examined by kb and Ksq values, indicating the activity of these compounds bind to DNA was on the series of complex II (kb = 2.38 × 103, Ksq = 0.0621) > complex I (kb = 2.02 × 103, Ksq = 0.0596). Cytotoxicity study showed the complexes have a certain inhibitory rate of cancer cell. And IC50 values exhibited that the anticancer activity series of these two complexes is consistent with their binding activity, and complex II exhibited similar cytotoxicity with cisplatin. Apoptosis experiments indicated that the obtained compounds possess the ability to induce apoptosis, which is an important reference on cytotoxic estimate. The two complexes in this paper deserve our continued research and are likely to become promising DNA-targeted agents.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
In the past few decades, coordination chemistry has expanded into many emerging fields, such as supramolecular chemistry [1–5], metal organic frameworks [6, 7], crystal engineering [8, 9], and therapeutic [10–13]. Since the late 1970s, the application of metals in the form of their complexes is achieving acceptance in anticancer properties because of the discovery and adhibition of cisplatin in medicinal [14]. Transition metal complexes including polydentate and aromatic ligands always have some suitable biological properties and with the development of bioinorganic chemistry, their use in various medication systems especially against tumors is well documented [15]. Most of the metal-based drugs are DNA-targeted agents and used as cleavage agents for DNA. Studying the interaction mode of complexes with DNA is an effective method for developing promising DNA structure probes and therapeutic agents.
Pyrazine-2,3-dicarboxylic acid (H2pyzdc), which is used as ligand in this paper, is fascinating that the dianion (pyzdc2–) form of it possesses two adjacent carboxylate groups which can provide four possible O-atom donors for coordination and meanwhile, the pyrazine ring contains two additional N-atom donors [16]. To the best of our knowledge, complexes derived from H2pyzdc ligand are usually synthesized to study their photoluminescence [17], catalytic activity [18], H2 absorption performance [19], magnetic and electrical conductance properties [16, 20], but rare on their bioactivity.
Based on this, we synthesized two transition metal complexes based on H2pyzdc ligand, namely [Co(pyzdc) ∙ (H2O)3]n (I) and [Cd2(pyzdc)2 ∙ (H2O)3]n (II). Their structures were characterized by X-ray single crystal diffraction. Furthermore, we investigated the interaction of synthesized complexes with DNA by absorption titration experiments, emission quenching study, electrophoresis and molecular docking, measured the cytotoxic by MTT assay and detected the property of the complexes against HeLa cells through apoptosis studies.
EXPERIMENTAL
Materials and measurements. All chemical reagents used were analytical grade and purchased directly with no further purified. Calf thymus DNA (CT-DNA) was purchased from Sinopharm Chemical Reagent Co., Ltd., pBR322 plasmid DNA was purchased from Takara Bio Inc. The human cervical cancer cell line HeLa was taken from American Type Culture Collection (ATCC). IR spectra were performed on the 4000–400 cm–1 region by KBr pellets on a Nicolet Nexus 470 FT-IR spectrometer. X-ray analysis was ran in a BRUKER SMART 1000 CCD diffractometer. The power X-ray diffraction (PXRD) data were obtained using a Bruker D8 Advance diffractometer. Electronic absorption spectroscopy was carried out in the 1 cm quartz cuvette on an ultraviolet-visible spectrophotometer (UV-2700, Shimadzu). Competitive binding experiments were recorded on a Perkin-Elmer LS55 fluorescence spectrophotometer.
Synthesis of [Co(pyzdc) ∙ (H2O)3]n(I). Co(ClO4) · 6H2O (11.00 mg, 0.03 mmol) and pyrazine-2,3-dicarboxylic acid (5.04 mg, 0.03 mmol) were dissolved in DMF–H2O (1 : 2, 3 mL) and stirred at ambient temperature for 2 h. Then the mixture was sealed in a 5 mL vial and heated at 90°C for 3 days, after the automatic cooling to ambient temperature, the orange block crystals were obtained by filtration.
For C6H2N2O7Co (I) | |||
Anal. calcd., % | C, 26.37 | H, 0.74 | N, 10.26 |
Found, % | C, 26.34 | H, 0.79 | N, 10.30 |
IR (KBr; ν, cm–1) 1658 s, 1584 m, 1433 w σ(C=C); 1345 s, σ(C–O); 1183 w δ(Ar–H); 1123 s ν(C–N); 854 m, 755 m δ(Ar–H).
Synthesis of [Cd2(pyzdc)2∙ (H2O)3]n(II). Cd(NO3)2 · 4H2O (9.25 mg, 0.03 mmol) and pyrazine-2,3-dicarboxylic acid (5.04 mg, 0.03 mmol) were dissolved in DMF–H2O (1 : 2, 3 mL), the mixture was adjusted to pH 4.0 with 0.05 M HNO3 solution and stirred for 2 h at room temperature. Then we use the same method as for complex I, and the turquoise flake crystals were obtained by filtration.
For C12H10N4O11Cd2 (II) | |||
Anal. calcd., % | C, 23.57 | H, 1.65 | N, 9.16 |
Found, % | C, 23.63 | H, 1.71 | N, 9.08 |
IR (KBr; ν, cm–1) 1637 s, 1596 s, 1572 m, 1466 w σ(C=C); 1387 s, 1364 s σ(C–O); 1169 w, 848 m, 749 m δ(Ar–H); 1123 s ν(C–N).
X-ray structure determination. Applicable single crystals of I and II for X-ray analysis were obtained directly from the above procedures. The diffraction data were collected using graphite-monochromatized MoKα radiation (λ = 0.71073 Å) on a BRUKER SMART 1000 CCD diffractometer. The structures of complexes I and II were analysed by direct method and refined by the full-matrix least-squares methods with SHELXTL-97 program package [21]. All non-hydrogen atoms were refined with anisotropic displacement parameters and hydrogen atoms were located from the Fourier mapsat. A summary of the crystallographic data and refinement parameters for I and II are given in Table 1. The selected bond lengths and angles of the crystal structures of 1 and 2 are given in Table 2.
Supplementary material for structure I and II has been deposited with the Cambridge Crystallographic Data Centre (CIF files CCDC nos. 1830635 and 1830636, respectivily; deposit@ccdc.cam.ac.uk or http://www. ccdc.cam.ac.uk).
DNA binding and cleavage.Electronic absorption experiment. UV-Vis absorption spectrum was used to examine the activity of DNA binding. The purity of CT-DNA was assessed by the ratio of UV absorbance at 260 nm versus 280 nm. Concentration of CT-DNA in this experiment was determined by UV spectroscopy according to the Lambert−Beer law. CT-DNA was added successively to 10 μM of complexes which dissolved in 10 mM Tris-HCl buffer (pH 7.2) and the spectrum recorded the absorbance of reaction mixtures after a 5-minutes preincubated.
Competitive binding experiments withethidiumbromide (EB). The fluorescence spectroscopy also researched interaction of the complexes to DNA. The competition experiments with EB and CT-DNA were performed as previously described [22–26]. Briefly, varying concentration of complexes (0–12 μM) was added into EB-DNA mixture where [DNA] = 2.5 μM and [EB] = 1 μM. The reaction was carried out in the Tris-HCl buffer (50 μM, pH 7.4) and reacted 2 h before recorded. For the experiments, the excitation and emission slit widths were remained at 10 nm and scan rate maintained unchanged.
Gel electrophoretic analysis. This assay was used to demonstrated the cleavage activity of the complexes to DNA. For the experiment, pBR322 DNA was treated with the prepared complexes solution and the mixture were reacted at room temperature for 2 h. The sample was electrophoresed for 2 h on 0.8% agarose gel in Tris-acetate buffer (50 mM Tris-acetate, 18 mM NaCl buffer, pH 7.2) at 90 V. Then 1.0 mg/mL EB was added to stain the gel, and the photograph was recorded by gel imaging analysis system.
Docking study. The binding mode of I and II with DNA was deeply verified by molecular docking. The study has been done by YASARA. PDB format of complexes I and II were obtained by converting CIF file using Mercury software. The structure of DNA used in this study was the human PARP-1 DNA binding domain complex (PDB ID: 4AV1) and obtained from the protein data bank at a resolution of 3.1 Å.
In vitro study.Culture of HeLa cells. Unless otherwise specified, the HeLa cells used in the experiment were routinely incubated at 37°C in 5% CO2 and grown in RPMI 1640 medium which contained 10% fetal bovine serum (FBS) and 1% penicillin.
Cytotoxicity test. The growth inhibitory effect of I and II and reference reagent cisplatin were investigated in HeLa cell line using an MTT assay [27–31]. Briefly, 1 × 105 cells were cultured into a 96-well plate and treated with complex I and complex II of serial concentrations for 24 h at 37°C in 5% CO2. After treatment, 20 μL MTT solution (5 mg/mL) was added to each well and further incubated for 4 h. The supernatants were carefully extracted, and 100 μL DMSO was added to dissolve formazan crystals. Following a five-minute vibration, a micro plate spectrophotometer was used to measured the absorbance of each well at a wavelength of 490 nm. The test was carried out at least three times to calculate IC50 values.
Flow cytometric analysis. The cultured cells were incubated with or without the tested compounds for 12 h at 37°C. Then attached cells were digested by trypsinization from adherent cultures and centrifugation at 1000 rpm for 5 min, the cells were washed twice time by cold phosphate-buffered saline (PBS) and following resuspended at a concentration of 1 × 106 cells/ mL in Binding Buffer. 100 μL of the solution (1 × 105 cells) were transferred to a 2 mL culture tube. 5 μL FITC Annexin-V and 5 μL propidine iodide (PI) were added as staining reagents. Gently shake cells in the tubes and incubate at ambient temperature for 15 min under dark condition, then each tube was added 400 μL Binding Buffer. The analyzed process were performed on flow cytometry (Accuri C6, USA) within 1 h.
Morphological analysis. HeLa cells (1 × 105) were treated with 6.25 μM complexes and incubated with routinely method for 12 h in culture dish. Then they were washed with cold PBS, and fixed by ethanol (95%, w/v). Following Annexin-V FITC and PI (FITC: 20 μg/mL, PI: 50 μg/mL) were added to counterstain cell nuclei and the cells were observed and imaged using Fluorescent Inverted microscope-XD30A-RFL.
RESULTS AND DISCUSSION
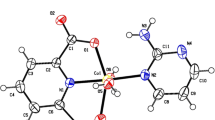
The molecular diagram of complex I with atomic notation scheme is presented in Fig. 1a. The central atom is coordinated to a nitrogen atom N(1A), two oxygen atoms (O(2A), O(3A)) from the organic ligand and other three oxygen atom from coordinated water molecules (O(5A), O(6A), and O(7A)) coordinate to form an octahedral structure. N(1A), O(3A), O(5A), and O(6A) form the equatorial plane, O(7A) and O(2A) are located in the axial position. The Co–N distance is 2.10(2) Å and the Co–O distance are in the range of 2.05(3) to 2.11(2) Å. As is shown in Fig. 2a, the most fascinating structure feature of complex I is that the single-helix-like structure which is constructed by the N(1A)–O(2A) bond (2.11 Å).
The unit of complex II contains two cadmium atoms, two repeated H2pyzdc ligand and three coordinated water molecules (Fig. 1b). Cd(1) is coordinated with one nitrogen atom (N(3)) from pyrazine ring, four oxygen atoms (O(1), 2 × O(5), and O(8)) from carboxyl groups and another oxygen atom from coordinated water molecules in the form of distorted octahedral geometry, N(3), O(5), O(8), and O(9) form the equatorial plane, O(1) and O(5) are located in the axial position. Cd(2) is coordinated with one nitrogen atom (N(1)) from pyrazine ring, three oxygen atoms (O(2), O(3), and O(4)) from carboxyl groups and two oxygen atoms from coordinated water molecules in the form of distorted octahedral geometry, N(1), O(2), O(3), and O(4) form the equatorial plane, O(10) and O(11) are located in the axial position. The Cd–N distance are 2.345(4) and 2.383(4) Å. The distance of Cd–O are in the range of 2.202(3) to 2.441(4) Å. Figure 2b shows the one-dimensional chain structure of II.
The sample purity of complexes I and II were tested by PXRD measurement. The sharp diffraction peaks of complexes I and II (Fig. 3) indicate that these particles are highly crystalline in nature. A good agreement between the experimental and theoretical single-crystal data of the study was obtained by comparing the powder diffraction pattern of the as-prepared complex with the calculated patterns generated from the single crystal X-ray diffraction data.
Electronic absorption spectra is an effective method for examining the interactions of complexes to DNA. In DNA binding studies, 5 × 10–5 M of the complexes were titrated against increasing concentrations of CT-DNA. The maximum absorption band of I and II exhibited at 280 nm and a lower energy MLCT band was monitored with additions of CT-DNA (Fig. 4), which means that the complexes interacted with CT-DNA. Absorbance of I and II all showed a certain decrease and a slightly bathochromic shift (2 nm). It suggested that the complexes may interact with DNA in a intercalation mode which always appear hypochromicity and bathochromic shift result from π–π stacking [32, 33].
The intrinsic binding constants (kb) of I and II were obtained by the following equation:
were, εa (the apparent absorption coefficients) refers to Aobs/[complex], εf is the extinction coefficient of free complex and εb corresponds to the extinction coefficient of the complex when it is fully bounded to DNA. The values of kb were caculated using the equation [DNA]/( εa – εf ) versus [DNA] which was exhibited in Fig. 5. According to calculation results, the binding activity to DNA of II (kb = 2.38 × 103) is stronger than that of I (kb = 2.02 × 103).
To further study the binding mode of I and II with CT-DNA, EB was used to carry out the competitive binding experiment for which mode of bindings are well established. Based on previous research, we can believed that the complex interacted with DNA in the same way with EB if any complex molecule replaces the bound EB from the CT-DNA [34, 35]. As a DNA-intercalator with planar phenanthridine ring, EB can insert itself between the DNA-base pairs. It binds to CT-DNA by an intercalative mode and causes a obvious increase in fluorescence emission intensity [36, 37]. It is clear in Fig. 6 that with the addition of I and II in the EB–DNA system, the fluorescence intensity exhibited a substantial change. This clearly confirms that these complexes interact with DNA in an intercalative mode.
The classical Stern−Volmer equation I0/I = 1 + Ksqr was used to calculated the quenching constant. In the equation, Ksq is a linear constant that depends on the ratio of the EB bound concentration to the DNA concentration. I0 and I figure the fluorescence intensities in the absence and presence of complexes, respectively, and r represents the concentration ratio of the complex to DNA [26, 38]. The quenching curves (Fig. 7) showed that the complexes interact with DNA are in the series of Ksq (I) (0.0596) < Ksq (II) (0.0621). This result indicated that the two complexes have different binding affinities with DNA, in the order complex II > complex I, which is consistent with electronic absorption assays.
The ability of DNA cleavage of I and II was studied using pBR322 plasmid DNA by agarose gel electrophoresis. Plasmids migrate at different rates on the gel and predominantly form three forms. From fastest to slowest these forms are: supercoiled circular conformation (Form 1), nicked circular (Form 2), which is the result of single-strand breaks, and linear conformation (Form 3), which is seen as a consequence of a double-strand break [39–41]. In Fig. 8, it is seen that with the concentration of complexes increases, the band intensity of the Form 1 decreases apparently, while that of Form 2 increases significantly. Among the two complexes, II exhibits the most effective activity on DNA-cleavage than I. This may be due to the complexes have different binding affinity for DNA and the similar observations for other complexes had also been reported [42, 43].
Docking study is a theoretical simulation method that can predict the interaction mode and affinity between small molecule and biomacromolecule [44]. Therefore, the docking studies were performed to further corroborate the experiment results of I and II interact with DNA. The part which containing only the ball-model is the complex molecules and others are the DNA in Fig. 9. The results of the docking studies reveal that the two compounds insert to the DNA double helix. It had been proved by previous report that most synthesized metal complexes adopted an intercalation mode to interact with DNA, which affects the stability of DNA and thus exhibiting the ability to cleave DNA [45–47].
MTT assay was carried out with reference to previous report to evaluated the activity against HeLa cells in this paper [48, 49]. The results were calculated and summarized in Table 3. According to the table, the ligand had poor inhibitory effect to HeLa cells that the IC50 value of it is above 100 μM/L. While it is surprisingly that the synthesized complexes which are derived from the ligand, exhibit much better tumor inhibitory effect. Among the two complexes, II exhibited the most active against HeLa cells in the test while I showed the least active. And compared to complex I (IC50 values: 23.57 ± 5.43), complex 2 (IC50 values: 8.05 ± 1.35) shows a similar IC50 value with cisplatin (IC50 values: 10.73 ± 2.93) [50–52].
Apoptosis is an autonomous and orderly death of cells that may occur in multicellular organisms. It can eliminate unwanted or abnormal cells and plays an important role in the evolution, development and homeostasis of multicellular organisms and has been proved to be a key cellular process for anti-tumor activity of metal anticancer drugs [53, 54]. The apoptosis study was carried out using Annexin V-FITC/PI dual staining to quantify apoptosis induced by obtained complexes and to distinguish the cell death caused by necrosis or apoptosis [55, 56]. As shown in Fig. 10, for these complexes, Annexin V positive cells were mostly appeared in the late apoptosis quadrant, which would imply that the time for early apoptosis might have been partly missed by 24 h [49]. The increase of apoptosis was most obvious following treatment with complex II, and less for complex I exposure after 24 h.
The HeLa cells cultured with or without 1000 μg/mL of the complexes I and II for 24 h were measured by surface expression of phosphatidyl serine using FITC-conjugated Annexin-V antibody. Membrane permeability was assessed by PI, analyzed by flow cytometry. Dot plots show percentages representing the population of cells that were non-apoptotic (lower left quadrant), early apoptotic (lower right quadrant) or late apoptotic/necrotic (upper right quadrant). Quadrants were established using controls.
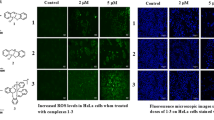
A number of biochemical and morphological changes are the features of apoptotic cell death [57]. The first change occurs in apoptotic is the shrink and deform of the cell. After loses its morphology, the cell detachment from its neighbors, undergoes chromatin condensation and DNA cleavage with formation of apoptotic bodies [58–61]. Changes in the cell morphological of complexes I and II were examined using a fluorescent microscopy. The result is shown in Fig. 11. These results suggested that these complexes significantly enhanced apotosis and supressed growth in HeLa cells.
In this paper, we synthesized and characterized the two complexes [Co(pyzdc)(H2O)3]n (I) and [Cd2-(pyzdc)2(H2O)3]n (II). The binding studies demonstrated that the obtained complexes could bind to DNA. The binding constant kb and quenching constant Ksq showed that complex II exhibited the better affinity with DNA among the two complexes. Agarose gel electrophoresis experiment described that I and II have the ability to cleavage DNA and complex II possessed the better cleavage activity. Apoptosis studies showed that the obtained complexes can induce apoptosis, which is an important reference on cytotoxic estimate. Cytotoxicity assay confirmed the two complexes could inhibit the proliferation of HeLa cell line, and IC50 value suggested that II possess the higher cytotoxicity among the two obtained complexes. In conclusion, all complexes in this paper are biologically active and the biological activity from strong to weak are complex II > complex I. Therefore, we can believe that the two complexes in this paper deserve our continued research and are likely to become promising DNA-targeted agents.
REFERENCES
Guo, L.L., Chen, F., Xie, N., et al., J. Colloid Interface Sci., 2018, vol. 528, p. 53.
Li, W. and Li, S., Chem. Engineering Sci., 2018, vol. 189, p. 65.
Li, X.R., Le, Z.Y., Chen, X.L., et al., Appl. Catal., 2018, vol. 236, p. 501.
Peng, J.X. and Wu, R.A., Anal. Chim. Acta, 2018, vol. 1027, p. 9.
Wu, G.G., Ma, J.P., Li, S., et al., J. Colloid Interface Sci., 2018, vol. 528, p. 360.
Cai, T.T., Huo, S.J., Wang, T., et al., Carbohydr. Polym., 2018, vol. 193, p. 54.
Wu, G.Y., Chen, L.J., Xu, L., et al., Coord. Chem. Rev., 2018, vol. 369, p. 39.
Ates, O.D., Zorlu, Y., Kanmazalp, S.D., et al., Cryst. Eng. Commun., 2018, vol. 20, p. 3858.
Bedekovic, N., Stilinovic, V., Friscic, T., and Cincic, D., New J. Chem., 2018, vol. 42, p. 10584.
Carpio, E.D., Hernandez, L., Ciangherotti, C., et al., Coord. Chem. Rev., 2018, vol. 372, p. 117.
Guo, D.B., Xu, S.T., Huang, Y., et al., Biomaterials, 2018, vol. 177, p. 67.
Mitra, K., Lyons, C.E., and Hartman, M.C.T., Angew. Chem. Int. Ed., 2018, vol. 57, p. 10263.
Yang, J., Cao, Q., Zhang, H., et al., Biomaterials, 2018, vol. 176, p. 94.
Konakanchi, R., Haribabu, J., Prashanth, J., et al., Appl. Organomet. Chem., 2018, vol. 32, p. 4415.
Jose, E.S., Philip, J.E., Shanty, A.A., et al., Inorg. Chim. Acta, 2018, vol. 478, p. 155.
Ghadermazi, M., Gharamaleki, J.A., Olmstead, M.M., and Almasi, M., Acta Crystallogr., Sect. C: Struct. Chem., 2015, vol. 71, p. 539.
Yang, L.R., Song, S., Zhang, W., et al., Synth. Met., 2011, vol. 161, p. 647.
Etaiw, S.E.H. and El-Bendary, M.M., J. Lumines., 2018, vol. 199, p. 232.
Andac, O., Gorduk, S., and Yilmaz, H., J. Iran. Chem. Soc., 2018, vol. 15, p. 1699.
Hassanpoor, A., Mirzaei, M., Eshtiagh-Hosseini, H., and Majcher, A., Cryst. Eng. Commun., 2018, vol. 20, p. 3711.
Zhuang, W., Ju, C.F., Zhang, X.Q., et al., Asian J. Chem., 2014, vol. 26, p. 3116.
Dehkordi, M.N. and Akerman, B., Chem. Biol. Interact., 2018, vol. 282, p. 55.
Jankovic, N., Muskinja, J., Ratkovic, Z., et al., RSC Adv., 2016, vol. 6, p. 39452.
Joksimovic, N., Baskic, D., Popovic, S., et al., Dalton Trans., 2016, vol. 45, p. 15067.
Kocak, I., Yildiz, U., Coban, B., and Sengul, A., J. Solid State Electr., 2015, vol. 19, p. 2189.
Milutinovic, M.M., Rilak, A., Bratsos, L., et al., J. Inorg. Biochem., 2017, vol. 169, p. 1.
Zeng, J., Liu, W., Fan, Y.Z., et al., Theranostics, 2018, vol. 8, p. 109.
Zhu, M.C., He, W.X., Gao, E.J., et al., Life Sci., 2012, vol. 90, p. 519.
Mandegani, Z., Asadi, Z., Asadi, M., et al., Dalton Trans., 2016, vol. 45, p. 6592.
Prosser, K.E., Chang, S.W., Saraci, F., et al., J. Inorg. Biochem., 2017, vol. 167, p. 89.
Abdullaziz, M.A., Abdel-Mohsen, H.T., El-Kerdawy, A.M., et al., Eur. J. Med. Chem., 2017, vol. 136, p. 315.
Follana-Berna, J., Seetharaman, S., Martin-Gomis, L., et al., Phys. Chem. Chem. Phys., 2018, vol. 20, p. 7798.
Thirunavukkarasu, T., Sparkes, H.A., Balachandran, C., et al., J. Photochem. Photobiol., B, 2018, vol. 181, p. 59.
Rendosova, M., Vargova, Z., Kuchar, J., et al., J. Inorg. Biochem., 2017, vol. 168, p. 1.
Colina-Vegas, L., Luna-Dulcey, L., Plutin, A.M., et al., Dalton Trans., 2017, vol. 46, p. 12865.
Manikandamathavan, V.M., Thangaraj, M., Weyhermuller, T., et al., Eur. J. Med. Chem., 2017, vol. 135, p. 434.
Posadas, I., Alonso-Moreno, C., Bravo, I., et al., J. Inorg. Biochem., 2017, vol. 168, p. 46.
Ou, Z., Xu, M.H., Gao, Y.Y., et al., New J. Chem., 2017, vol. 41, p. 9397.
Perontsis, S., Tialiou, A., Hatzidimitriou, A.G., et al., Polyhedron, 2017, vol. 138, p. 258.
Kim, J., Cardenal, A.D., Greve, H.J., et al., Inorg. Chim. Acta, 2018, vol. 469, p. 484.
Kumar, P., Dasari, S., and Patra, A.K., Eur. J. Med. Chem., 2017, vol. 136, p. 52.
Dustkami, M. and Mansouri-Torshizi, H., Int. J. Biol. Macromol., 2017, vol. 99, p. 319.
Das, M., Kundu, B.K., Tiwari, R., et al., Inorg. Chim. Acta, 2018, vol. 469, p. 111.
Mallela, R., Konakanchi, R., Guda, R., et al., Inorg. Chim. Acta, 2018, vol. 469, p. 66.
Jose, G., Kumara, T.H.S., Sowmya, H.B.V., et al., Eur. J. Med. Chem., 2017, vol. 131, p. 275.
Tumer, Y., Asmafiliz, N., Zeyrek, C.T., et al., New J. Chem., 2018, vol. 42, p. 1740.
Zhu, M.C., Cui, X.T., Zhang, S.Z., et al., J. Inorg. Biochem., 2016, vol. 157, p. 34.
Reddy, T.S., Priver, S.H., Mirzadeh, N., and Bhargava, S.K., Eur. J. Med. Chem., 2018, vol. 145, p. 291.
Acilan, C., Cevatemre, B., Adiguzel, Z., et al., BBA-Gen.Subjects, 2017, vol. 1861, p. 218.
Zhang, J.C., Zhang, F.F., Wang, L.W., et al., J. Coord. Chem., 2012, vol. 65, p. 2159.
Zhu, M.C., Song, D., Liu, N., et al., Appl. Organomet. Chem., 2019, vol. 33, p. 13.
Zafar, H., Kareem, A., Sherwani, A., et al., J. Photochem. Photobiology, B, 2015, vol. 142, p. 8.
Stojkovic, D.L., Jevtic, V.V., Radic, G.P., et al., New J. Chem., 2018, vol. 42, p. 3924.
Yan, Q., Li, R.X., Xin, A.Y., et al., Bioorg. Med. Chem., 2017, vol. 25, p. 6542.
Kumar, R.R., Ramesh, R., and Malecki, J.G., New J. Chem., 2017, vol. 41, p. 9130.
Capilla, A.S., Soucek, R., Grau, L., et al., Eur. J. Med. Chem., 2018, vol. 145, p. 51.
Nehru, S., Veeralakshmi, S., and Arunachalam, S., New J. Chem., 2017, vol. 41, p. 13830.
Munzeiwa, W.A., Omondi, B., and Nyamori, V.O., Polyhedron, 2017, vol. 138, p. 295.
Acilan, C., Cevatemre, B., Adiguzel, Z., et al., Eur. J. Cancer, 2016, vol. 61, p. 137.
Kaulage, M.H., Maji, B., Pasadi, S., et al., Eur. J. Med. Chem., 2017, vol. 139, p. 1016.
Bildziukevich, U., Rarova, L., Saman, D., and Wimmer, Z., Eur. J. Med. Chem., 2018, vol. 1945, p. 41.
Funding
We sincerely acknowledge the Natural Science Foundation of China (nos. U1608224, 21171118 and 21671138), the Distinguished Professor Project of Liaoning province, the Liaoning Provincial Natural Science Foundation of China (no. 20180551108), and the Department of Education Basic Research Project of Liaoning Provincial (LJ2019010).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gao, E.J., Hui, Y., Wang, N. et al. Synthesis, Crystal Structures, Interaction with DNA, Cytotoxicity, and Apoptosis Studies of Co(II), Cd(II) Complexes Bearing Pyrazine-2,3-dicarboxylic Acid. Russ J Coord Chem 46, 573–583 (2020). https://doi.org/10.1134/S1070328420080035
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1070328420080035