Abstract
Over the decades, extensive studies have been performed to elucidate the molecular mechanisms underlying the floral transition process in model plants, as well as in crop plants. It has been demonstrated that floral integrator genes, such as FLOWERING LOCUS T and SUPPRESSOR OF OVEREXPRESSION OF CO 1, are highly conserved in most of the flowering plants. This finding has accelerated the identification and functional analyses of these orthologues involved in floral transition in flowering plant species. Even though the upstream regulator networks of the floral integrator genes seem to be quite diverged among plant species, they share four conserved flowering pathways, including the photoperiod, autonomous, gibberellin, and vernalization pathways. The comprehensive knowledge of the molecular mechanisms underlying floral transitions in the model plant Arabidopsis thaliana has helped us explore and elucidate the molecular mechanisms controlling floral transitions in other crop plants. This review highlights the current understandings of the flowering pathways elucidated in Arabidopsis, and mainly focuses on understanding the vernalization pathway in Arabidopsis as well as in several horticultural crop plants, including those of the genus Brassica.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
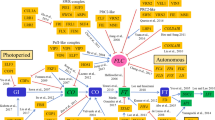
In the life cycle of a plant, the floral transition is important for survival as well as reproductive success. Environmental cues, such as photoperiod (day/night) and temperature, trigger the floral transition. Environmental signals are interpreted by multiple regulatory networks in plants (Fig. 1). Intensive genetic and molecular analyses have identified four major flowering pathways, namely the photoperiod, autonomous, gibberellin, and vernalization pathways, in plants (Kim et al. 2009; Amasino and Michaels 2010; Blazquez et al. 2001; Song et al. 2013; Capovilla et al. 2015). Even though the recent integration of several additional pathways, such as sugar-, hormone-, and ambient temperature-dependent pathways, have been reported (Bolouri Moghaddam and Van den Ende 2013; Wahl et al. 2013; Rolland et al. 2002; Conti 2017; Seo et al. 2011; D’Aloia et al. 2011; Tsai and Gazzarrini 2012; Li et al. 2016; Susila et al. 2018), it is beyond the scope of this review to discuss the details of these pathways. This review discusses the current understanding of each flowering pathway operating in the model plant Arabidopsis thaliana. Furthermore, the current understanding of the vernalization pathway is provided in the latter part of this review.
Schematic of the flowering pathways in Arabidopsis plants. The components involved in each flowering pathway are shown below the corresponding pathway name. The floral integrator genes, including FT and SOC1, act as floral inducers in plants. The leaf-derived FT protein interacts with FD to form an FT-FD dimer (Turck et al. 2008), which activates SOC1 and several meristem identity genes, such as SEP3, FUL, AP1, and LFY, to initiate the floral transition in the Arabidopsis plants (Abe et al. 2005). FLC suppresses floral transition by inhibiting the transcription of FT and SOC1 (Kim et al. 2009; Helliwell et al. 2006). Meanwhile, CO acts to activate the FT and SOC1 genes (Hepworth et al. 2002; Wenkel et al. 2006; Samach et al. 2000). The photoperiod pathway involves the circadian clock that comprises four complicated and intricate complexes and is tightly connected to the light quality pathway (Gardner et al. 2006; Guerriero et al. 2014; Huang and Nusinow 2016). GI acts in both CO-dependent (i.e., by repressing CDF1) and CO-independent ways (i.e., by suppressing the SVP and TEM factors or by promoting miR172 expression) to control the flowering program. The light quality pathway also operates in both CO-dependent and CO-independent ways to regulate of FT expression (Cerdan and Chory 2003; Backstrom et al. 2007). PHYB acts to suppress CO protein activity, whereas PHYA, CRY1, and CRY2 function to enhance the activity of CO. In parallel, PHYB affects FT transcription by suppressing PFT1, an upstream activator of FT (Inigo et al. 2012). The autonomous pathway contains eight components and a recently-identified antisense non-coding RNA (ncRNA) called COOLAIR (Swiezewski et al. 2009). The plant hormone, gibberellic acid (GA), was shown to directly affect the transcription of SOC1 and LFY. Before vernalization, the level of FLC mRNA is highly activated by three cooperative complexes named as the SWR1-complex, PAF1-complex, and FRI-complex (Choi et al. 2011). The vernalization pathway includes two PHD-finger domain proteins, VIN3 and VIL1, and two polycomb complexes, PRC1 and PRC2 (Kim and Sung 2014b). The ncRNAs, COOLAIR and COLDWRAP, were also shown to be involved in the vernalization pathway (Heo and Sung 2011; Kim and Sung 2017). The PHD-PRC2 complex was also shown to associate with other transcriptional regulators, such as VAL1 and VAL2 (Yuan et al. 2016; Questa et al. 2016). The red bar indicates the suppression or negative effect, and the blue arrows indicate the activation or positive effect
2 Floral induction by the photoperiod pathway
Plants utilize photoperiod signals to perceive a seasonal change. Photoperiod (or day length) regulates flowering time in many plants. The core components involved in the photoperiod pathway, such as CONSTANS (CO) and FLOWERING LOCUS T (FT), are well conserved among many plant species (Kobayashi and Weigel 2007). In Arabidopsis, the level of FT mRNA expression determines the timing of bolting (Turck et al. 2008; Corbesier et al. 2007). As a facultative long-day (LD) plant, Arabidopsis exhibits accelerated expression of FT under LD conditions, but lowered expression under short-day (SD) conditions. Thus, the transcriptional activation of FT is critical for the induction of floral transition in the photoperiod pathway (Searle and Coupland 2004).
The photoperiodic induction of flowering appears to operate via a system in which the CO expression levels are affected by day length (Corbesier et al. 2007; Kobayashi and Weigel 2007; Notaguchi et al. 2008). The CO gene, which encodes a BBX domain protein, is a main upstream activator of FT in Arabidopsis (Adrian et al. 2010). In LD conditions, the Arabidopsis CO expression extends to the daytime phase, and light enhances CO protein stability (Yanovsky and Kay 2002; Valverde et al. 2004). The stabilized CO protein directly binds to the proximal promoter region of FT and stimulates the transcription of FT in the inductive LD condition (Hepworth et al. 2002; Wenkel et al. 2006; Samach et al. 2000) (Fig. 1).
The levels of both CO transcripts and CO protein are tightly coordinated by several signaling systems, such as circadian clock and light signaling, including photoreceptors (Fig. 1). In light signaling, the transcription of CO is directly repressed by the upstream regulator, CYCLING DOF FACTOR 1 (CDF1). Blue light promotes the transcriptional activation of CO with the help of a blue-light photoreceptor F-box protein, FLAVIN-BINDING, KELCH REPEAT, F BOX 1 (FKF1) and GIGANTEA (GI) protein. The transcript and protein levels of both FKF1 and GI display a diurnal rhythm and show the highest abundance at the afternoon, which coincides with the peak time of CO protein expression. Abundant amounts of FKF1 or GI physically interact with CDF1, subsequently triggering CDF1 degradation (Fig. 1). Consequently, the reduction of CDF1 releases CO from repression under LD conditions, thus inducing the floral transition (Fornara et al. 2009; Imaizumi et al. 2005; Song et al. 2012b).
In the dark (night) condition, CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1) and members of the SUPPRESSOR OF PHYA-105 (SPA) family (SPA1, SPA3, and SPA4) are abundant and physically interact with one another to form the COP1/SPA complex, which acts to destabilize CO by ubiquitination in the dark condition (Laubinger et al. 2006; Liu et al. 2008). In the blue-light condition, however, two blue-light receptors, CRYPTOCHROME 1 (CRY1) and CRY2, interact with COP1 and SPA and prevent the physical interaction of COP1/SPA with CO, thus stabilizing the CO protein in the blue-light condition (Fig. 1).
In Arabidopsis plants, there are five red/far-red light photoreceptors, called phytochromes (PHYA–PHYE) (Quail 2010), which are produced in the cytosol of plant cells. They can be reversibly converted from an inactive form (Pr) to an active form (Pfr) and vice versa, depending on the presence of light. The inactive Pr form of PHYs in the dark can be converted into their active Pfr form in the light, which is subsequently translocated into the nucleus to trigger light-induced photomorphogenesis (Bae and Choi 2008). Among the red-light receptors, while PHYB acts to delay floral transition by destabilizing the CO protein through ubiquitination, PHYA has an opposite function to enhance the stability of CO by inhibiting the COP1/SPA1-destabilizing activity (Valverde et al. 2004). As a result, the phyA mutants exhibit late flowering, whereas the phyB mutants show early flowering compared to the wild-type plants. Unlike the case of PHYA, the PHYB-mediated control of CO stability does not involve the COP1/SPA1 components (Valverde et al. 2004; Jang et al. 2008). Instead, it was recently proposed that a RING-finger-containing E3-ligase protein, HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE 1 (HOS1) is involved in the PHYB-mediated destabilization of CO (Lazaro et al. 2015). PHYB was shown to physically interact with HOS1 and CO in vivo and in planta, implying that they might form a complex and function to coordinate the precise abundance of CO in the light condition in Arabidopsis. Interestingly, PHYB functions not only in a CO-dependent manner, but also in a CO-independent manner. For instance, PHYB suppresses PHYTOCHROME AND FLOWERING TIME 1 (PFT1)/MEDIATOR 25 (MED25), which acts to activate FT expression in optimal light conditions (Cerdan and Chory 2003; Backstrom et al. 2007).
In several recent studies, it has been proposed that microRNAs (miRNAs) are involved in floral transition in a CO-independent manner (Hong and Jackson 2015; Zhu and Helliwell 2011; Teotia and Tang 2015; Aukerman and Sakai 2003). Two AP2-type transcription factors, TARGET OF EARLY ACTIVATION TAGGED 1 (TOE1) and TOE2, directly bind and repress FT expression. As plants age, both TOE1 and TOE2 are post-transcriptionally inhibited by the action of miR172 (Aukerman and Sakai 2003). TOE1 was shown to block the direct binding of CO to the promoter of FT (Zhang et al. 2015). TOE1 acts together with the transcriptional repressor TOPLESS (TPL) to suppress the FT gene (Fig. 1). In addition, another AP2 transcription factor, SCHLAFMUTZE (SMZ), and its paralogous protein, SNARCHZAPFEN (SNZ), function to suppress FT expression. In a similar way, these two factors were also targeted by miRNA172 in an age-dependent manner (Mathieu et al. 2009; Wu et al. 2009). These data imply that the expression of FT is not only controlled by a light signal, but also by the age-dependent flowering pathway.
Light signaling is indeed connected to the intrinsic circadian clock (Gardner et al. 2006; Guerriero et al. 2014). Different light wavelengths, including red, far-red, blue, and UV, can give rise to different developmental outputs. Light information perceived by photoreceptors is delivered to the circadian clock system to modulate gene sets involved in the floral transition process in Arabidopsis plants (Nakamichi 2011; Imaizumi 2010). Most of the components of the circadian clock act as transcriptional repressors. The circadian clock system in plants comprises four distinctive, but interconnected regulatory complexes: (1) morning complex, (2) middle complex, (3) evening complex, and (4) night complex (Huang and Nusinow 2016; Hsu and Harmer 2014; Shim and Imaizumi 2015; Seo and Mas 2014) (Fig. 1). At the dawn stage, two morning complex components, CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), are abundant and directly suppress the other three complex genes (Adams et al. 2015; Gendron et al. 2012). The middle complex contains several PSEUDO RESPONSE REGULATOR (PRR) genes. The evening complex genes include EARLY FLOWERING 3 (ELF3), ELF4, and LUX ARRYTHMO (LUX) (Kamioka et al. 2016; Nusinow et al. 2011). The night complex includes TIMING OF CAB EXPRESSION 1 (TOC1, also known as PRR1), a transcriptional repressor acting on genes belonging to the other three complexes (Gendron et al. 2012). These components in the circadian clock system are interconnected and form a feedback regulation loop with one another at the transcriptional level.
In early morning, the expression of CCA1 and LHY is robust and directly suppresses the other middle, evening, and night complex genes. CCA1 and LHY were previously shown to recognize and bind to the cis-acting Evening Element (EE) and CCA1-binding sites (CBS). In addition, a genome-wide chromatin immunoprecipitation sequencing (ChIP-seq) assay found that the CCA1-containing complex binds approximately 449 loci and recognizes additional DNA elements, such as G-box and CT repeats, which might be recognized with the help of a binding partner interacting with CCA1 (Kamioka et al. 2016).
From morning to the early evening stage, the PRR proteins PRR7, PRR9, and PRR5 are highly expressed and act to suppress CCA1 and LHY expression (Nakamichi et al. 2010; Liu et al. 2016). This repressive action of the PRR proteins on the CCA1 and LHY genes allows the evening complex genes to be eventually accumulated in the evening time. As a result, the evening complex proteins become dominant at the evening time and then the ELF3-ELF4-LUX evening complex suppresses the middle complex genes through direct binding to the promoter regions of PRR9 and LUX (Dixon et al. 2011; Helfer et al. 2011; Nusinow et al. 2011). At night time, the level of TOC1 becomes abundant and it acts to repress CCA1 and LHY as well as other complex genes (Gendron et al. 2012; Huang et al. 2012). As time progresses toward the end of night, the TOC1-mediated repression is abolished by the action of E3-ubiquitin ligases, ZEITLUPE (ZTL), FKF1, and LOV KELCH PROTEIN 2 (LKP2) (Baudry et al. 2010; Mas et al. 2003). These ZTL family E3-ubiquitin ligases target TOC1 for ubiquitin-mediated degradation at night. GI plays an important role in the ZTL-mediated degradation of TOC1. Indeed, there is a complicated and intricate feedback loop regulation among these circadian clock components. Since it is beyond the scope of this review to describe the details of this network, please refer to other recent reviews (Shim et al. 2017).
During daytime, GI physically interacts with ZTL and protects ZTL from degradation by sequestering the ZTL protein in a GI-ZTL complex (Kim et al. 2007; Kiba et al. 2007; Mas et al. 2003; Pokhilko et al. 2010). At night, ZTL is released from the GI-ZTL complex and then TOC1 and PRR5 are targeted for degradation. As mentioned above, GI interacts with FKF1 and the GI-FKF1 complex acts to increase CO transcription by triggering the degradation of CDF1, the upstream transcriptional repressor of the CO gene. In parallel, GI can regulate the transcription of FT independent of CO in at least two different ways. First, GI physically interacts with a couple of repressors of FT, such as SHORT VEGETABLE PHASE (SVP), TEMPRANILLO 1 (TEM1), and TEM2 (Sawa and Kay 2011). The GI and FT repressors (SVP-TEM1-TEM2) proteins were shown to compete for binding to the promoter site of FT. Thus, it is likely that GI can directly bind and activate FT expression by out-competing the binding of FT repressors to the promoter of FT during the daytime. The second way involves miRNA172, which, as mentioned above, inhibits the expression of AP2-like transcription factors, such as TOE1, TOE2, SMZ, and SNZ, acting to repress FT transcription (Zhang et al. 2015; Mathieu et al. 2009). Interestingly, GI has been shown to enhance the expression of miRNA172, which can subsequently cause a reduction in the AP2 repressor gene transcripts, thus resulting in a switch from transcriptional repression to activation of FT (Jung et al. 2007; Mathieu et al. 2009). These data indicate that there are multiple layers of regulation via a CO-dependent or CO-independent pathway to correctly respond to the photoperiod to optimize plant growth according to the environmental conditions. The information on the individual components involved in the photoperiod pathway is provided in Supplementary Table 1.
In conclusion, light signaling and the circadian clock are integrated to ensure that the CO and FT proteins accumulate only in optimal light conditions (LD), thus allowing plants to correctly respond to a seasonal change.
3 Floral induction by gibberellic acids (GAs)
Gibberellins (GAs) are plant hormones, which are involved in many aspects of plant growth and development, including seed germination, hypocotyl elongation, chlorophyll biosynthesis, and flowering induction (Yamaguchi 2008). GAs are biosynthesized from an initial compound, ent-kaurenoic acid, and the activity of GIBBERELLIN 3-beta-DIOXYGENASE (GA3ox) is important for the production of bioactive GAs, such as GA4, whereas GA2ox converts the bioactive GA4 into its inactive form in Arabidopsis plants (Olszewski et al. 2002). Treatment with GA can initiate the floral transition in many plant species, including Arabidopsis. Mutations in the genes involved in either GA biosynthesis or the GA signaling pathway result in alterations in flowering time (Blazquez et al. 1998, 2002). For instance, the mutant ga1-3, which does not produce GA, fails to flower under SD conditions and displays a delay in flowering under LD conditions (Mutasa-Gottgens and Hedden 2009). GA seems to act independently of the photoperiod pathway because the delayed flowering in ga1 mutants is relatively minor under LD conditions compared to SD conditions. Furthermore, a double mutant of ga1 with the photoperiod mutant co displays an additive phenotype of extremely delayed flowering in LD conditions (Reeves and Coupland 2001). However, GA seems to be not inherently required for vernalization because when the ga1 mutation was introduced into vernalization-requiring plants, the plants still retained a complete vernalization response in LD conditions (Borner et al. 2000; Michaels and Amasino 1999). Instead, it is suggested that GA promotes flowering by promoting the expression of the floral integrators, such as LEAFY(LFY) and SUPPRESSOR OF OVEREXPRESSION OF CO 1 (SOC1), via a DELLA-dependent mechanism (Moon et al. 2003; Achard et al. 2004). Thus, the photoperiod- and GA-induced flowering pathways act independently to promote floral transition by activating the expression of floral integrators such as LFY and SOC1 (Fig. 1).
4 Floral induction by the autonomous pathway (AP)
A handful of late-flowering mutants were derived from the summer annual, rapid-flowering Arabidopsis accessions, like Columbia-0 (Col-0) plants, and were referred to as defining the autonomous pathway (AP) of floral promotion (Simpson 2004; Amasino and Michaels 2010) (Fig. 1). The AP promotes flowering independently of environmental conditions (Amasino and Michaels 2010). The AP mutants, characterized by delayed flowering in both LD and SD conditions, are distinctive from the photoperiod pathway mutants, which exhibit delayed flowering only under inductive LD photoperiods in Arabidopsis. To date, eight AP genes have been identified: LUMINIDEPENDENS (LD), FLOWERING CONTROL LOCUS A (FCA), FLOWERING LOCUS Y (FY), FPA, FLOWERING LOCUS D (FLD), FVE/MIS4 (MULTICOPY SUPPRESSOR OF IRA1 4), FLOWERING LOCUS K (FLK), and RELATIVE OF EARLY FLOWERING 6 (REF6) (Simpson 2004; Lim et al. 2004; Schomburg et al. 2001; Noh et al. 2004). FCA, FPA, FLK, and FY encode proteins that are predicted to be involved in RNA metabolism (Macknight et al. 1997; Schomburg et al. 2001; Simpson et al. 2003; Lim et al. 2004). FVE, FLD, LD, and REF6 have domains commonly found in chromatin-modifying proteins: FVE encodes a WD-repeat protein, which is found in various chromatin-remodeling complexes (Ausin et al. 2004), and FLD and REF6 have been predicted to encode two different types of histone demethylases (He et al. 2003; Jiang et al. 2007; Noh et al. 2004). A major target of these AP proteins is FLOWERING LOCUS C (FLC), a floral repressor. For example, the delayed flowering phenotypes of the AP mutants were completely suppressed in the flc null mutant background, indicating that AP acts to repress a common target, the FLC gene, which inhibits floral transition (Michaels and Amasino 2001; Sheldon et al. 1999). Thus, it is likely that they are involved in floral transition by regulating FLC expression via RNA processing and/or chromatin modification of FLC.
Some of the AP group members were shown to control the expression of FLC by modulating the expression of the antisense non-coding RNA (ncRNA), COOLAIR, which is derived from the 3′ region of the sense FLC locus (Swiezewski et al. 2009). For example, both FCA and FPA contain a plant-specific RNA-recognition motif (RRM)-type RNA-binding domain and regulate alternative polyadenylation of the antisense RNA, COOLAIR (Liu et al. 2010). For example, FCA and FPA were shown to promote the cleavage and polyadenylation event at the proximal sites of the COOLAIR antisense RNA at the end of the sense FLC transcript. In the absence of FPA and FCA, the 3′-end formation occurs at the distal sites of COOLAIR, reaching the proximal promoter region of the sense FLC gene.
FCA was shown to physically interact with another RNA-processing factor, FY. This interaction is mediated by the PPLPP motifs in FY and the WW domain within FCA (Henderson et al. 2005). In addition, FCA and FPA require FLD, a histone H3K4 demethylase, to downregulate the FLC mRNA level, showing a link between RNA processing and chromatin modification (Liu et al. 2007).
FLK encodes a plant-specific K-homology (KH) RNA-binding protein (Lim et al. 2004). The mutants of the FLC gene displayed a severe late flowering phenotype under both LD and SD conditions. Consistent with this, FLC expression was highly upregulated in the FLC mutants. The delayed flowering phenotype was suppressed by the GA and vernalization treatments, indicating that FLK acts in the autonomous pathway. However, the molecular function of FLK in FLC expression is not clearly determined.
FVE, also referred to as MULTICOPY SUPPRESSOR OF IRA1 4 (MSI4), acts in a large chromatin-modifying complex, POLYCOMB REPRESSIVE COMPLEX 2 (PRC2), which catalyzes a repressive histone mark, the tri-methylation of histone H3-lysine 27 (H3K27me3). The PRC2 complex is described in detail in the section describing the vernalization pathway in this review.
The LD protein contains a homeodomain-like domain and is localized in the nucleus (Aukerman et al. 1999; Lee et al. 1994). It negatively inhibits the transcriptional activity of SUPPRESSOR OF FRIGIDA 4 (SUF4), which activates the transcription of FLC in a FRIGIDA (FRI)-containing protein complex in Arabidopsis. Even though it is obvious that LD acts to repress FLC expression, its detailed mechanism remains to be clarified.
REF6 belongs to a group of jumonji (Jmj)-domain family proteins (Noh et al. 2004). The Jmj-domain proteins act in a large complex, the chromatin demethylase complex, which functions to remove a certain methyl group from the target gene chromatins. It has been recently demonstrated that REF6 is an H3K27me3 demethylase acting on FLC in Arabidopsis plants (Lu et al. 2011; Li et al. 2016). Another recent study suggested that REF6 might act primarily on antisense RNAs with some other AP RNA-binding proteins (Hornyik et al. 2010), confirming that REF6 might link chromatin demethylation and RNA processing in the regulation of FLC expression. However, it is still not clear how a chromatin demethylase, REF6, can participate in the regulation of the transcript levels of the antisense RNA, COOLAIR. The H4 acetylation levels in the FLC chromatin decreased in the ref6 mutants compared to the wild-type plants, indicating that REF6 enables a close relationship between histone demethylation and acetylation. Thus, it might be an interesting topic to identify the chromatin regulatory proteins that interact with REF6 for histone demethylation and acetylation of FLC by biochemical analyses, such as an immunoprecipitation (IP) assay. The individual components of the autonomous pathway are described in Supplementary Table 1.
It is evident that the AP group genes act primarily on FLC because a mutation in the FLC gene completely suppresses the late flowering phenotype. However, recent studies reported that some AP genes are also involved in other developmental processes, including seed germination (Baurle et al. 2007; Veley and Michaels 2008; Auge et al. 2018). For instance, the mutants of some AP genes displayed abnormal seed germination frequencies (Auge et al. 2018). In addition, FCA and FPA were shown to be, at least partly, involved in RNA-mediated transposon silencing. Thus, it is likely that there might be still unidentified functions of the AP genes in plant developmental programs.
5 Floral induction by the vernalization pathway
Vernalization is a process in which plants acquire the competence to flower in the following spring through exposure to long-term cold temperatures (Kim et al. 2009; Amasino and Michaels 2010; Amasino 2004; Kim and Sung 2014a; Song et al. 2012a). Unlike the cold acclimation response, vernalization is not immediately triggered by a short-term cold stimulus (Amasino 2005; Sung and Amasino 2005; Amasino 2004). Rather, as a result of vernalization, accelerated flowering appears when the original stimulus (low temperatures) is removed, in other words, when plants are re-exposed to warm temperatures in the following spring. This epigenetic nature of vernalization indicates that low temperatures during winter establish stable changes that last until the following spring to evoke floral transition (Kim and Sung 2014a; Amasino et al. 2017; Amasino 2018).
Arabidopsis plants can be grouped into summer-annual and winter-annual plants based on their vernalization requirement (Amasino 2004, 2018). Previous genetic and molecular studies identified that two major factors, FLC and FRI, provide the vernalization requirement in the winter-annual ecotype plants. Both FLC and FRI repress flowering in Arabidopsis plants (Henderson et al. 2003). FRI acts to upregulate the expression of the floral repressor FLC in winter-annual plants, thus inducing delayed flowering, whereas summer-annual plants harbor a genomic deletion in the FRI allele, failing on the upregulation of FLC and consequently showing an early flowering phenotype (Amasino 2005; Sheldon et al. 1999; Michaels and Amasino 2001; Johanson et al. 2000). FLC, a MADS-box protein, functions to suppress flowering by directly inhibiting the expression of the floral integrator genes, such as FT and SOC1 (Kim et al. 2009; Helliwell et al. 2006). Prior to vernalization, FLC is highly expressed to prevent the floral transition. The FLC-mediated inhibition of the floral transition is most pronounced in winter-annual plants because of the presence of the functional FRI allele in Arabidopsis. A prolonged exposure to cold (i.e., the winter season) eventually represses FLC expression. The cold-triggered repressed state of FLC is stably maintained throughout subsequent mitotic cell divisions even in warm growth temperatures of the following spring season. The repression of FLC releases the repression of FT and SOC1 to initiate the floral transition after return to warm conditions (Bastow et al. 2004; De Lucia et al. 2008). Therefore, the vernalization-mediated stable repression of FLC is a prerequisite for floral transition in the next spring season, wherein the CO-mediated photoperiod pathway strongly promotes the expression of the floral integrator genes, FT and SOC1 (Fig. 1).
6 Polycomb group proteins (PcG): PRC1 and PRC2
Polycomb group proteins (PcG) are evolutionarilly conserved multi-protein complexes that play important roles in the epigenetic control of gene expression in plants as well as other eukaryotes (Kim et al. 2009; Molitor and Shen 2013). The PcG genes were first isolated from the genetic mutant screening of Drosophila to identify the genes involved in controlling homeotic gene expression (Simon and Kingston 2013). To date, 18 PcG proteins have been identified in Drosophila and 18–37 homologs were shown to exist by multiple duplication events in mammals. An increasing number of proteins exist as members of the PcG complexes, providing an additional layer of complexity in their function (Di Croce and Helin 2013). These PcG proteins have long been one of the excellent models to elucidate the epigenetic mechanisms of cell development (Schwartz and Pirrotta 2007; Simon and Kingston 2013). They are classified into two different groups of multi-protein complexes, polycomb repressive complex 2 (PRC2) and polycomb repressive complex 1 (PRC1) (Schatlowski et al. 2008; Margueron and Reinberg 2011). The PRC2 and PRC1 complexes repress gene expression through covalent histone modifications H3 methylation and H2A ubiquitination, respectively.
7 Polycomb components involved in vernalization
The Arabidopsis core PRC2 complex containing CURLY LEAF (CLF) regulate floral transition by silencing the expression of the floral repressor FLC (De Lucia et al. 2008; Kim and Sung 2013; Wood et al. 2006). Through vernalization, the PRC2 complex is substantially enriched at FLC chromatin (Fig. 2). Through genetic screening, which was aimed to isolate the mutants resistant to the vernalization treatment, several components have been identified to date. One of these components is VERNALIZATION INSENSITIVE 3 (VIN3), which encodes a PLANT HOMEODOMAIN (PHD) finger domain protein (Bastow et al. 2004; Sung and Amasino 2004). VIN3 is temporarily expressed upon long-term exposure to cold and rapidly disappears upon return to warm growth conditions, implying that VIN3 might be one of the early factors triggering the vernalization response. A PHD finger domain within the VIN3 protein was shown to bind preferentially to H3K9me2, a repressive histone mark, in an in vitro histone peptide assay (Kim and Sung 2013). The enrichments of H3K9me2 and H3K27me3, which accumulate during vernalization, were significantly impaired in the vin3 mutants. The repression of the expression of FLC was alleviated in the vin3 mutants during and after vernalization, indicating that VIN3 is essentially required for the proper repression of FLC during vernalization. These data suggest that VIN3 plays a pivotal role in the reduction of FLC expression in vernalization via modulating epigenetic histone modifications.
Schematic model of transcriptional repression by polycomb complexes, PRC1 and PRC2. Upon exposure to long-term cold, a potent floral repressor, FLC, is repressed by both multi-protein complexes, PRC1 and PRC2. Each polycomb complex has several core components, which are highly conserved among higher eukaryotes. In addition, each polycomb complex physically associates with several other regulators. For instance, two PHD-finger domain proteins (VIN3 and VIL1), two B3 domain proteins (VAL1 and VAL2), and several HDAC proteins, including HDA19, were reported to associate with the PRC2 core complex. Furthermore, the PRC1 core complex components associate with LHP1, VRN1, and EMF1 to completely suppress FLC expression in Arabidopsis
The biochemical and genetic analyses demonstrated that another PHD finger protein, VIN3-LIKE 1 (VIL1)/VERNALIZATION5 (VRN5), physically interacts with VIN3 in vitro, as shown a by yeast two-hybrid assay, and associates with the VIN3-containing PRC2 complex (Fig. 2) (Sung et al. 2006b; Greb et al. 2007). Like the case of VIN3, VIL1/VRN5 is also involved in the vernalization-mediated repression of FLC. The vil1/vrn5 mutants displayed the de-repressed expression of FLC and a delayed flowering time after vernalization. Thus, these two PHD-finger domain proteins, VIN3 and VIL1/VRN5, act together in a PRC2 complex (often referred to as PHD-PRC2) for repressing FLC expression during vernalization.
The Arabidopsis PRC1 complex comprises two core members, RING1 proteins and Arabidopsis homolog of B Lymphoma Mo-MLV Insertion Region 1(AtBMI1) proteins (Fig. 2) (Kim and Sung 2014b; Merini et al. 2017). Two E3-ubiquitin ligases, RING1A and RING1B, catalyze monoubiquitylation at histone H2A lysine 121 (H2AK121ub) of target chromatins in Arabidopsis (Bratzel et al. 2010). AtRING1A (AT5G44280) and AtRING1B (AT1G03770), the homologs of human RING1A and RING1B, respectively (Kim and Sung 2014b; Sanchez-Pulido et al. 2008), were shown to redundantly regulate key developmental genes related to embryo development and shoot and root meristem development (Molitor and Shen 2013; Chen et al. 2016; Xu and Shen 2008). While AtRING1A/B executes the enzymatic activity for H2AK121ub modification, three Arabidopsis homologs of human BMI1, AtBMI1A (At2g30580), AtBMI1B (At1g06770), and AtBMI1C (At3g23060), act to stimulate the enzymatic activity of RING1A/B in the Arabidopsis PRC1 complex. The double or triple mutants of the AtBMI1A/B/C genes displayed detrimental defects in a diversity of plant developmental programs, including embryogenesis, seed dormancy, flower and root development, among others (Chen et al. 2010; Merini et al. 2017; Pico et al. 2015).
LIKE-HETEROCHROMATIN PROTEIN 1(LHP1)/TERMINAL FLOWER2 (TFL2) was shown to accumulate at the FLC chromatin as a result of vernalization (Fig. 2) (Sung et al. 2006a; Mylne et al. 2006). In the lhp1 mutants, the vernalization-mediated repression of FLC is not stably maintained because of the impaired enrichment of the H3K27me3 mark on the FLC chromatin. Recently, it was reported that LHP1 interacts with two B3 domain DNA-binding proteins, VP1/ABI3-LIKE 1 (VAL1) and VAL2, to achieve the stable repression of FLC in vernalization (Fig. 1) (Yuan et al. 2016; Questa et al. 2016). Like LHP1, VAL1 and VAL2 were also shown to preferentially bind to the H3K27me2/3 histone mark via its PHD-L-domain. VAL1 and VAL2 were shown to bind to the Sph/RY-like (-TTCTGCATGG-) motifs located in the first intron of the FLC genomic region (Questa et al. 2016; Yuan et al. 2016). They were also co-purified with the VIN3-containing PRC2 complex and HDA19-containing HDAC (histone deacetylase) complex, indicative of their linker role in connecting the LHP1-PRC1, PHD-PRC2, and HDAC complexes (Fig. 2). Most of all, the loss of the VAL1 gene resulted in a de-repressed level of FLC expression and a lower level of the H3K27me3 mark deposited by PRC2 and a higher level of the H3K27ac mark after vernalization when compared to the wild-type plants. Thus, it is likely that VAL1 acts as a recruiter of three distinct repressive complexes, LHP1-PRC1, PHD-PRC2, and HDAC, which coordinate the proper repression of FLC via H3K27me3 deposition and H3K27ac removal. However, even though VAL1, potentially with VAL2, is suggested to work as a sequence-specific repressor in the PRC2 complex for FLC, it is still ambiguous how VAL1/2 induces vernalization-mediated FLC repression because both VAL1 and VAL2 are consistently expressed, and they appear to bind to the FLC chromatin, irrespective of the cold condition. Further studies are needed to address this question.
Another B3 domain protein, VERNLAIZATION 1 (VRN1), is considered as a member of the non-canonical PRC1 complex in Arabidopsis and contributes to vernalization-mediated repression of FLC in Arabidopsis (Fig. 2) (Levy et al. 2002). VRN1 was shown to bind DNA in a non-sequence specific manner. In vrn1 mutants, the enrichment of H3K9me2 on FLC chromatin is severely compromised, but the histone modification of H3K27me3 was not impaired, indicating that VRN1 might be specifically involved in H3K9me2 deposition on target chromatin. Even though the vrn1 mutants displayed significantly delayed flowering time in vernalization, its function in vernalization is not well addressed to date. Another component of core PRC1, AtBMI1C was shown to be involved in the repression of FLC through its H2Aub1 activity (Li et al. 2011). However, it is not clarified yet whether AtBMI1C is involved in the vernalization-mediated suppression of FLC. Detailed information on the individual components of these polycomb complexes is provided in Supplementary Table 1. In conclusion, the Arabidopsis core PRC1 and PRC2 complex closely interacts with other chromatin regulators (e.g., HDA19) and transcription factors (e.g., VAL1) to precisely repress FLC in vernalization.
8 Vernalization response in other flowering plants
Although Arabidopsis plants have served as an excellent model system to understand the molecular mechanisms of the vernalization response, it has not been clearly understood whether other vernalization-required species might use similarly conserved or different gene regulatory circuitries for their vernalization response. Here, I briefly describe the current understanding on molecular circuitries working in the vernalization response in other flowering plant species.
8.1 Alphine rock-cress (Arabis alpina L.)
Arabis alpina, a perennial relative of Arabidopsis, is distinctive in its vernalization response compared to the annual/biennial Arabidopsis accessions (Ansell et al. 2008; Koch et al. 2006). In contrast to the annual Arabidopsis plants, the A. alpina plants do not require an inductive photoperiod (Wang et al. 2009; Bergonzi et al. 2013). Thus, young, juvenile-stage A. alpina plants, which have only immature meristems, do not respond to vernalization for floral transition (Bergonzi et al. 2013).
Annual plants initiate floral transition in all apical meristems at the same time during their life time; this phenomenon is known as monocarpy. In contrast, perennial plants bloom in spring and summer seasons, but arrest flowering in the later seasons. Perennial plants resume vegetative growth in the fall and repeatedly undergo vernalization. Therefore, perennial plants flower and set seeds many times in their life time (known as polycarpy). The A. alpina plants repeat the cycle of vegetative and reproductive growth phases. Similar to Arabidopsis, an ortholog of FLC, PERPETUAL FLOWERING 1 (PEP1), functions as a major floral repressor in A. alpina plants (Wang et al. 2009). The expression of PEP1 is repressed by vernalization, thus allowing plants to bloom (Fig. 3a). However, unlike annual Arabidopsis plants, PEP1 is de-repressed when plants are returned to warm growth temperatures. Reactivated PEP1 in warm conditions represses A. alpina FT orthologs (AaFT1 and AaFT3), which promotes flowering in a way similar to annual Arabidopsis plants (Hyun et al. 2019). This fluctuating nature of the repression of PEP1 allows A. alpina plants to display polycarpic flowering behavior in their lifespan. Consistent with the fluctuating pattern of PEP1 mRNA expression, a repressive histone mark, H3K27me3, is enriched at the PEP1 chromatin during the cold exposure, but depleted when plants are exposed to warm temperatures.
Outline of flowering pathways in several crop plants. aArabis alpina is a perennial species related to Arabidopsis. In its young vegetative stage (upper panel), the juvenile-to-adult vegetative transition is strictly prohibited by miRNA156 action. miRNA156 is abundantly expressed in the early seedling stage and blocks the juvenile-to-adult vegetative transition by degrading the mRNA of a group of SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes, which play a role in developmental transition from the vegetative to reproductive stage via activating the floral activator, Arabis alpina FLOWERING LOCUS T (AaFT) (Wang et al. 2009; Hyun et al. 2019; Bergonzi et al. 2013). Thus, the miRNA156-targeted silencing of SPL15 plays a major role in inhibiting floral transition in the early vegetative stage of the A. alpina plants. As time progresses, the levels of miRNA156 decrease and, conversely, SPL accumulates abundantly and provides partial competence to flowering plants. However, even in the adult vegetative stage (bottom panel), a MADS-box domain protein, named PERPETUAL FLOWERING 1 (PEP1), which is a homolog of Arabidopsis FLC, actively acts to prohibit floral transition. The A. alpina ortholog of Arabidopsis AP2, PERPETUAL FLOWERING 2 (PEP2), enhances the expression of PEP1 in the vernalization pathway, thus blocking floral transition. Vernalization triggers the silencing of PEP1 to induce the floral transition by, at least in part, activation of AaFT. b In beet, the regulation of floral transition is shown to be controlled by the antagonistic interplay of two homologs of the Arabidopsis FT (Pin et al. 2010). BvFT2 acts as a floral activator, similar to the function of Arabidopsis FT. In contrast, BvFT1 functions to repress floral transition by suppressing BvFT2 expression. Thus, the vernalization pathway in beets targets BvFT1 for downregulation in order to induce flowering. c In lettuce, high temperature triggers the expression of two heat-shock transcription factors, HsfA1e and HsfA4c, which directly bind to the promoter region of Lactuca sativa SOC1 (LsSOC1) and induce heat-promoted bolting. LsSOC1 encodes a MADS-box protein and acts as a floral activator in lettuce (Chen et al. 2018). Two other MADS-box domain proteins, LsAGL6 and LsAGL24, physically interact with LsSOC1, forming a multi-protein complex that plays a key role in floral induction in lettuce. d In Brassica species, including B. rapa, the vernalization pathway acts to reduce the expression of BrFLC2, as well as other BrFLC paralogous genes, via epigenetic histone modifications in a manner similar to that in Arabidopsis plants (Kawanabe et al. 2016). Active histone marks, H3K4me3 and/or H3K36me3, were highly enriched at the BrFLC chromatin before vernalization, providing robust BrFLC expressions to block floral transition. The vernalization treatment removes the active histone marks from the BrFLC chromatin, and promotes the high-level accumulation of repressive histone marks, such as H3K27me3, at the BrFLC region, which is stably maintained even after return to the normal growth temperature. This indicates that epigenetic histone modifications play an important role in changing the expression levels of the BrFLC genes in B. rapa
In Arabidopsis plants, TERMINAL FLOWER1 (TFL1) represses the floral transition and regulates inflorescence architecture. In a similar way, the ortholog of TFL1 (AaTFL1) in A. alpina is also shown to be involved in the response to the vernalization-mediated floral transition (Wang et al. 2011a). A mutation in AaTFL1 resulted in early flowering and determinate inflorescences, which prematurely terminate the flowering program after the formation of a few flower buds. The silencing of AaTFL1 in A. alpina did not eliminate the vernalization requirement for floral transition, but allowed the plants to respond to a relatively shorter duration of cold. These data indicate the genetic connection between the duration of the vernalization requirement and inflorescence determinacy in A. alpina plants. In addition, AaTFL functions as a repressor of flowering to guarantee that plants spend a certain period of developmental growth before floral transition in A. alpina plants.
An APETALA2 (AP2)-type transcription factor, PEP2 (Arabis ortholog of Arabidopsis AP2), also functions to repress flowering in A. alpina plants (Bergonzi et al. 2013). PEP2 acts to up-regulate PEP1 in order to prevent flowering prior to vernalization. Interestingly, A. alpina plants respond to vernalization only when they reach a certain mature age. This age-dependent response to vernalization is achieved by a miRNA group termed as miRNA156 (Bergonzi et al. 2013). When plants are at the young and juvenile vegetative stage, miRNA156 is abundantly expressed and inhibits flowering by degrading the transcripts of the SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) floral activators, which induce the expression of AaFT, the A. alpina homolog of FT. However, as plants age, the miRNA156 levels eventually decline, resulting in an increase in the expression levels of the SPL floral activators. The increased amounts of SPL protein acts to upregulate the level of miRNA172, which targets the mRNA of a group of floral repressors, including TOE1 and TOE2, thus providing competence to flowering at the adult vegetative stage (Fig. 3a). Thus, the miRNA156-targeted SPL repression module allows only adult vegetative-stage plants to respond to vernalization.
Previously, a genome-wide analysis found that PEP1 directly binds to the promoter of SPL15 in A. alpina plants (Mateos et al. 2017). Most recently, SPL15 was shown to play a key role in the age-dependent vernalization response in perennial A. alpina plants (Hyun et al. 2019). The activity of SPL15 is uniquely confined to older shoots and branches during vernalization, allowing only adult-stage meristems to respond to cold. The annual vernalization response was recapitulated in A. alpina perennial plants by using the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) genome editing tools and interspecific gene transfer methods, confirming that the age-dependent vernalization response can be conferred by PEP1-mediated repression of the SPL15 module in perennial A. alpina plants.
Altogether, two tightly interlocked modules, vernalization (PEP2-PEP1 module) and juvenility (miRNA156-mediated repression of the SPL15 module) function cooperatively to ensure that A. Alpina plants become competent to flower only when they have reached the appropriate vegetative stage and have been exposed to vernalization (Fig. 3a).
8.2 Beet (Beta vulgaris L.)
In beet, the FT homologs, BvFT1 and BvFT2, which encode the proteins of the phosphatidylethanolamine-binding protein (PEBP) family, act antagonistically in flowering (Fig. 3b). While BvFT1 represses the floral transition, BvFT2 activates flowering (Pin et al. 2010). It has been shown that vernalization acts to decrease the expression of the floral repressor, BvFT1. The vernalization-triggered repression of BvFT1 is consistently maintained even after plants are returned to warm growth temperatures, indicating that BvFT1 is a functional equivalent of the Arabidopsis FLC gene. The vernalization requirement in beet is provided by one dominant allele named BvBTC1, which regulates the downstream targets, BvFT1 and BvFT2 (Pin et al. 2012) (Fig. 3b). Some annual beets not requiring vernalization have a dominant allele of BvBTC1, whose expression is increased by long days and which promotes flowering by reducing the expression of BvFT1 and activating the transcription of BvFT2 under LD conditions. As a result, the annual beet plants carrying a functional BvBTC1 gene exhibit rapid flowering and do not require vernalization for flowering. Meanwhile, the biennial beet plants possess a partial loss-of-function allele of BvBTC1, Bvbtc1, which is not substantially induced under LD conditions without the vernalization treatment. The Bvbtc1 allele is only gradually activated by vernalization and is able to reach to the level sufficient to suppress the floral repressor gene, BvFT1, and activate the floral activator gene, BvFT2.
8.3 Lettuce (Lactuca sativa L.)
A cold-temperature (4 °C) treatment does not promote the floral transition in lettuce, whereas a high-temperature treatment at the late seedling stage accelerates this progress (Fukuda et al. 2011). Thus, lettuce plants are considered as so-called “non-low temperature vernalization” plants. Floral transition in lettuce is promoted under high-temperature conditions. Upon bolting, lettuce loses its economic value because its leaves accumulate a bitter taste. Thus, understanding the mechanism of bolting in lettuce is important for breeding for high-value traits. Recently, the molecular mechanism underlying the floral transition in lettuce was elucidated (Chen et al. 2018). A homolog of Arabidopsis SOC1, LsSOC1, was identified as one of the key factors responsible for heat-promoted floral transition (Fig. 3c). LsSOC1 encodes a MADS-box protein, which acts as a floral activator in lettuce. In addition, it was shown that two other MADS-box domain proteins, lettuce homolog of Arabidopsis AGAMOUS-LIKE 6 (LsAGL6) and LsAGL24, physically interact with LsSOC1, forming a multi-protein complex for floral induction (Fig. 3c). Furthermore, two heat-shock transcription factors, HsfA1e and HsfA4c, were shown to directly bind to the promoter region of LsSOC1 and induce heat-promoted bolting.
8.4 Brassica rapa and B. oleracea
Brassica is a genus belonging to the family Brassicaceae and contains 37 flowering plant species, including Brassica rapa and B. oleracea. Similar to Arabidopsis, they flower early under LD conditions (Leijten et al. 2018). They commonly have spring- and winter-type plants. The spring-type plants do not require vernalization and display early flowering (Qi et al. 2015; Yi et al. 2014). They have been grown in geographical areas with severe winter climates or in subtropical climates. The winter-type plants require vernalization for inducing flowering and have been grown in areas with moderate winter climates. A comparative phylogenic analysis of the Brassica plants identified three FLC clades, which reflect the occurrence of genome-wide triplication events during the evolution of their genomes (Razi et al. 2008; Schranz et al. 2002; Zou et al. 2012).
The B. rapa crops, including Chinese cabbage, pak choi, and turnip, are cultivated worldwide and are most popular in Asian countries (Wang et al. 2011b; Leijten et al. 2018; Lee et al. 2015; Kim et al. 2014; Takada et al. 2019). Several quantitative trait loci (QTL) influencing flowering time in B. rapa were identified using the F2 mapping population between an annual and a biennial cultivar (Teutonico and Osborn 1995; Osborn et al. 1997). In the genome of the B. rapa cultivar ‘Chiifu-401-42,’ four FLC homologs were identified and named as BrFLC1 (Bra009055), BrFLC2 (Bra028599), BrFLC3 (Bra006051), and BrFLC5 (Bra022771). BrFLC1 and BrFLC2 were shown to be linked to the QTLs controlling flowering time using an F2 mapping population (Li et al. 2009; Yuan et al. 2009). In addition, a QTL analysis using another F2 mapping population between early-flowering and late-flowering cultivars reported that BrFLC2 was co-localized to a major QTL (Zhao et al. 2010; Xiao et al. 2013; Wu et al. 2012). In a different F2 population (Early × Tsukena No. 2), BrFLC2 and BrFLC3 were co-localized to the QTLs affecting flowering time after the vernalization treatment (Kakizaki et al. 2011). These two genes were detected to have large insertions in the first intron, suggesting that the sequence element in the first intron might be responsible for the repression of the BrFLC2 and BrFLC3 genes upon vernalization (Kitamoto et al. 2014). It was shown that the protein-coding sequences of these four BrFLC genes are highly conserved among the Brassica species, although BrFLC5 is considered to be a pseudogene because it lacks two exons. However, the genomic sequence of the upstream part and intron region of the FLC homologs were relatively divergent among the Brassica species (Zou et al. 2012), suggesting that the sequence divergence of these non-coding regions might account for different expression patterns in the vernalization response of the B. rapa species. The expression of all four BrFLC homologs was decreased by vernalization and stably maintained at low levels even after plants were exposed to warm temperatures (Fig. 3d) (Kawanabe et al. 2016). Active histone marks, H3K4me3 and/or H3K36me3, were highly enriched at the BrFLC chromatins before vernalization, providing robust BrFLC expressions to block the floral transition (Fig. 3d). The vernalization treatment removes the active histone marks from the BrFLC chromatin, and represses histone marks, such as H3K27me3, which are highly accumulated at the BrFLC region, which is stably maintained even after return to normal growth temperatures. This indicates that similar to the case of Arabidopsis, epigenetic histone modifications play an important role in changing the expression levels of the BrFLC genes in B. rapa varieties.
The B. oleracea plants include many commercially important vegetables and can be categorized according to their edible parts. For example, cabbage, kohlrabi, and kale are harvested at the vegetative stage, while broccoli and cauliflower are harvested for their curd (edible flower head part of the plant) after bolting. Therefore, the regulation of the flowering time of these Brassica species is of great interest and importance. Owing to the limitation of space, this review focuses on the vernalization of B. oleracea plants, such as cabbage (B. oleracea L. var. capitata), cauliflower (B. oleracea L. var. botrytis), and broccoli (B. oleracea L. var. italica).
The orthologs of FLC were also identified from B. oleracea plants and reported to be involved in floral transition (Lin et al. 2005; Leijten et al. 2018). Through intensive QTL analyses, BoFLC4 (also known as BoFLC2) was identified as a major locus conferring the vernalization requirement in broccoli (Okazaki et al. 2007; Irwin et al. 2016), cabbage (Okazaki et al. 2007), and cauliflower (Ridge et al. 2015). A genomic fragment containing the whole sequence of the BoFLC4 gene was transformed into the Arabidopsis FRI_flc2 mutant background (Michaels and Amasino 1999). This heterologous transformation of BoFLC4 complemented an early flowering phenotype of FRI_flc2, suggesting that BoFLC4 might contribute to provide the vernalization requirement in B. oleracea plants (Irwin et al. 2016). In another QTL analysis, BoFLC3, BoFLC5, and BoFLC1 were also found to co-localize with a QTL (Razi et al. 2008). Recently, BoFLC3 was shown to be involved in curd induction variation in the subtropical broccoli breeding lines under a subtropical environment (Lin et al. 2018). Interestingly, another recent study reported that a FLC homolog, named the BoFLC1.C9 locus, contains an insertion of 67 nucleotides in the second intron in late-flowering cultivars, which seems to be originated from two DNA fragments of the Arabidopsis FLC sequence (Abuyusuf et al. 2019). Two FT loci (BoFT.C2 and BoFT.C6) have been identified and shown to exhibit an increased expression pattern after the vernalization treatment, in a manner similar to Arabidopsis FT (Lin et al. 2005; Ridge et al. 2015; Irwin et al. 2016). Even though the BoFLC clade genes are well conserved and demonstrated to be involved in flowering time control in B. oleracea, a detailed understanding of the role of these BoFLC genes is still lacking. A method using the CRISPR/Cas9 genome editing tools might be a good approach to study the detailed role of each BoFLC member involved in flowering time control of B. oleracea plants. Additionally, through F2 mapping and a candidate gene approach, a recent paper reported that a gene with a peroxidase domain, named BolPrx.2, contributes to the variation of flowering time in cabbage (Abuyusuf et al. 2018). Interestingly, an intron of BolPrx.2 displayed 76% sequence similarity to the Arabidopsis FLC sequence. In addition, an early flowering accession used in this study showed a 27-bp insertion and a 2-bp deletion in the intron region, which might result in the variation of bolting time. It might be an interesting topic to investigate the biological role of these inserted/deleted DNA fragments in terms of their recruitment of the polycomb complex and epigenetic gene silencing processes on flowering-related genes in cabbage.
In Arabidopsis plants, the proximal promoter and the first intron region of FLC are important for its stable repression (Sheldon et al. 2002; Helliwell et al. 2011). More recently, two Arabidopsis non-coding RNAs, COLDAIR and COLDWRAP, were shown to originate from the first intron and the proximal promoter region of FLC, respectively. They are involved in the vernalization-mediated intragenic gene loop formation (Heo and Sung 2011; Kim and Sung 2017). As mention above, a large insertion in the first intron of BrFLC2 and BrFLC3 caused a defect in vernalization-mediated repression, suggesting that the sequence elements in the first intron of BrFLC2 and BrFLC3 might be required for the vernalization-mediated repression of the BrFLC genes. However, the sequences similar to COLDAIR or COLDWRAP have not been identified yet in Chinese cabbage (B. rapa subsp. pekinensis) and remain to be further studied. The absence or mutation of the COLDAIR and COLDWRAP sequences in A. thaliana resulted in a defect in intragenic loop formation between the proximal promoter and the end of the first intron region. It resulted in the de-repression of Arabidopsis FLC and displayed an extremely late-flowering phenotype (Kim and Sung 2017). Thus, it would be an interesting topic to investigate whether the BrFLC genes also undergo an intragenic chromatin conformational change because of the vernalization treatment. Another antisense ncRNA group, COOLAIR, which originates from the 3′ region of FLC, was also suggested to be involved in the regulation of FLC in Arabidopsis (Swiezewski et al. 2009). In B. rapa, COOLAIR-like transcripts were detected in the BrFLC2 gene. Because overexpression of the COOLAIR-like transcripts resulted in the reduced expression of FLC and an early-flowering phenotype, the COOLAIR-like transcripts in B. rapa might be involved in the repression of BrFLC2 and possibly other BrFLC genes. Taken together, BrFLC2 might play a major role in the vernalization response. DNA element(s) responsible for the stable repression of BrFLC2 in the vernalization response remain to be further identified.
9 Conclusion
Flowering time is an agriculturally important trait. Knowledge of the mechanisms underlying flowering time control in plants can be applied to improve important crop traits. For example, the leafy vegetables of the genus Brassica, such as Chinese cabbage, eventually lose their commercial value after bolting because energy and metabolites are reallocated to reproductive tissues, causing the devaluation of their leafy tissues. In contrast, canola (B. napus) is mainly cultured to harvest seeds for oil. Therefore, an appropriate control of flowering time can maximize the productivity and quality of leaf, flower, or seed tissues in crop plants. In this regard, understanding the molecular mechanisms underlying floral transition is of particular interest in agricultural breeding programs.
This review describes the current understanding of the molecular mechanisms revealed in the model plant Arabidopsis and several crop plants. Intensive studies on this topic greatly increased our knowledge on the genes involved in these pathways, protein–protein interactions, and the molecular interaction networks among these genes. In addition, the epigenetic chromatin regulators (i.e., PRC complexes) of the key genes involved in flowering time expanded our understanding of how plants optimize their growth and development according to changing environmental cues. However, some areas still remain unclear. For example, it is not fully understood how plants accurately measure the duration of cold in the winter season and prevent premature flowering even in fluctuating temperature variations during winter. The mechanisms by which plants can count the length of the vernalizing cold temperatures should be independent of the cold acclimation pathway.
Even though our understanding of flowering pathways in crop plants has been greatly enhanced, the molecular mechanisms underlying these pathways remain poorly understood. Therefore, it is required to explore and elucidate the molecular mechanisms controlling flowering pathways in crop plants. We expect that the comparative study based on the model plant Arabidopsis will help us acquire more comprehensive understanding on the molecular details underlying plant flowering programs. Especially, the computation analysis using the next-generation sequencing data and genomics tools for map-based cloning will highly accelerate the identification of agriculturally important loci and genes in many crop species. Thus, we highly expect that these current approaches will help us identify essential DNA elements required for the vernalization-mediated floral transition. Furthermore, a recently developed genome-editing tool, CRISPR-Cas9, can be utilized to modify the identified DNA elements to engineer crop traits, including flowering time, to enhance the commercial value of crop plants.
References
Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309:1052–1056. https://doi.org/10.1126/science.1115983
Abuyusuf M, Nath UK, Kim HT, Biswas MK, Park JI, Nou IS (2018) Intronic sequence variations in a gene with peroxidase domain alter bolting time in cabbage (Brassica oleracea var. capitata). Plant Mol Biol Rep 36:725–737. https://doi.org/10.1007/s11105-018-1113-z
Abuyusuf M, Nath UK, Kim HT, Islam MR, Park JI, Nou IS (2019) Molecular markers based on sequence variation in BoFLC1.C9 for characterizing early- and late-flowering cabbage genotypes. BMC Genet 20:42. https://doi.org/10.1186/s12863-019-0740-1
Achard P, Herr A, Baulcombe DC, Harberd NP (2004) Modulation of floral development by a gibberellin-regulated microRNA. Development 131:3357–3365. https://doi.org/10.1242/dev.01206
Adams S, Manfield I, Stockley P, Carre IA (2015) Revised morning loops of the arabidopsis circadian clock based on analyses of direct regulatory interactions. PLoS ONE 10:e0143943. https://doi.org/10.1371/journal.pone.0143943
Adrian J, Farrona S, Reimer JJ, Albani MC, Coupland G, Turck F (2010) cis-Regulatory elements and chromatin state coordinately control temporal and spatial expression of FLOWERING LOCUS T in Arabidopsis. Plant Cell 22:1425–1440. https://doi.org/10.1105/tpc.110.074682
Amasino R (2004) Vernalization, competence, and the epigenetic memory of winter. Plant Cell 16:2553–2559. https://doi.org/10.1105/tpc.104.161070
Amasino RM (2005) Vernalization and flowering time. Curr Opin Biotechnol 16:154–158. https://doi.org/10.1016/j.copbio.2005.02.004
Amasino R (2018) A path to a biennial life history. Nat Plants 4:752–753. https://doi.org/10.1038/s41477-018-0265-z
Amasino RM, Michaels SD (2010) The timing of flowering. Plant Physiol 154:516–520. https://doi.org/10.1104/pp.110.161653
Amasino RM, Cheung AY, Dresselhaus T, Kuhlemeier C (2017) Focus on flowering and reproduction. Plant Physiol 173:1–4. https://doi.org/10.1104/pp.16.01867
Ansell SW, Grundmann M, Russell SJ, Schneider H, Vogel JC (2008) Genetic discontinuity, breeding-system change and population history of Arabis alpina in the Italian Peninsula and adjacent Alps. Mol Ecol 17:2245–2257. https://doi.org/10.1111/j.1365-294X.2008.03739.x
Auge GA, Blair LK, Karediya A, Donohue K (2018) The autonomous flowering-time pathway pleiotropically regulates seed germination in Arabidopsis thaliana. Ann Bot 121:183–191. https://doi.org/10.1093/aob/mcx132
Aukerman MJ, Sakai H (2003) Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 15:2730–2741. https://doi.org/10.1105/tpc.016238
Aukerman MJ, Lee I, Weigel D, Amasino RM (1999) The Arabidopsis flowering-time gene LUMINIDEPENDENS is expressed primarily in regions of cell proliferation and encodes a nuclear protein that regulates LEAFY expression. Plant J 18:195–203
Ausin I, Alonso-Blanco C, Jarillo JA, Ruiz-Garcia L, Martinez-Zapater JM (2004) Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nat Genet 36:162–166. https://doi.org/10.1038/ng1295
Backstrom S, Elfving N, Nilsson R, Wingsle G, Bjorklund S (2007) Purification of a plant mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol Cell 26:717–729. https://doi.org/10.1016/j.molcel.2007.05.007
Bae G, Choi G (2008) Decoding of light signals by plant phytochromes and their interacting proteins. Annu Rev Plant Biol 59:281–311. https://doi.org/10.1146/annurev.arplant.59.032607.092859
Bastow R, Mylne JS, Lister C, Lippman Z, Martienssen RA, Dean C (2004) Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427:164–167. https://doi.org/10.1038/nature02269
Baudry A, Ito S, Song YH, Strait AA, Kiba T, Lu S, Henriques R, Pruneda-Paz JL, Chua NH, Tobin EM, Kay SA, Imaizumi T (2010) F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant Cell 22:606–622. https://doi.org/10.1105/tpc.109.072843
Baurle I, Smith L, Baulcombe DC, Dean C (2007) Widespread role for the flowering-time regulators FCA and FPA in RNA-mediated chromatin silencing. Science 318:109–112. https://doi.org/10.1126/science.1146565
Bergonzi S, Albani MC, van Themaat EVL, Nordstrom KJ, Wang R, Schneeberger K, Moerland PD, Coupland G (2013) Mechanisms of age-dependent response to winter temperature in perennial flowering of Arabis alpina. Science 340:1094–1097. https://doi.org/10.1126/science.1234116
Blazquez MA, Green R, Nilsson O, Sussman MR, Weigel D (1998) Gibberellins promote flowering of arabidopsis by activating the LEAFY promoter. Plant Cell 10:791–800
Blazquez M, Koornneef M, Putterill J (2001) Flowering on time: genes that regulate the floral transition. In: Workshop on the molecular basis of flowering time control. EMBO Rep, vol 2(12), pp 1078–1082. https://doi.org/10.1093/embo-reports/kve254
Blazquez MA, Trenor M, Weigel D (2002) Independent control of gibberellin biosynthesis and flowering time by the circadian clock in Arabidopsis. Plant Physiol 130:1770–1775. https://doi.org/10.1104/pp.007625
Bolouri Moghaddam MR, Van den Ende W (2013) Sugars, the clock and transition to flowering. Front Plant Sci 4:22. https://doi.org/10.3389/fpls.2013.00022
Borner R, Kampmann G, Chandler J, Gleissner R, Wisman E, Apel K, Melzer S (2000) A MADS domain gene involved in the transition to flowering in Arabidopsis. Plant J 24:591–599
Bratzel F, Lopez-Torrejon G, Koch M, Del Pozo JC, Calonje M (2010) Keeping cell identity in Arabidopsis requires PRC1 RING-finger homologs that catalyze H2A monoubiquitination. Curr Biol 20:1853–1859. https://doi.org/10.1016/j.cub.2010.09.046
Capovilla G, Schmid M, Pose D (2015) Control of flowering by ambient temperature. J Exp Bot 66:59–69. https://doi.org/10.1093/jxb/eru416
Cerdan PD, Chory J (2003) Regulation of flowering time by light quality. Nature 423:881–885. https://doi.org/10.1038/nature01636
Chen D, Molitor A, Liu C, Shen WH (2010) The Arabidopsis PRC1-like ring-finger proteins are necessary for repression of embryonic traits during vegetative growth. Cell Res 20:1332–1344. https://doi.org/10.1038/cr.2010.151
Chen D, Molitor AM, Xu L, Shen WH (2016) Arabidopsis PRC1 core component AtRING1 regulates stem cell-determining carpel development mainly through repression of class I KNOX genes. BMC Biol 14:112. https://doi.org/10.1186/s12915-016-0336-4
Chen ZJ, Han YY, Ning K, Ding YY, Zhao WS, Yan SS, Luo C, Jiang XT, Ge DF, Liu RY, Wang Q, Zhang XL (2018) Inflorescence development and the role of LsFT in regulating bolting in lettuce (Lactuca sativa L.). Front Plant Sci 8:2248. https://doi.org/10.3389/fpls.2017.02248
Choi K, Kim J, Hwang HJ, Kim S, Park C, Kim SY, Lee I (2011) The FRIGIDA complex activates transcription of FLC, a strong flowering repressor in Arabidopsis, by recruiting chromatin modification factors. Plant Cell 23:289–303. https://doi.org/10.1105/tpc.110.075911
Conti L (2017) Hormonal control of the floral transition: Can one catch them all? Dev Biol 430:288–301. https://doi.org/10.1016/j.ydbio.2017.03.024
Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, Coupland G (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316:1030–1033. https://doi.org/10.1126/science.1141752
D’Aloia M, Bonhomme D, Bouche F, Tamseddak K, Ormenese S, Torti S, Coupland G, Perilleux C (2011) Cytokinin promotes flowering of Arabidopsis via transcriptional activation of the FT paralogue TSF. Plant J 65:972–979. https://doi.org/10.1111/j.1365-313X.2011.04482.x
De Lucia F, Crevillen P, Jones AM, Greb T, Dean C (2008) A PHD-polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc Natl Acad Sci U S A 105:16831–16836. https://doi.org/10.1073/pnas.0808687105
Di Croce L, Helin K (2013) Transcriptional regulation by polycomb group proteins. Nat Struct Mol Biol 20:1147–1155. https://doi.org/10.1038/nsmb.2669
Dixon LE, Knox K, Kozma-Bognar L, Southern MM, Pokhilko A, Millar AJ (2011) Temporal repression of core circadian genes is mediated through EARLY FLOWERING 3 in Arabidopsis. Curr Biol 21:120–125. https://doi.org/10.1016/j.cub.2010.12.013
Fornara F, Panigrahi KC, Gissot L, Sauerbrunn N, Ruhl M, Jarillo JA, Coupland G (2009) Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev Cell 17:75–86. https://doi.org/10.1016/j.devcel.2009.06.015
Fukuda M, Matsuo S, Kikuchi K, Kawazu Y, Fujiyama R, Honda I (2011) Isolation and functional characterization of the FLOWERING LOCUS T homolog, the LsFT gene, in lettuce. J Plant Physiol 168:1602–1607. https://doi.org/10.1016/j.jplph.2011.02.004
Gardner MJ, Hubbard KE, Hotta CT, Dodd AN, Webb AA (2006) How plants tell the time. Biochem J 397:15–24. https://doi.org/10.1042/BJ20060484
Gendron JM, Pruneda-Paz JL, Doherty CJ, Gross AM, Kang SE, Kay SA (2012) Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc Natl Acad Sci U S A 109:3167–3172. https://doi.org/10.1073/pnas.1200355109
Greb T, Mylne JS, Crevillen P, Geraldo N, An H, Gendall AR, Dean C (2007) The PHD finger protein VRN5 functions in the epigenetic silencing of Arabidopsis FLC. Curr Biol 17:73–78. https://doi.org/10.1016/j.cub.2006.11.052
Guerriero ML, Akman OE, van Ooijen G (2014) Stochastic models of cellular circadian rhythms in plants help to understand the impact of noise on robustness and clock structure. Front Plant Sci 5:564. https://doi.org/10.3389/fpls.2014.00564
He Y, Michaels SD, Amasino RM (2003) Regulation of flowering time by histone acetylation in Arabidopsis. Science 302:1751–1754. https://doi.org/10.1126/science.1091109
Helfer A, Nusinow DA, Chow BY, Gehrke AR, Bulyk ML, Kay SA (2011) LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr Biol 21:126–133. https://doi.org/10.1016/j.cub.2010.12.021
Helliwell CA, Wood CC, Robertson M, James Peacock W, Dennis ES (2006) The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J 46:183–192. https://doi.org/10.1111/j.1365-313X.2006.02686.x
Helliwell CA, Robertson M, Finnegan EJ, Buzas DM, Dennis ES (2011) Vernalization-repression of Arabidopsis FLC requires promoter sequences but not antisense transcripts. Plos One 6:e21513. https://doi.org/10.1371/journal.pone.0021513
Henderson IR, Shindo C, Dean C (2003) The need for winter in the switch to flowering. Annu Rev Genet 37:371–392. https://doi.org/10.1146/annurev.genet.37.110801.142640
Henderson IR, Liu F, Drea S, Simpson GG, Dean C (2005) An allelic series reveals essential roles for FY in plant development in addition to flowering-time control. Development 132:3597–3607. https://doi.org/10.1242/dev.01924
Heo JB, Sung S (2011) Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331:76–79. https://doi.org/10.1126/science.1197349
Hepworth SR, Valverde F, Ravenscroft D, Mouradov A, Coupland G (2002) Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J 21:4327–4337
Hong Y, Jackson S (2015) Floral induction and flower formation–the role and potential applications of miRNAs. Plant Biotechnol J 13:282–292. https://doi.org/10.1111/pbi.12340
Hornyik C, Duc C, Rataj K, Terzi LC, Simpson GG (2010) Alternative polyadenylation of antisense RNAs and flowering time control. Biochem Soc Trans 38:1077–1081. https://doi.org/10.1042/BST0381077
Hsu PY, Harmer SL (2014) Wheels within wheels: the plant circadian system. Trends Plant Sci 19:240–249. https://doi.org/10.1016/j.tplants.2013.11.007
Huang H, Nusinow DA (2016) Into the evening: complex interactions in the Arabidopsis circadian clock. Trends Genet 32:674–686. https://doi.org/10.1016/j.tig.2016.08.002
Huang W, Perez-Garcia P, Pokhilko A, Millar AJ, Antoshechkin I, Riechmann JL, Mas P (2012) Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336:75–79. https://doi.org/10.1126/science.1219075
Hyun Y, Vincent C, Tilmes V, Bergonzi S, Kiefer C, Richter R, Martinez-Gallegos R, Severing E, Coupland G (2019) A regulatory circuit conferring varied flowering response to cold in annual and perennial plants. Science 363:409–412. https://doi.org/10.1126/science.aau8197
Imaizumi T (2010) Arabidopsis circadian clock and photoperiodism: time to think about location. Curr Opin Plant Biol 13:83–89. https://doi.org/10.1016/j.pbi.2009.09.007
Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA (2005) FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309:293–297. https://doi.org/10.1126/science.1110586
Inigo S, Alvarez MJ, Strasser B, Califano A, Cerdan PD (2012) PFT1, the MED25 subunit of the plant mediator complex, promotes flowering through CONSTANS dependent and independent mechanisms in Arabidopsis. Plant J 69:601–612. https://doi.org/10.1111/j.1365-313X.2011.04815.x
Irwin JA, Soumpourou E, Lister C, Ligthart JD, Kennedy S, Dean C (2016) Nucleotide polymorphism affecting FLC expression underpins heading date variation in horticultural brassicas. Plant J 87:597–605. https://doi.org/10.1111/tpj.13221
Jang S, Marchal V, Panigrahi KC, Wenkel S, Soppe W, Deng XW, Valverde F, Coupland G (2008) Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J 27:1277–1288. https://doi.org/10.1038/emboj.2008.68
Jiang D, Yang W, He Y, Amasino RM (2007) Arabidopsis relatives of the human lysine-specific Demethylase1 repress the expression of FWA and FLOWERING LOCUS C and thus promote the floral transition. Plant Cell 19:2975–2987. https://doi.org/10.1105/tpc.107.052373
Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C (2000) Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290:344–347
Jung JH, Seo YH, Seo PJ, Reyes JL, Yun J, Chua NH, Park CM (2007) The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 19:2736–2748. https://doi.org/10.1105/tpc.107.054528
Kakizaki T, Kato T, Fukino N, Ishida M, Hatakeyama K, Matsumoto S (2011) Identification of quantitative trait loci controlling late bolting in Chinese cabbage (Brassica rapa L.) parental line Nou 6 gou. Breed Sci 61:151–159. https://doi.org/10.1270/jsbbs.61.151
Kamioka M, Takao S, Suzuki T, Taki K, Higashiyama T, Kinoshita T, Nakamichi N (2016) Direct repression of evening genes by CIRCADIAN CLOCK-ASSOCIATED1 in the Arabidopsis circadian clock. Plant Cell 28:696–711. https://doi.org/10.1105/tpc.15.00737
Kawanabe T, Osabe K, Itabashi E, Okazaki K, Dennis ES, Fujimoto R (2016) Development of primer sets that can verify the enrichment of histone modifications, and their application to examining vernalization-mediated chromatin changes in Brassica rapa L. Genes Genet Syst 91:1–10. https://doi.org/10.1266/ggs.15-00058
Kiba T, Henriques R, Sakakibara H, Chua NH (2007) Targeted degradation of PSEUDO-RESPONSE REGULATOR5 by an SCFZTL complex regulates clock function and photomorphogenesis in Arabidopsis thaliana. Plant Cell 19:2516–2530. https://doi.org/10.1105/tpc.107.053033
Kim DH, Sung S (2013) Coordination of the vernalization response through a VIN3 and FLC gene family regulatory network in Arabidopsis. Plant Cell 25:454–469. https://doi.org/10.1105/tpc.112.104760
Kim DH, Sung S (2014a) Genetic and epigenetic mechanisms underlying vernalization. Arabidopsis Book 12:e0171. https://doi.org/10.1199/tab.0171
Kim DH, Sung S (2014b) Polycomb-mediated gene silencing in Arabidopsis thaliana. Mol Cells 37:841–850. https://doi.org/10.14348/molcells.2014.0249
Kim DH, Sung S (2017) Vernalization-triggered intragenic chromatin loop formation by long noncoding RNAs. Dev Cell 40:302–312. https://doi.org/10.1016/j.devcel.2016.12.021
Kim WY, Fujiwara S, Suh SS, Kim J, Kim Y, Han L, David K, Putterill J, Nam HG, Somers DE (2007) ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449:356–360. https://doi.org/10.1038/nature06132
Kim DH, Doyle MR, Sung S, Amasino RM (2009) Vernalization: winter and the timing of flowering in plants. Annu Rev Cell Dev Biol 25:277–299. https://doi.org/10.1146/annurev.cellbio.042308.113411
Kim HI, Hong CP, Im S, Choi SR, Lim YP (2014) Development of molecular markers and application for breeding in Chinese cabbage. Korean J Hortic Sci 32:745–752. https://doi.org/10.7235/hort.2014.12203
Kitamoto N, Yui S, Nishikawa K, Takahata Y, Yokoi S (2014) A naturally occurring long insertion in the first intron in the Brassica rapa FLC2 gene causes delayed bolting. Euphytica 196:213–223. https://doi.org/10.1007/s10681-013-1025-9
Kobayashi Y, Weigel D (2007) Move on up, it’s time for change–mobile signals controlling photoperiod-dependent flowering. Genes Dev 21:2371–2384. https://doi.org/10.1101/gad.1589007
Koch MA, Kiefer C, Ehrich D, Vogel J, Brochmann C, Mummenhoff K (2006) Three times out of Asia Minor: the phylogeography of Arabis alpina L. (Brassicaceae). Mol Ecol 15:825–839. https://doi.org/10.1111/j.1365-294X.2005.02848.x
Laubinger S, Marchal V, Le Gourrierec J, Wenkel S, Adrian J, Jang S, Kulajta C, Braun H, Coupland G, Hoecker U (2006) Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development 133:3213–3222. https://doi.org/10.1242/dev.02481
Lazaro A, Mouriz A, Pineiro M, Jarillo JA (2015) Red light-mediated degradation of CONSTANS by the E3 ubiquitin ligase HOS1 regulates photoperiodic flowering in Arabidopsis. Plant Cell 27:2437–2454. https://doi.org/10.1105/tpc.15.00529
Lee I, Aukerman MJ, Gore SL, Lohman KN, Michaels SD, Weaver LM, John MC, Feldmann KA, Amasino RM (1994) Isolation of LUMINIDEPENDENS: a gene involved in the control of flowering time in Arabidopsis. Plant Cell 6:75–83. https://doi.org/10.1105/tpc.6.1.75
Lee SG, Lee HJ, Kim SK, Choi CS, Park ST, Jang YA, Do KR (2015) Effects of vernalization, temperature, and soil drying periods on the growth and yield of Chinese cabbage. Korean J Hortic Sci 33:820–828. https://doi.org/10.7235/hort.2015.15076
Leijten W, Koes R, Roobeek I, Frugis G (2018) Translating flowering time from Arabidopsis thaliana to Brassicaceae and Asteraceae Crop Species. Plants (Basel). https://doi.org/10.3390/plants7040111
Levy YY, Mesnage S, Mylne JS, Gendall AR, Dean C (2002) Multiple roles of Arabidopsis VRN1 in vernalization and flowering time control. Science 297:243–246. https://doi.org/10.1126/science.1072147
Li F, Kitashiba H, Inaba K, Nishio T (2009) A Brassica rapa linkage map of EST-based SNP markers for identification of candidate genes controlling flowering time and leaf morphological traits. DNA Res 16:311–323. https://doi.org/10.1093/dnares/dsp020
Li W, Wang Z, Li J, Yang H, Cui S, Wang X, Ma L (2011) Overexpression of AtBMI1C, a polycomb group protein gene, accelerates flowering in Arabidopsis. PLoS ONE 6:e21364. https://doi.org/10.1371/journal.pone.0021364
Li C, Gu L, Gao L, Chen C, Wei CQ, Qiu Q, Chien CW, Wang S, Jiang L, Ai LF, Chen CY, Yang S, Nguyen V, Qi Y, Snyder MP, Burlingame AL, Kohalmi SE, Huang S, Cao X, Wang ZY, Wu K, Chen X, Cui Y (2016) Concerted genomic targeting of H3K27 demethylase REF6 and chromatin-remodeling ATPase BRM in Arabidopsis. Nat Genet 48:687–693. https://doi.org/10.1038/ng.3555
Lim MH, Kim J, Kim YS, Chung KS, Seo YH, Lee I, Kim J, Hong CB, Kim HJ, Park CM (2004) A new Arabidopsis gene, FLK, encodes an RNA binding protein with K homology motifs and regulates flowering time via FLOWERING LOCUS C. Plant Cell 16:731–740. https://doi.org/10.1105/tpc.019331
Lin SI, Wang JG, Poon SY, Su CL, Wang SS, Chiou TJ (2005) Differential regulation of FLOWERING LOCUS C expression by vernalization in cabbage and Arabidopsis. Plant Physiol 137:1037–1048. https://doi.org/10.1104/pp.104.058974
Lin YR, Lee JY, Tseng MC, Lee CY, Shen CH, Wang CS, Liou CC, Shuang LS, Paterson AH, Hwu KK (2018) Subtropical adaptation of a temperate plant (Brassica oleracea var. italica) utilizes non-vernalization-responsive QTLs. Sci Rep 8:13609. https://doi.org/10.1038/s41598-018-31987-1
Liu F, Quesada V, Crevillen P, Baurle I, Swiezewski S, Dean C (2007) The Arabidopsis RNA-binding protein FCA requires a lysine-specific demethylase 1 homolog to downregulate FLC. Mol Cell 28:398–407. https://doi.org/10.1016/j.molcel.2007.10.018
Liu LJ, Zhang YC, Li QH, Sang Y, Mao J, Lian HL, Wang L, Yang HQ (2008) COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20:292–306. https://doi.org/10.1105/tpc.107.057281
Liu F, Marquardt S, Lister C, Swiezewski S, Dean C (2010) Targeted 3′ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science 327:94–97. https://doi.org/10.1126/science.1180278
Liu TL, Newton L, Liu MJ, Shiu SH, Farre EM (2016) A G-box-like motif is necessary for transcriptional regulation by circadian pseudo-response regulators in Arabidopsis. Plant Physiol 170:528–539. https://doi.org/10.1104/pp.15.01562
Lu F, Cui X, Zhang S, Jenuwein T, Cao X (2011) Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat Genet 43:715–719. https://doi.org/10.1038/ng.854
Macknight R, Bancroft I, Page T, Lister C, Schmidt R, Love K, Westphal L, Murphy G, Sherson S, Cobbett C, Dean C (1997) FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell 89:737–745
Margueron R, Reinberg D (2011) The Polycomb complex PRC2 and its mark in life. Nature 469:343–349. https://doi.org/10.1038/nature09784
Mas P, Kim WY, Somers DE, Kay SA (2003) Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426:567–570. https://doi.org/10.1038/nature02163
Mateos JL, Tilmes V, Madrigal P, Severing E, Richter R, Rijkenberg CWM, Krajewski P, Coupland G (2017) Divergence of regulatory networks governed by the orthologous transcription factors FLC and PEP1 in Brassicaceae species. Proc Natl Acad Sci U S A 114:E11037–E11046. https://doi.org/10.1073/pnas.1618075114
Mathieu J, Yant LJ, Murdter F, Kuttner F, Schmid M (2009) Repression of flowering by the miR172 target SMZ. PLoS Biol 7:e1000148. https://doi.org/10.1371/journal.pbio.1000148
Merini W, Romero-Campero FJ, Gomez-Zambrano A, Zhou Y, Turck F, Calonje M (2017) The Arabidopsis polycomb repressive complex 1 (PRC1) components AtBMI1A, B, and C impact gene networks throughout all stages of plant development. Plant Physiol 173:627–641. https://doi.org/10.1104/pp.16.01259
Michaels SD, Amasino RM (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11:949–956
Michaels SD, Amasino RM (2001) Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13:935–941
Molitor A, Shen WH (2013) The polycomb complex PRC1: composition and function in plants. J Genet Genomics 40:231–238. https://doi.org/10.1016/j.jgg.2012.12.005
Moon J, Suh SS, Lee H, Choi KR, Hong CB, Paek NC, Kim SG, Lee I (2003) The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J 35:613–623
Mutasa-Gottgens E, Hedden P (2009) Gibberellin as a factor in floral regulatory networks. J Exp Bot 60:1979–1989. https://doi.org/10.1093/jxb/erp040
Mylne JS, Barrett L, Tessadori F, Mesnage S, Johnson L, Bernatavichute YV, Jacobsen SE, Fransz P, Dean C (2006) LHP1, the Arabidopsis homologue of HETEROCHROMATIN PROTEIN1, is required for epigenetic silencing of FLC. Proc Natl Acad Sci U S A 103:5012–5017. https://doi.org/10.1073/pnas.0507427103
Nakamichi N (2011) Molecular mechanisms underlying the Arabidopsis circadian clock. Plant Cell Physiol 52:1709–1718. https://doi.org/10.1093/pcp/pcr118
Nakamichi N, Kiba T, Henriques R, Mizuno T, Chua NH, Sakakibara H (2010) PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22:594–605. https://doi.org/10.1105/tpc.109.072892
Noh B, Lee SH, Kim HJ, Yi G, Shin EA, Lee M, Jung KJ, Doyle MR, Amasino RM, Noh YS (2004) Divergent roles of a pair of homologous jumonji/zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time. Plant Cell 16:2601–2613. https://doi.org/10.1105/tpc.104.025353
Notaguchi M, Abe M, Kimura T, Daimon Y, Kobayashi T, Yamaguchi A, Tomita Y, Dohi K, Mori M, Araki T (2008) Long-distance, graft-transmissible action of Arabidopsis FLOWERING LOCUS T protein to promote flowering. Plant Cell Physiol 49:1645–1658. https://doi.org/10.1093/pcp/pcn154
Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farre EM, Kay SA (2011) The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475:398–402. https://doi.org/10.1038/nature10182
Okazaki K, Sakamoto K, Kikuchi R, Saito A, Togashi E, Kuginuki Y, Matsumoto S, Hirai M (2007) Mapping and characterization of FLC homologs and QTL analysis of flowering time in Brassica oleracea. Theor Appl Genet 114:595–608. https://doi.org/10.1007/s00122-006-0460-6
Olszewski N, Sun TP, Gubler F (2002) Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell 14:S61–S80
Osborn TC, Kole C, Parkin IAP, Sharpe AG, Kuiper M, Lydiate DJ, Trick M (1997) Comparison of flowering time genes in Brassica rapa, B-napus and Arabidopsis thaliana. Genetics 146:1123–1129
Pico S, Ortiz-Marchena MI, Merini W, Calonje M (2015) Deciphering the role of POLYCOMB REPRESSIVE COMPLEX1 variants in regulating the acquisition of flowering competence in Arabidopsis. Plant Physiol 168:1286–1297. https://doi.org/10.1104/pp.15.00073
Pin PA, Benlloch R, Bonnet D, Wremerth-Weich E, Kraft T, Gielen JJ, Nilsson O (2010) An antagonistic pair of FT homologs mediates the control of flowering time in sugar beet. Science 330:1397–1400. https://doi.org/10.1126/science.1197004
Pin PA, Zhang W, Vogt SH, Dally N, Buttner B, Schulze-Buxloh G, Jelly NS, Chia TY, Mutasa-Gottgens ES, Dohm JC, Himmelbauer H, Weisshaar B, Kraus J, Gielen JJ, Lommel M, Weyens G, Wahl B, Schechert A, Nilsson O, Jung C, Kraft T, Muller AE (2012) The role of a pseudo-response regulator gene in life cycle adaptation and domestication of beet. Curr Biol 22:1095–1101. https://doi.org/10.1016/j.cub.2012.04.007
Pokhilko A, Hodge SK, Stratford K, Knox K, Edwards KD, Thomson AW, Mizuno T, Millar AJ (2010) Data assimilation constrains new connections and components in a complex, eukaryotic circadian clock model. Mol Syst Biol 6:416. https://doi.org/10.1038/msb.2010.69
Qi XH, Townsley B, Aguilar-Martinez JA, Yin LH, Gao XY, Hou LP, Gao MY, Li ML (2015) Cloning and characterization of the LFY homologue from Chinese cabbage (Brassica rapa subsp pekinensis). Hortic Environ Biotechnol 56:821–829. https://doi.org/10.1007/s13580-015-0066-5
Quail PH (2010) Phytochromes. Curr Biol 20:R504–R507. https://doi.org/10.1016/j.cub.2010.04.014
Questa JI, Song J, Geraldo N, An H, Dean C (2016) Arabidopsis transcriptional repressor VAL1 triggers polycomb silencing at FLC during vernalization. Science 353:485–488. https://doi.org/10.1126/science.aaf7354
Razi H, Howell EC, Newbury HJ, Kearsey MJ (2008) Does sequence polymorphism of FLC paralogues underlie flowering time QTL in Brassica oleracea? Theor Appl Genet 116:179–192. https://doi.org/10.1007/s00122-007-0657-3
Reeves PH, Coupland G (2001) Analysis of flowering time control in Arabidopsis by comparison of double and triple mutants. Plant Physiol 126:1085–1091
Ridge S, Brown PH, Hecht V, Driessen RG, Weller JL (2015) The role of BoFLC2 in cauliflower (Brassica oleracea var. botrytis L.) reproductive development. J Exp Bot 66:125–135. https://doi.org/10.1093/jxb/eru408
Rolland F, Moore B, Sheen J (2002) Sugar sensing and signaling in plants. Plant Cell 14:S185–S205
Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288:1613–1616
Sanchez-Pulido L, Devos D, Sung ZR, Calonje M (2008) RAWUL: a new ubiquitin-like domain in PRC1 ring finger proteins that unveils putative plant and worm PRC1 orthologs. BMC Genomics 9:308. https://doi.org/10.1186/1471-2164-9-308
Sawa M, Kay SA (2011) GIGANTEA directly activates Flowering Locus T in Arabidopsis thaliana. Proc Natl Acad Sci U S A 108:11698–11703. https://doi.org/10.1073/pnas.1106771108
Schatlowski N, Creasey K, Goodrich J, Schubert D (2008) Keeping plants in shape: polycomb-group genes and histone methylation. Semin Cell Dev Biol 19:547–553. https://doi.org/10.1016/j.semcdb.2008.07.019
Schomburg FM, Patton DA, Meinke DW, Amasino RM (2001) FPA, a gene involved in floral induction in Arabidopsis, encodes a protein containing RNA-recognition motifs. Plant Cell 13:1427–1436
Schranz ME, Quijada P, Sung SB, Lukens L, Amasino R, Osborn TC (2002) Characterization and effects of the replicated flowering time gene FLC in Brassica rapa. Genetics 162:1457–1468
Schwartz YB, Pirrotta V (2007) Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet 8:9–22. https://doi.org/10.1038/nrg1981
Searle I, Coupland G (2004) Induction of flowering by seasonal changes in photoperiod. EMBO J 23:1217–1222. https://doi.org/10.1038/sj.emboj.7600117
Seo PJ, Mas P (2014) Multiple layers of posttranslational regulation refine circadian clock activity in Arabidopsis. Plant Cell 26:79–87. https://doi.org/10.1105/tpc.113.119842
Seo PJ, Ryu J, Kang SK, Park CM (2011) Modulation of sugar metabolism by an INDETERMINATE DOMAIN transcription factor contributes to photoperiodic flowering in Arabidopsis. Plant J 65:418–429. https://doi.org/10.1111/j.1365-313X.2010.04432.x
Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES (1999) The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11:445–458
Sheldon CC, Conn AB, Dennis ES, Peacock WJ (2002) Different regulatory regions are required for the vernalization-induced repression of FLOWERING LOCUS C and for the epigenetic maintenance of repression. Plant Cell 14:2527–2537. https://doi.org/10.1105/tpc.004564
Shim JS, Imaizumi T (2015) Circadian clock and photoperiodic response in Arabidopsis: from seasonal flowering to redox homeostasis. Biochemistry 54:157–170. https://doi.org/10.1021/bi500922q
Shim JS, Kubota A, Imaizumi T (2017) Circadian clock and photoperiodic flowering in Arabidopsis: CONSTANS is a hub for signal integration. Plant Physiol 173:5–15. https://doi.org/10.1104/pp.16.01327
Simon JA, Kingston RE (2013) Occupying chromatin: polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol Cell 49:808–824. https://doi.org/10.1016/j.molcel.2013.02.013
Simpson GG (2004) The autonomous pathway: epigenetic and post-transcriptional gene regulation in the control of Arabidopsis flowering time. Curr Opin Plant Biol 7:570–574. https://doi.org/10.1016/j.pbi.2004.07.002
Simpson GG, Dijkwel PP, Quesada V, Henderson I, Dean C (2003) FY is an RNA 3′ end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell 113:777–787
Song J, Angel A, Howard M, Dean C (2012a) Vernalization—a cold-induced epigenetic switch. J Cell Sci 125:3723–3731. https://doi.org/10.1242/jcs.084764
Song YH, Smith RW, To BJ, Millar AJ, Imaizumi T (2012b) FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336:1045–1049. https://doi.org/10.1126/science.1219644
Song YH, Ito S, Imaizumi T (2013) Flowering time regulation: photoperiod- and temperature-sensing in leaves. Trends Plant Sci 18:575–583. https://doi.org/10.1016/j.tplants.2013.05.003
Sung S, Amasino RM (2004) Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427:159–164. https://doi.org/10.1038/nature02195
Sung S, Amasino RM (2005) Remembering winter: toward a molecular understanding of vernalization. Annu Rev Plant Biol 56:491–508. https://doi.org/10.1146/annurev.arplant.56.032604.144307
Sung S, He Y, Eshoo TW, Tamada Y, Johnson L, Nakahigashi K, Goto K, Jacobsen SE, Amasino RM (2006a) Epigenetic maintenance of the vernalized state in Arabidopsis thaliana requires LIKE HETEROCHROMATIN PROTEIN 1. Nat Genet 38:706–710. https://doi.org/10.1038/ng1795
Sung S, Schmitz RJ, Amasino RM (2006b) A PHD finger protein involved in both the vernalization and photoperiod pathways in Arabidopsis. Genes Dev 20:3244–3248. https://doi.org/10.1101/gad.1493306
Susila H, Nasim Z, Ahn JH (2018) Ambient temperature-responsive mechanisms coordinate regulation of flowering time. Int J Mol Sci 19:3196. https://doi.org/10.3390/ijms19103196
Swiezewski S, Liu F, Magusin A, Dean C (2009) Cold-induced silencing by long antisense transcripts of an Arabidopsis polycomb target. Nature 462:799–802. https://doi.org/10.1038/nature08618
Takada S, Akter A, Itabashi E, Nishida N, Shea DJ, Miyaji N, Mehraj H, Osabe K, Shimizu M, Takasaki-Yasuda T, Kakizaki T, Okazaki K, Dennis ES, Fujimoto R (2019) The role of FRIGIDA and FLOWERING LOCUS C genes in flowering time of Brassica rapa leafy vegetables. Sci Rep 9:1–11. https://doi.org/10.1038/s41598-019-50122-2
Teotia S, Tang G (2015) To bloom or not to bloom: role of microRNAs in plant flowering. Mol Plant 8:359–377. https://doi.org/10.1016/j.molp.2014.12.018
Teutonico RA, Osborn TC (1995) Mapping loci controlling vernalization requirement in Brassica rapa. Theor Appl Genet 91:1279–1283. https://doi.org/10.1007/Bf00220941
Tsai AY, Gazzarrini S (2012) AKIN10 and FUSCA3 interact to control lateral organ development and phase transitions in Arabidopsis. Plant J 69:809–821. https://doi.org/10.1111/j.1365-313X.2011.04832.x
Turck F, Fornara F, Coupland G (2008) Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol 59:573–594. https://doi.org/10.1146/annurev.arplant.59.032607.092755
Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G (2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303:1003–1006. https://doi.org/10.1126/science.1091761
Veley KM, Michaels SD (2008) Functional redundancy and new roles for genes of the autonomous floral-promotion pathway. Plant Physiol 147:682–695. https://doi.org/10.1104/pp.108.118927
Wahl V, Ponnu J, Schlereth A, Arrivault S, Langenecker T, Franke A, Feil R, Lunn JE, Stitt M, Schmid M (2013) Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science 339:704–707. https://doi.org/10.1126/science.1230406
Wang R, Farrona S, Vincent C, Joecker A, Schoof H, Turck F, Alonso-Blanco C, Coupland G, Albani MC (2009) PEP1 regulates perennial flowering in Arabis alpina. Nature 459:423–427. https://doi.org/10.1038/nature07988
Wang R, Albani MC, Vincent C, Bergonzi S, Luan M, Bai Y, Kiefer C, Castillo R, Coupland G (2011a) Aa TFL1 confers an age-dependent response to vernalization in perennial Arabis alpina. Plant Cell 23:1307–1321. https://doi.org/10.1105/tpc.111.083451
Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, Bai Y, Mun JH, Bancroft I, Cheng F, Huang S, Li X, Hua W, Wang J, Wang X, Freeling M, Pires JC, Paterson AH, Chalhoub B, Wang B, Hayward A, Sharpe AG, Park BS, Weisshaar B, Liu B, Li B, Liu B, Tong C, Song C, Duran C, Peng C, Geng C, Koh C, Lin C, Edwards D, Mu D, Shen D, Soumpourou E, Li F, Fraser F, Conant G, Lassalle G, King GJ, Bonnema G, Tang H, Wang H, Belcram H, Zhou H, Hirakawa H, Abe H, Guo H, Wang H, Jin H, Parkin IA, Batley J, Kim JS, Just J, Li J, Xu J, Deng J, Kim JA, Li J, Yu J, Meng J, Wang J, Min J, Poulain J, Wang J, Hatakeyama K, Wu K, Wang L, Fang L, Trick M, Links MG, Zhao M, Jin M, Ramchiary N, Drou N, Berkman PJ, Cai Q, Huang Q, Li R, Tabata S, Cheng S, Zhang S, Zhang S, Huang S, Sato S, Sun S, Kwon SJ, Choi SR, Lee TH, Fan W, Zhao X, Tan X, Xu X, Wang Y, Qiu Y, Yin Y, Li Y, Du Y, Liao Y, Lim Y, Narusaka Y, Wang Y, Wang Z, Li Z, Wang Z, Xiong Z, Zhang Z, Brassica rapa Genome Sequencing Project C (2011b) The genome of the mesopolyploid crop species Brassica rapa. Nat Genet 43:1035–1039
Wenkel S, Turck F, Singer K, Gissot L, Le Gourrierec J, Samach A, Coupland G (2006) CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 18:2971–2984. https://doi.org/10.1105/tpc.106.043299
Wood CC, Robertson M, Tanner G, Peacock WJ, Dennis ES, Helliwell CA (2006) The Arabidopsis thaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. Proc Natl Acad Sci U S A 103:14631–14636. https://doi.org/10.1073/pnas.0606385103
Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS (2009) The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138:750–759. https://doi.org/10.1016/j.cell.2009.06.031
Wu J, Wei KY, Cheng F, Li SK, Wang Q, Zhao JJ, Bonnema G, Wang XW (2012) A naturally occurring InDel variation in BraA.FLC.b(BrFLC2) associated with flowering time variation in Brassica rapa. BMC Plant Biol 12:151. https://doi.org/10.1186/1471-2229-12-151
Xiao D, Zhao JJ, Hou XL, Basnet RK, Carpio DPD, Zhang NW, Bucher J, Lin K, Cheng F, Wang XW, Bonnema G (2013) The Brassica rapa FLC homologue FLC2 is a key regulator of flowering time, identified through transcriptional co-expression networks. J Exp Bot 64:4503–4516. https://doi.org/10.1093/jxb/ert264
Xu L, Shen WH (2008) Polycomb silencing of KNOX genes confines shoot stem cell niches in Arabidopsis. Curr Biol 18:1966–1971. https://doi.org/10.1016/j.cub.2008.11.019
Yamaguchi S (2008) Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59:225–251. https://doi.org/10.1146/annurev.arplant.59.032607.092804
Yanovsky MJ, Kay SA (2002) Molecular basis of seasonal time measurement in Arabidopsis. Nature 419:308–312. https://doi.org/10.1038/nature00996
Yi G, Park H, Kim JS, Chae WB, Park S, Huh JH (2014) Identification of three FLOWERING LOCUS C genes responsible for vernalization response in radish (Raphanus sativus L). Hortic Environ Biotechnol 55:548–556. https://doi.org/10.1007/s13580-014-1151-x
Yuan YX, Wu J, Sun RF, Zhang XW, Xu DH, Bonnema G, Wang XW (2009) A naturally occurring splicing site mutation in the Brassica rapa FLC1 gene is associated with variation in flowering time. J Exp Bot 60:1299–1308. https://doi.org/10.1093/jxb/erp010
Yuan W, Luo X, Li Z, Yang W, Wang Y, Liu R, Du J, He Y (2016) A cis cold memory element and a trans epigenome reader mediate polycomb silencing of FLC by vernalization in Arabidopsis. Nat Genet 48:1527–1534. https://doi.org/10.1038/ng.3712
Zhang B, Wang L, Zeng L, Zhang C, Ma H (2015) Arabidopsis TOE proteins convey a photoperiodic signal to antagonize CONSTANS and regulate flowering time. Genes Dev 29:975–987. https://doi.org/10.1101/gad.251520.114
Zhao JJ, Kulkarni V, Liu NN, Del Carpio DP, Bucher J, Bonnema G (2010) BrFLC2 (FLOWERING LOCUS C) as a candidate gene for a vernalization response QTL in Brassica rapa. J Exp Bot 61:1817–1825. https://doi.org/10.1093/jxb/erq048
Zhu QH, Helliwell CA (2011) Regulation of flowering time and floral patterning by miR172. J Exp Bot 62:487–495. https://doi.org/10.1093/jxb/erq295
Zou XX, Suppanz I, Raman H, Hou JN, Wang J, Long Y, Jung C, Meng JL (2012) Comparative analysis of FLC homologues in Brassicaceae provides insight into their role in the evolution of oilseed rape. PLoS ONE 7:e45751. https://doi.org/10.1371/journal.pone.0045751
Acknowledgements
This work was supported by a National Research Foundation of Korea Grant (NRF, 2018R1D1A1B07049214) provided to Dong-Hwan Kim.
Author information
Authors and Affiliations
Contributions
DHK planned and wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The author declare that I have no conflict of interest.
Additional information
Communicated by Tae-Ho Han, Ph.D.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kim, DH. Current understanding of flowering pathways in plants: focusing on the vernalization pathway in Arabidopsis and several vegetable crop plants. Hortic. Environ. Biotechnol. 61, 209–227 (2020). https://doi.org/10.1007/s13580-019-00218-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13580-019-00218-5