Abstract
The Mildew resistance locus o (Mlo) is a plant-specific gene family that encodes a protein with seven-transmembrane (TM) domains that play an important role in plant resistance to powdery mildew (PM). PM caused by Podosphaera xanthii is a widespread plant disease and represents the major fungal threat for many Cucurbits. The recently reported Cucurbita maxima genome sequence data provides an opportunity to identify and characterize the MLO gene family in this species. A total of twenty genes designated CmaMLO1 to CmaMLO20 have been identified using an in silico cloning method with the MLO gene sequences of Cucumis sativus, Cucumis melo, Citrullus lanatus, and Cucurbita pepo as probes. These CmaMLOs were evenly distributed on 15 of the 20 C. maxima chromosomes without any obvious clustering. Multiple sequence alignment showed that common structural features of MLO gene family, such as TM domains, a calmodulin-binding domain, and 30 important amino acid residues for MLO function, were well conserved. Phylogenetic analysis of the CmaMLO genes and other plant species revealed seven different clades (I through VII); however, clade IV is specific to monocots (rice, barley and wheat). Expression analysis showed that four of the five CmaMLO genes that belonged to clade V were up-regulated in pumpkins infected with P. xanthii. Phylogenetic and expression analysis provided preliminary evidence that these five genes could be susceptibility genes that are important for PM resistance. This study is the first comprehensive report on MLO genes in C. maxima to our knowledge. These findings will facilitate the functional analysis of MLOs related to PM susceptibility and are valuable resources for the development of disease resistance in pumpkin.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Powdery mildew (PM) caused by Podosphaera xanthii is a widespread plant disease and considered to be a major fungal threat for cucurbits (McGrath and Thomas 1996). Since resistance (R) genes confer specific resistance to diseases in plants (Hammond-Kosack and Jones 2000; Taler et al. 2004), dominant R genes are generally used to breed crops resistant to diseases. However, the effect of R genes is limited due to the emergence of virulent pathogen strains that can rapidly overcome resistance over time (Pessina et al. 2014). An alternative strategy for improving host plant resistance is the repression of susceptibility (S) genes, whose loss-of-function gives long-term and broad-spectrum resistance in plants (Pavan et al. 2010). The most well known examples of S-gene loss-of-function alleles are the barley mlo mutants, which displayed near complete resistance to the powdery mildew pathogen, Blumeria graminis f.sp. hordei (Bgh) and those mutants carried recessively inherited loss-of-function mutations in the gene Mildew resistance locus o (MLO) (Acevedo-Garcia et al. 2014).
The mildew resistance locus O (MLO) is a plant-specific gene family that encodes a protein that contains seven transmembrane (TM) domains, a unique intracellular C-terminal calmodulin-binding domain (CaMBD), and an extracellular N-terminus (Buschges et al. 1997; Devoto et al. 1999; Kim et al. 2002a). Members of the MLO gene family participate in various developmental pathways and response against foreign biotic and abiotic stresses in plants (Deshmukh et al. 2014). The MLO gene was first identified as a loss-of-function mutation that causes lasting broad-spectrum resistance to the barley PM pathogen, Blumeria graminis f. sp. hordei (Bgh) (Buschges et al. 1997; Devoto et al. 1999; Kim et al. 2002a). The inactivation of specific MLO protein family homologs is associated with the enhancement of exocytic defense pathways at plant-pathogen interaction sites, which might contribute to the prevention of fungal penetration of host cells (Assaad et al. 2004). Initially discovered in barley, mlo resistance has been later discovered in other monocots including OsMLO3 in rice (Devoto et al. 2003) and TaMLO_A1 and TaMLO_B1 in wheat (Devoto et al. 2003; Várallyay et al. 2012. Such genes have also been identified in dicots; AtMLO2, AtMLO6 and AtMLO12 in Arabidopsis (Consonni et al. 2006), SlMLO1 in tomato (Bai et al. 2008), PsMLO1 in pea (Humphry et al. 2011), CaMLO2 in pepper (Kim and Hwang 2012; Zheng et al. 2013), LjMLO1 in lotus (Humphry et al. 2011), MtMLO1 in barrel clover (Humphry et al. 2011), and CsMLO1 in cucumber (Nie et al. 2015). The inactivation of MLO susceptibility genes has become an alternative resistance model for breeding approaches for several cultivated species affected by PM disease (Dangl et al. 2013; Pavan et al. 2010).
Pumpkin (Cucurbita maxima), an economically important crop belonging to the Cucurbitaceae family; it is commonly known as winter squash and its mature fruits consumed as vegetables in most of the world, especially in Asia (primarily China and India) and Africa (Zhang et al. 2015). Powdery mildew is one of the major diseases that reduce the yield of susceptible pumpkins (McGrath and Thomas 1996). However, there are very limited genetic resistance resources available in C. maxima against PM, unlike other cucurbits such as cucumber, melon watermelon, C. pepo, and C. moschata, for which several putative disease resistance genes (R-genes) (Xu et al. 2016, 2017; Adeniji and Coyne 1983) and susceptibility genes (S-genes) (Schouten et al. 2014; Garcia-Mas et al. 2012; Lovieno et al. 2015) have been identified. Recently, a high-density genetic map and whole genome sequence data became available for C. maxima Duch. (Zhang et al. 2015) and have provided powerful tools for the genetic and genomic exploration of powdery mildew resistance-related genes for pumpkin. No member of the pumpkin MLO gene families has been isolated to date; however, loss-of-function mutations in barley (HvMLO) and pea (PsMLO1) have been successfully employed in barley and pea breeding for long time, suggesting the same might be possible for pumpkin. In this study, we identified the corresponding MLO family genes in C. maxima and analyzed their structural features, homology, and distribution to evaluate the phylogenetic relationship between MLOs from C. maxima and other plant species. Moreover, we examined the expression patterns of CmaMLO genes in clade V to investigate their response to the PM fungus Podosphaera xanthii. This study facilitates the functional characterization of the MLOs related to PM susceptibility and contributes to molecular breeding approaches that could introduce PM resistance using reverse genetics strategies.
2 Materials and methods
2.1 Identification and annotation of C. maxima MLOs
In order to identify the MLO gene family in C. maxima, we performed BLAST searches using MLO sequences from various species in the Cucurbitaceae family against C. maxima genome sequence data provided by “The International C. maxima Genome Initiative” (http://www.icugi.org/cgi-bin/ICuGI/genome/maxima.cgi). Sequences of MLO encoding proteins for these species were downloaded from various databases. Cucumis melo MLO proteins were retrieved from the Melonomics melon genomic database (http://melonomics.net). The sequences for Cucumis sativus and Citrullus lanatus MLO proteins were retrieved from the Cucurbit Genomic Database (http://www.icugi.org/cgi-bin/ICuGI/index.cgi). Sequences from Cucurbita pepo MLO proteins were reported by Lovieno et al. (2015) and retrieved from the C. pepo scaffolds (http://cucurbigene.upv.es/genome-v3.2/). All potential C. maxima MLO sequences were systematically validated with BLAST on the C. maxima genome sequence data using Geneious v 9.1 (http://www.geneious.com; Kearse et al. 2012). The full length CDS and encoded protein sequences for C. maxima MLOs were predicted with the Fgenesh using the “Cucumis melo gene model” (http://www.softberry.com/berry.phtml?topic=index&group=programs&subgroup=gfind). Each C. maxima MLO protein sequence was submitted to the Pfam database (the protein families database) to verify the presence of an MLO domain and retrieve conserved MLO domain sequences in the Pfam database (ID: PF03094). Chromosomal locations for each C. maxima MLO gene were determined according to chromosomal information from the C. maxima genome sequence database.
2.2 Protein structure analysis and motif prediction
The deduced amino acid sequences for the potential MLO proteins from C. maxima were subjected to several prediction programs, including the online transmembrane helix prediction sever TMHMM v2.0 (www.cbs.dtu.dk/services/TMHMM/), the consensus prediction of membrane protein topology TOPCONS (http://old.topcons.net/), and InterProScan5 (http://www.ebi.ac.uk/interpro/search/sequence-search) to determine their sub-cellular localization and protein topologies. To highlight the conserved amino acid residues in the seven TM domains, a TM domain sequence logo was generated via the online logo tool WebLogo (http://weblogo.berkeley.edu/logo.cgi) (Crooks et al. 2004). The conserved motifs were generated using the Multiple Expectation Maximization for Motif Elicitation (MEME) online program (http://meme.sdsc.edu/meme/intro.html). The optimized parameters of MEME were set as a maximum of 10 motifs with a motif width between 10 and 50 residues.
2.3 Multiple sequence alignments and phylogenetic analysis
Multiple protein sequence alignments were performed with multiple sequence comparisons by log-expectation (MUSCLE) multiple alignment using the conserved MLO domain sequences of C. maxima as inputs in Geneious v 9.1. Phylogenetic analysis was performed using newly identified CmaMLOs containing at least 50% of the full-length MLO-Pfam domain. In addition, to understand the phylogenetic relationship of MLO genes in plants, we included the whole Arabidopsis (Arabidopsis thaliana) MLO protein family and the MLO proteins previously reported with PM susceptibility in barley (Hordeum vulgare), rice (Oryza sativa), wheat (Triticum aestivum), pea (Pisum sativum), tomato (Solanum lycopersicum), barrel clover (Medicago truncatula), lotus (Lotus japonicus), and pepper (Capsicum annuum). All of these sequences were extracted from the NCBI database (http://www.ncbi.nlm.nih.gov). Phylogenetic relationships between MLO proteins were inferred using the maximum likelihood method based on the Whelan and Goldman model in the MEGA7 software (http://www.megasoftware.net). Bootstrapping of 1000 replicates was used to evaluate the degree of support for a particular grouping pattern in the phylogenetic tree (Felsenstein 1985). A full-length multiple alignment was conducted to identify the conserved amino acids and motifs using 20 C. maxima MLO proteins and 12 reference MLO proteins characterized in other plant species.
2.4 Plant materials and inoculation of pathogen
A PM susceptible C. maxima accession (SJ-1, South Korea) and a moderately resistant accession (PI 169404, Turkey) were grown in plastic pots in a greenhouse at Sejong University, Seoul, South Korea in the spring of 2017. Podosphaera xanthii was collected from naturally infected leaves of pumpkin plants in the greenhouse at Asia Seed Co. (Icheon, Gyeonggi-do, South Korea). We inoculated pumpkin seedlings at the three-leaf-stage by dusting and touching the plants with heavily infected pumpkin leaves. Control plants were not inoculated and maintained separately from the inoculated plants in the same greenhouse. The inoculated plants were wrapped with polythene to maintain a high humidity level until the disease symptoms developed. The leaf samples were collected at 4 d post inoculation (dpi) for both the inoculated and uninoculated control plants and collection continued for 5, 6, and 8 dpi in the inoculated plants from both accessions. Three plants from each treatment were used as three independent replicates per time point. The collected samples were frozen immediately in liquid nitrogen and kept at − 80 °C for RNA isolation.
2.5 RNA isolation and expression analysis
Total RNA was extracted from control and treated frozen leaf samples using the Plant RNA Purification Reagent (Invitrogen, Waltham, MA, USA). The RNA quality was determined using a Nanodrop ND-2000 spectrophotometer (Nanodrop Technologies, Waltham, MA, USA) and only high-quality RNA samples (A260/A230 > 2.0 and A260/A280 > 1.8) were used for subsequent experiments. Then, first-strand cDNA was synthesized from 5 μg of the total RNA following the protocol for the ReverTra Ace qPCR RT Master Mix kit (Toyobo, Osaka, Japan). Real-time PCR was conducted on a CFX real-time system (Bio-Rad, Hercules, CA, USA) using THUNDERBIRD SYBR qPCR mix (Toyobo, Osaka, Japan). Samples were amplified in the following method: 95 °C for 5 min, followed by 40 cycles of 95 °C for 30 s, 59 °C for 30 s, and 72 °C for 30 s. Three technical replications were used per sample. The transcript levels were normalized against expression of the C. maxima actin gene (GenBank Accession no. KF831060). The actin reference sample was defined as a 1 × expression level and the relative expression in our experimental samples was defined as the fold increase in mRNA level over the reference sample. The gene specific primers used for this analysis are shown in Table 4.
3 Results and discussion
3.1 Identification of the MLO gene family in C. maxima
In order to identify MLO genes in C. maxima, 16 MLO sequences from C. melo, 14 from C. lanatus, 13 from C. sativus, and 18 from C. pepo were used to query a BLASTn search on the C. maxima genome sequence data using Geneious v 9.1. As a result of this homology search, twenty genes were identified as potential members of the CmaMLO family and designated CmaMLO1 to CmaMLO20 according to their chromosomal location (Supplemental Data 1). As illustrated in Fig. 1, the 20 CmaMLO genes were evenly distributed on fifteen chromosomes of the twenty chromosomes that make up the C. maxima genome, but there were no CmaMLO genes on chromosomes 10, 11, 12, 14, and 19. Chromosomes 2, 3, 4, 13, and 20 had two CmaMLO genes each, while the other remaining chromosomes have only one CmaMLO gene. We also validated the absence of conserved MLO domains on chromosomes 10, 11, 12, 14, and 19 using the NCBI conserved domain search program. The pumpkin MLO genes had a scattered distribution pattern across its chromosomes, which could have arisen through segmental duplication; a clustered occurrence, which can occur due to tandem duplication of gene families, was not observed (Schauser et al. 2005). The total number of MLO genes identified in the C. maxima was the highest number among the MLO family genes have been reported in other species of Cucurbitaceae, for which the number of MLO genes ranged between 13 and 18 (Zhou et al. 2013; Lovieno et al. 2015). This result is consistent with previous reports of genome-wide surveys that suggest the presence of a number of MLO homologs that can vary from 13 to 20 in this plant family (Devoto et al. 2003; Feechan et al. 2008; Pessina et al. 2014; Appiano et al. 2015b; Rispail and Rubiales 2016).
The genomic and the predicted protein size of CmaMLO genes were further identified (Table 1) using the Fgenesh automatic annotation (Solovyev et al. 2006). Large variation was detected in the sizes of MLO genes in C. maxima. The genomic length of CmaMLO genes varied from 2.97 to 7.07 kb, and the length of their coding regions varied from 1.07 to 1.76 kb with average of 9 to 16 exons. This is comparable to the mean genomic length (3.79 to 9.95 kb), the length of coding regions (1.32 to 1.82 kb), and the number of exons (11 to 17) in other Cucurbits including cucumber, melon, and watermelon (Lovieno et al. 2015; Schouten et al. 2014). Accordingly, the protein size predicted to be encoded by CmaMLO genes varied from 354 to 595 amino acids (41.28 to 68.56 kDa) and all CmaMLO genes except CmaMLO6 and CmaMLO20 had amino acid lengths comparable to the Arabidopsis AtMLO homologs, which range from 460 to 593 residues in length (Devoto et al. 2003). The predicted isoelectric points of the CmaMLO-deduced polypeptides showed basic residues, which are common features in MLO proteins (Chen et al. 2014).
3.2 Structural features of CmaMLO proteins
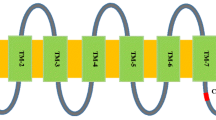
MLO proteins are characterized as an integral plasma membrane protein with seven transmembrane (TM) helices, an extracellular N-terminus, and a cytoplasmic C-terminus (Devoto et al. 1999). The TMs of CmaMLO genes in this study were predicted with different prediction servers and the number of TMs varied between the prediction servers, ranging from two (CmaMLO6) to eight (CmaMLO9) (Table 1). Among the twenty CmaMLO genes, nine genes (CmaMLO1, CmaMLO3, CmaMLO7, CmaMLO8, CmaMLO12, CmaMLO13, CmaMLO15, CmaMLO18 and CmaMLO19) were consistently found to have seven TMs (TM1 through TM7), which is a unique characteristic of the MLO gene family. However, sequence alignment further revealed that CmaMLO18 lacked the TM2 domain and had an additional domain between TM3 and TM4 (Fig. 2a). A BLASTp search in NCBI showed that the additional TM domain in CmaMLO18 had high sequence similarity with CsMLO5 from C. sativus and CmMLO5 from C. melo. CmaMLO9 not only contained all seven TMs but also had an additional domain in front of TM1 (Fig. 2a). Moreover, the BLASTp search showed that the additional TM domain in CmaMLO9 had high sequence similarity with C. sativus CsMLO2 and C. melo CmMLO6. The ten remaining CmaMLO genes showed various degrees of conservation of the seven TMs (Figs. 2a, 3). The TM domain sequence logo construction revealed that most of the amino acid residues of all seven TM domains were highly conserved (Fig. 2b).
The number and distribution of transmembrane (TM) domains of CmaMLO predicted by TOPCONS membrane protein topology prediction software (a) and Logos of amino acid sequences for the seven TMs (b) in the MLO-like C. maxima genes. Fully conserved amino acid residues in five TM domains (TM1, TM2, TM5, TM6 and TM7) was shown as red colored arrowheads in (b)
Multiple sequence alignment of 20 predicted MLO-like proteins from C. maxima and the barley MLO protein. The bars indicate the positions of the seven TM domains, and CaMBD was inferred from HvMLO (Devoto et al. 1999; Panstruga 2005). The open boxes indicate the two conserved peptide domains identified by Panstruga (2005) within the highly polymorphic C-terminus using Roman numerals I and II
3.3 Sequence alignment and conserved motif analysis of CmaMLO proteins
To analyze the sequence characteristics of the CmaMLO proteins, a multiple alignment of the candidate proteins including the barley MLO protein was conducted using ClustalW in Geneious v 9.1. The results indicated the presence of seven TM domains and 30 invariable amino acid residues identified in 38 MLOs from various species (Elliott et al. 2005; Fig. 3). Thirteen of these amino acid residues were located in six of the seven TM domains. We further analyzed the conservation of these 30 important amino acid residues in each CmaMLO protein. Four CmaMLOs (CmaMLO3, CmaMLO9, CmaMLO13 and CmaMLO19) conserved all 30 amino acid residues (Table 2). Eight CmaMLO proteins (CmaMLO1, CmaMLO4, CmaMLO8, CmaMLO10, CmaMLO12, CmaMLO14, CmaMLO15 and CmaMLO20) lost one or more amino acid residues, while two (CmaMLO7 and CmaMLO17) had mutations in some amino acid residues. The remaining six CmaMLO proteins had both deletions and mutations in some of the 30 invariable amino acid residues. All CmaMLOs except for CmaMLO20 preserved four extracellular cysteine residues (C86, C98, C114, and C367), which were considered essential for its function in barley (Elliott et al. 2005; Table 2).
A calmodulin-binding (CaMBD) domain in the C-terminus of MLO proteins is necessary and sufficient for Ca2+-dependent CaM complex formation, which is required for full activity of MLO to negatively regulate plant defense against powdery mildew in vivo and it contains three hydrophobic amino acid residues and a conserved tryptophan residue (Kim et al. 2002a, b; Bhat et al. 2005). Sequence alignment indicated a putative CaMBD domain in most of CmaMLO proteins that was composed of hydrophobic amino acid residues at positions 420, 427 and 433, and a highly conserved tryptophan residue at position 423, which corresponded to the position in HvMLO (Fig. 3). Panstruga (2005) reported two conserved peptide domains (I and II) in the C-terminal of several MLO proteins that may modulate powdery mildew infection in various plants including Arabidopsis, cabbage, tomato, pepper, barley, maize, rice, and wheat. Peptide domain I is located approximately 15–20 residues downstream of the CaMBD and characterized by conserved serine and threonine residues; the peptide domain II is characterized by the consensus sequence D/E–F–S/T–F (Panstruga 2005). The sequence alignment revealed that among twenty CmaMLO proteins, seven proteins (CmaMLO3, CmaMLO4, CmaMLO7, CmaMLO9, CmaMLO10, CmaMLO13, CmaMLO18, and CmaMLO19) had both peptide domains I and II in their C-terminal regions (Fig. 3).
Subsequently, we used the MEME online analysis tool to search conserved motifs in the CmaMLOs. There were ten conserved motifs ranging from 19 to 42 amino acid residues in twenty CmaMLO proteins (Fig. 4; Table 3). This result is consistent with the occurrence of ten conserved motifs in tomato MLO proteins (Chen et al. 2014), Cucumis sativus Mlo proteins (Zhou et al. 2013) and Vitis flexuosa (Islam and Yun 2016). Among the twenty CmaMLO proteins, five (CmaMLO4, CmaMLO7, CmaMLO9, CmaMLO13 and CmaMLO19) contained ten motifs, while four (CmaMLO2, CmaMLO3, CmaMLO14 and CmaMLO18) and seven CmaMLOs (CmaMLO1, CmaMLO5, CmaMLO8, CmaMLO11, CmaMLO12, CmaMLO15 and CmaMLO17) had nine and eight motifs, respectively. CmaMLO16 and CmaMLO20 had six motifs; CmaMLO10 and CmaMLO6 have seven and five motifs, respectively. Six of the identified conserved motifs (1, 2, 3, 6, 7 and 8) were located in six of the seven TM domains and motif 1 also included the CaMBD domain, while the remaining four motifs were located in other regions of the predicted CmaMLO proteins. These results are similar to previous reports that seven of the ten conserved motifs are located in the TM domains and the CaMBD domain, while the remaining three motifs are distributed in other regions of the predicted CmaMLO proteins (Zhou et al. 2013; Islam and Yun 2016).
3.4 Phylogenetic relationship of MLO proteins in C. maxima with other species
A phylogenetic tree containing 44 MLO proteins from dicots and monocots was constructed using the MLO-Pfam domain to illustrate the phylogenetic relationship of the sequences of CmaMLO proteins with other plant MLO proteins (Fig. 5). The MLO members collapsed in seven distinct clades (I through VII) with designations based on the classification of Arabidopsis and monocot MLO homologs (Devoto et al. 2003). Phylogenetic analysis showed that the CmaMLO distributed in six of the seven clades. Clade I included five CmaMLO proteins (CmamLO1, CmaMLO6, CmaMLO12, CmaMLO15 and CmaMLO16) while clade II comprised of three CmaMLO proteins (CmaMLO11, CmaMLO17 and CmaMLO20). Clade III contained four CmaMLO proteins (CmaMLO4, CmaMLO8, CmaMLO9 and CmaMLO10) and Clade V contained five CmaMLO proteins (CmaMLO3, CmaMLO7, CmaMLO13, CmaMLO18 and CmaMLO19). CmaMLO14 was clustered to clade VI while CmaMLO2 and CmaMLO5 were clustered to newly identified clade VII. Clades IV and V were previously reported to be involved in PM susceptibility (Jørgensen 1992; Elliott et al. 2002; Kim et al. 2002b; Consonni et al. 2006; Berg et al. 2015; Nie et al. 2015). HvMLO was the first identified MLO protein that causes PM susceptibility and clustered in clade IV with rice (OsMLO2) and wheat (TaMLO-A1 and TaMLO-B1) MLO proteins. Devoto et al. (2003) also identified this clade as a monocot-specific group. In clade V, five CmaMLOs were clustered with AtMLO2, AtMLO6 and AtMLO12 from Arabidopsis, MtMLO from barrel clover, PsMLO from pea, LiMLO from lotus, CaMLO from pepper, SlMLO from tomato, which are functionally characterized as being related to PM resistance. Recently, Appiano et al. (2015a, b) reported in transgenic complementation tests that all the monocot and dicot MLO homologs required for PM susceptibility were in clade V. In this study, five CmaMLOs was sorted into clade V and were further analyzed with three MLOs from Arabidopsis and barley by multiple sequence alignment to examine the amino acid sequence conservation in clade V (Fig. 6). Except for the absence of the TM2 domain in CsaMLO18, all of the CmaMLO proteins had a high degree of conservation at the predicted TM domain positions (Devoto et al. 2003). Moreover, a CaMBD in the C-terminus of MLO proteins was highly conserved in all the tested CmaMLO proteins. Additionally, all five CmaMLO proteins had two conserved peptide domains in their C-terminus (Fig. 6). Since these five CmaMLO proteins clustered into clade V and included AtMLO2, AtMLO6 and AtMLO12, which are considered PM susceptibility factors, their conserved sequence suggests that CaMBD and the two the peptide domains in C-terminus are potentially functional domains conferring the PM susceptibility in C. maxima plants. Expression analysis under pathogenic infection would need to be conducted to investigate and verify the patterns of CmaMLO gene expression.
Phylogenetic relationship between CmaMLO and the 24 reference MLO proteins already characterized in other plant species. The tree was generated using the MEGA7 software and maximum likelihood method based on Whelan and Goldman model. Bootstrap values (above 50%) from 1000 replicates are indicated above the branches
Multiple sequence alignment of CmaMLO proteins in clade V with selected MLO proteins involved in PM susceptibility in barley (HvMLO) and Arabidopsis (AtMLO2, AtMLO6 and AtMLO12). The bars indicate the positions of the seven TM domains, and CaMBD was inferred from HvMLO (Devoto et al. 1999; Panstruga 2005). The open boxes indicate the two conserved peptide domains identified by Panstruga (2005) within the highly polymorphic C-terminus represented by the Roman numerals I and II
3.5 Expression analysis of CmaMLO genes
Although the MLO protein family is divided into seven phylogenetic clades (Acevedo-Garcia et al. 2014; Pessina et al. 2014), all MLO proteins that are functionally related to PM susceptibility appear to be restricted to two clades; clade V for dicot species (Consonni et al. 2006; Bai et al. 2008; Feechan et al. 2008; Appiano et al. 2015a) and clade IV for monocots (Panstruga 2005; Reinstadler et al. 2010). Previous studies reported that tomato SlMLO1, pepper CaMLO2, eggplant SmMLO1, potato StMLO1 and tobacco NtMLO1 were causally associated with PM susceptibility and they clustered into clade V based on protein sequence like other dicot species (Panstruga 2005; Appiano et al. 2015a). To investigate the expression patterns of CmaMLO genes in clade V after pathogen inoculation, expression analysis was performed by real-time quantitative PCR using gene specific primers (Table 4). The mRNA transcripts of CmaMLO3, CmaMLO7, CmaMLO18, and CmaMLO19 but not CmaMLO13 were observed after inoculation with the PM pathogen P. xanthii. The four MLO genes (CmaMLO3, CmaMLO7, CmaMLO18 and CmaMLO19) showed distinctive expression patterns based on time points measured after PM pathogen inoculation and the compatible or incompatible plant-pathogen interaction type. In PI 169404 (incompatible interaction), CmaMLO3, CmaMLO18 and CmaMLO19 showed highly expression on 4 dpi and 5 dpi; their expression then decreased on 6 dpi but increased again on 8 dpi. The expression of CmaMLO7 showed significant up-regulation only on 8 dpi. Conversely, in SJ-1 (compatible interaction) only CmaMLO7 had significant up-regulation on 4 dpi and 5 dpi, with the highest expression measured on 8 dpi, whereas other three genes, CmaMLO3, CmaMLO18, and CmaMLO19 showed the highest expression and significant up-regulation only on 8 dpi. All four expressed genes (CmaMLO3, CmaMLO7, CmaMLO18 and CmaMLO19) are orthologous to the AtMLO2, AtMLO6 and AtMLO12 from A. thaliana (Fig. 5). Chen et al. (2006) reported that AtMLO2, AtMLO3, AtMLO6 and AtMLO12 were up-regulated by the biotrophic PM pathogen Erysiphe cichoracearum. Other studies also reported that the grapevine and apple MLO family genes that are orthologous to the AtMLO2, AtMLO6, and AtMLO12 were up-regulated after inoculation with the PM pathogens Vitis vinifera L. and Vitis flexuosa (Feechan et al. 2008; Islam and Yun 2016; Pessina et al. 2014). In addition, up-regulation of the MLO gene under P. xanthii was often associated with susceptibility to the P. xanthii pathogen in cucumber, barley, and apple (Berg et al. 2015; Pessina et al. 2014; Piffanelli et al. 2002; Schouten et al. 2014). In this our gene expression analysis, four MLO genes belonged to clade V were specifically induced by the PM pathogen. Furthermore two distinctive expression patterns were observed; the three MLO genes (CmaMLO3, CmaMLO18, and CmaMLO19) were specifically induced in the incompatible reaction of PI 169404 accession. In contrast, one MLO gene (CmaMLO7) was specifically induced in the compatible reaction of SJ-1 accession. This result speculates that different functional types of MLO genes could be differently involved in specific pumpkin-P. xanthii interactions. Conclusively, based on phylogenetic and expression analysis, these four MLO genes may be involved in both compatible and incompatible responses to the PM pathogen (Fig. 7).
Expression pattern of four MLO genes in C. maxima ‘SJ-1’ and ‘PI 169404’ lines after powdery mildew inoculation. The error bars represent the standard error of the means from three independent replicates. Significant differences between inoculated samples and control samples are indicated with an asterisk (p < 0.05)
4 Conclusion
MLO genes are plant specific and some MLOs are well documented in PM susceptibility. Loss-of-function of susceptibility genes could be an alternative approach for achieving lasting resistance against PM; the most well known examples are barley mlo and pea mlo mutants, which have been successfully employed to develop PM resistant lines. In the Cucurbitaceae family, loss-of-function mutations of CsMLO1 confer lasting PM resistance in cucumber (Nie et al. 2015). In this study, we identified twenty non-duplicated MLO genes in C. maxima using bioinformatics tools and structural features, as well as analyzed phylogenetic relationships based on protein sequences. Among the twenty MLO genes, five genes (CmaMLO3, CmaMLO7, CmaMLO13, CmaMLO18 and CmaMLO19) in clade V were significantly related to PM susceptibility, consistent with previous work that has shown that MLO genes involved in PM susceptibility are present in clade V. Structural analysis and comparison studies confirmed that these genes encode typical MLO proteins that have seven-transmembrane, a CaMBD domain, and two conserved peptide domains in the C-terminus, all of which are common structural features of other plant MLO proteins. Moreover, all of the predicted proteins had a high degree of homology with MLO proteins of other Cucurbits. We investigated the expression patterns of CmaMLO genes from clade V after PM pathogen infection and identified four MLO genes in clade V (CmaMLO3, CmaMLO7, CmaMLO18 and CmaMLO19) that were induced by PM pathogen inoculation. CmaMLO7 was specifically induced during the compatible reactions in the SJ-1 accession; CmaMLO3, CmaMLO18, and CmaMLO19 were specifically induced during incompatible reactions in the PI 169404 accession. This is the first comprehensive report of MLO genes in C. maxima to our knowledge. These findings provide valuable information of the functional characterization of MLOs related to PM susceptibility/resistance and can assist in the development of PM resistant pumpkin.
References
Acevedo-Garcia J, Kusch S, Panstruga R (2014) Magical mystery tour: MLO proteins in plant immunity and beyond. New Phytol. https://doi.org/10.1111/nph.12889
Adeniji AA, Coyne DP (1983) Genetics and nature of resistance to powdery mildew in crosses of Butternut with Calabaza squash and ‘Seminole Pumpkin’. J Am Soc Hortic Sci 108:360–368
Appiano M, Catalano D, Martı´nez MS, Lotti C, Zheng Z, Visser RGF, Ricciardi L, Bai Y, Pavan S (2015a) Monocot and dicot Mlo powdery mildew susceptibility factors are functionally conserved in spite of the evolution of class-specific molecular features. BMC Plant Biol 15:257
Appiano M, Pavan S, Catalano D, Zheng Z, Bracuto V, Lotti C, Visser RGF, Ricciardi L, Bai Y (2015b) Identification of candidate MLO powdery mildew susceptibility genes in cultivated Solanaceae and functional characterization of tobacco NtMLO1. Transgenic Res 24:847–858
Assaad FF, Qiu J-L, Youngs H, Ehrhardt D, Zimmerli L, Kalde M, Wanner G, Peck SC, Edwards H et al (2004) The PEN1 syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Mol Biol Cell 15:5118–5129
Bai Y, Pavan S, Zheng Z, Zappel NF, Reinstadler A, Lotti C, Giovanni C, Ricciardi L, Lindhout P et al (2008) Naturally occurring broad-spectrum powdery mildew resistance in a Central American tomato accession is caused by loss of Mlo function. Mol Plant-Microbe Interact 21:30–39
Berg J, Appiano M, Santillán Martínez M, Hermans F, Vriezen W, Visser R, Bai Y, Schouten HJ (2015) A transposable element insertion in the susceptibility gene CsaMLO8 results in hypocotyl resistance to powdery mildew in cucumber. BMC Plant Biol 15:243. https://doi.org/10.1186/s12870-015-0635-x
Bhat RA, Miklis M, Schmelzer E, Schulze-Lefert P, Panstruga R (2005) Recruitment and interaction dynamics of plant penetration resistance components in a plasma membrane microdomain. Proc Natl Acad Sci USA 102:3135–3140
Buschges R, Hollricher K, Panstruga R, Simons G, Wolter M, Frijters A, Van-Daelen R, Van-Der-Lee T, Diergaade P et al (1997) The barley Mlo gene: a novel control element of plant pathogen resistance. Cell 88:695–705
Chen Z, Hartmann HA, Wu M, Friedman EJ, Chen J-G, Pulley M et al (2006) Expression analysis of the AtMLO gene family encoding plant-specific seven-transmembrane domain proteins. Plant Mol Biol 60:583–597
Chen YB, Wang Y, Zhang H (2014) Genome-wide analysis of the mildew resistance locus o (MLO) gene family in tomato (Solanum lycopersicum L.). Plant Omics J 7:87–93
Consonni C, Humphry ME, Hartmann HA, Livaja M, Durner J, Westphal L, Vogel J, Lipka V, Kemmwelinf B et al (2006) Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat Genet 38:716–720
Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14:1188–1190
Dangl JL, Horvath DM, Staskawicz BJ (2013) Pivoting the plant immune system from dissection to deployment. Science 341:746–751
Deshmukh R, Singh VK, Singh BD (2014) Comparative phylogenetic analysis of genome-wide Mlo gene family members from Glycine max and Arabidopsis thaliana. Mol Genet Genomics 289:345–359
Devoto A, Piffanelli P, Nilsson I, Wallin E, Panstruga R, Von-Heijne G, Schulze-Lefert P (1999) Topology, subcellular localization, and sequence diversity of the Mlo family in plants. J Biol Chem 274:34993–35004
Devoto A, Hartmann HA, Piffanelli P, Elliott C, Simmons C, Taramino G, Goh CS, Cohen FE, Emerson BS et al (2003) Molecular phylogeny and evolution of the plant-specific seven-transmembrane MLO family. J Mol Evol 56:77–88
Elliott C, Zhou F, Spielmeyer W, Panstruga R, Schulze-Lefert P (2002) Functional conservation of wheat and rice Mlo orthologs in defense modulation to the powdery mildew fungus. Mol Plant Microbe Interact 15:1069–1077
Elliott C, Muller J, Miklis M, Bhat RA, Schulze-Lefert P, Panstruga R (2005) Conserved extracellular cysteine residues and cytoplasmic loop-loop interplay are required for functionality of the heptahelical MLO protein. Biochem J 385:243–254
Feechan A, Jermakow AM, Torregrosa L, Panstruga R, Dry IB (2008) Identification of grapevine MLO gene candidates involved in susceptibility to powdery mildew. Funct Plant Biol 35:1255–1266
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Garcia-Mas J, Benjak A, Sanseverino W, Bourgeois M, Mir G, Gonzalez VM, Henaff E, Camara F, Cozzuto L et al (2012) The genome of Melon (Cucumis melo L.). Proc Natl Acad Sci USA 109:11872–11877
Hammond-Kosack KE, Jones JD (2000) Response to plant pathogen. In: Buchanan BB, Gruissem W, Jones RL (eds) Biochemistry and molecular biology of plants. American Society of Plant Physiologists, Rockville, pp 1102–1156
Humphry M, Reinstädlerm A, Ivanov S, Bisseling T, Panstruga R (2011) Durable broad-spectrum powdery mildew resistance in pea er1 plants is conferred by natural loss-of-function mutations in PsMLO1. Mol Plant Pathol 12:866–878
Islam MZ, Yun H (2016) Characterization of nine Mlo family genes and analysis of their expression against pathogen infections in Vitis flexuosa. Euphytica 211:379–394
Jørgensen IH (1992) Discovery, characterization and exploitation of Mlo powdery mildew resistance in barley. Euphytica 63:141–152
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S et al (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649
Kim DS, Hwang BK (2012) The pepper MLO gene, CaMLO2, is required for susceptibility cell death response and bacterial and oomycete proliferation. Plant J 72:843–855
Kim MC, Lee SH, Kim JK, Chun HJ, Choi MS, Chung WS, Moon BC, Kang CH, Park CY et al (2002a) Mlo, a modulator of plant defense and cell death, is a novel calmodulin-binding protein. Isolation and characterization of a rice Mlo homologue. J Biol Chem 277:19304–19314
Kim MC, Panstruga R, Elliott C, Muller J, Devoto A, Yoon HW, Park HC, Cho MJ, Schulze-Lefert P (2002b) Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature 416:447–451
Lovieno P, Andolfo G, Schiavulli A, Catalano D, Ricciardi L, Frusciante L, Ercolano MR, Pavan S (2015) Structure, evolution and functional inference on the Mildew Locus O (MLO) gene family in three cultivated Cucurbitaceae spp. BMC Genomics 16:1112. https://doi.org/10.1186/s12864-015-2325-3
McGrath MT, Thomas CE (1996) Powdery mildew. In: Zitter TA, Hopins DL, Thomas CE (eds) Compendium of cucurbit diseases. APS Press, St. Paul, pp 28–30
Nie J, Wang Y, He H, Guo C, Zhu W, Pan J, Li D, Lian H, Pan J et al (2015) Loss-of-function mutations in CsMLO1 confer durable powdery mildew resistance in cucumber (Cucumis sativus L.) Front. Plant Sci 6:1155. https://doi.org/10.3389/fpls.2015.01155
Panstruga R (2005) Serpentine plant MLO proteins as entry portals for powdery mildew fungi. Biochem Soc Trans 33:389–392
Pavan S, Jacobsen E, Visser RGF, Bai Y (2010) Loss of susceptibility as a novel breeding strategy for durable and broad-spectrum resistance. Mol Breed 25:1–12
Pessina S, Pavan S, Catalano D, Gallotta A, Visser RGF, Bai Y, Malnoy M, Schouten HJ (2014) Characterization of Mlo gene family in Rosaceae and gene expression analysis in Malus domestica. BMC Genomics 15:618
Piffanelli P, Zhou F, Casais C, Orme J, Schaffrath U, Collins N, Panstruga R, Schulze-Lefert P (2002) The barley MLO modulator of defense and cell death is responsive to biotic and abiotic stress stimuli. Plant Physiol 129:1076–1085
Reinstadler A, Muller J, Czembor JH, Piffanelli P, Panstruga R (2010) Novel induced mlo mutant alleles in combination with site-directed mutagenesis reveal functionally important domains in the heptahelical barley Mlo protein. BMC Plant Biol 10:31
Rispail N, Rubiales D (2016) Genome-wide identification and comparison of legume MLO gene family. Sci Rep 6:32673. https://doi.org/10.1038/srep32673
Schauser L, Wieloch W, Stougaard J (2005) Evolution of NIN-like proteins in Arabidopsis, rice, and Lotus japonicus. J Mol Evol 60:229–237
Schouten HJ, Krauskopf J, Visser RG, Bai Y (2014) Identification of candidate genes required for susceptibility to powdery or downy mildew in cucumber. Euphytica 200:475–486
Solovyev V, Kosarev P, Seledsov I, Vorobyev D (2006) Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol 7:1–12
Taler D, Galperin M, Benjamin I, Cohen Y, Kenigsbuch D (2004) Plant R genes that encode photorespiratory enzyme confer resistance against disease. Plant Cell 16:172–184
Várallyay É, Giczey G, Burgyán J (2012) Virus-induced gene silencing of Mlo genes induces powdery mildew resistance in Triticum aestivum. Arch Virol 157:1345–1350
Xu X, Yu T, Xu R, Shi Y, Lin X, Xu Q, Qi X, Weng Y, Chen X (2016) Fine mapping of a dominantly inherited powdery mildew resistance major-effect QTL, Pm1.1, in cucumber identifies a 41.1 kb region containing two tandemly arrayed cysteine-rich receptor-like protein kinase genes. Theor Appl Genet 129:507–516. https://doi.org/10.1007/s00122-015-2644-4
Xu Q, Xu X, Shi Y, Qi X, Chen X (2017) Elucidation of the molecular responses of a cucumber segment substitution line carrying Pm5.1 and its recurrent parent triggered by powdery mildew by comparative transcriptome profiling. BMC Genomics 18:21. https://doi.org/10.1186/s12864-016-3438-z
Zhang G, Ren Y, Sun H, Guo S, Zhang F, Zhang J, Zhang H, Jia Z, Fei Z et al (2015) A high-density genetic map for anchoring genome sequences and identifying QTLs associated with dwarf vine in pumpkin (Cucurbita maxima Duch.). BMC Genomics 16:1101
Zheng Z, Nonomura T, Appiano M, Pavan S, Matsuda Y, Toyoda H, Wolters A-MA, Visser RGF, Bai Y (2013) Loss of function of Mlo orthologs reduces susceptibility of pepper and tomato to powdery mildew disease caused by Leveillula taurica. PLoS ONE 8:e70723
Zhou SJ, Jing Z, Shi JL (2013) Genome-wide identification, characterization, and expression analysis of the MLO gene family in Cucumis sativus. Genet Mol Res 12:6565–6578
Acknowledgements
This work was supported by grants from the Bio-industry Technology Development Program (111057-5 and 316087-4) of iPET (Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry). We are grateful to Asia Seed Company, South Korea for providing the susceptible C. maxima accession and powdery mildew-infected pumpkin leaves. PI 169404 was kindly supplied by United States Department of Agriculture.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Win, K.T., Zhang, C. & Lee, S. Genome-wide identification and description of MLO family genes in pumpkin (Cucurbita maxima Duch.). Hortic. Environ. Biotechnol. 59, 397–410 (2018). https://doi.org/10.1007/s13580-018-0036-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13580-018-0036-9