Abstract
Colon cancer is one of the most prevalent malignant tumors across the world. Increasing studies have demonstrated that long non-coding RNAs (lncRNAs) take part in colon cancer development. Our study intends to explore the expression characteristics of LBX2-AS1, a novel lncRNA, in colon cancer and its underlying mechanisms. The results illustrated that LBX2-AS1 level was substantially increased in colon cancer tissues and was obviously correlated with the tumor volume and early distant metastasis of patients. Besides, overexpression of LBX2-AS1 remarkably boosted growth, proliferation, and metastasis and restrained apoptosis in colon cancer cells, whereas LBX2-AS1 knockdown produced the opposite effect. On the other hand, miR-627-5p, down-regulated in colon cancer tissues, was negatively associated with LBX2-AS1 expression. Functional experiments showed that miR-627-5p suppressed colon cancer growth. Mechanistically, LBX2-AS1, as an endogenous competitive RNA, targeted miR-627-5p and restrained its expression, while miR-627-5p targeted and negatively regulated the RAC1/PI3K/AKT axis. Collectively, this study has revealed that LBX2-AS1 is a poor prognostic factor of colon cancer and can regulate colon cancer progression by regulating the miR-627-5p/RAC1/PI3K/AKT pathway.
Similar content being viewed by others

Avoid common mistakes on your manuscript.
Introduction
Colon cancer is a prevalent malignancy in the gastrointestinal tract [1]. People’s dietary structure and living habits changes also have potential role in colon cancer development by altering intestinal microbiome [2]. Clinically, colon cancer can be treated through surgery, chemotherapy, radiotherapy, and biological targeting [3]. However, a probe into effective molecular markers is necessary because they can assist in diagnosis and prognosis. Therefore, our paper revolves around the molecular mechanism of colon cancer growth and metastasis with a view to improving the diagnosis and prognosis.
Long non-coding RNAs (lncRNAs), a class of RNAs larger than 200 nucleotides, are not translated into proteins [4]. Studies have confirmed the cancer-promoting effects of lncRNAs in various malignancies [5]. For instance, lncRNA PVT1 facilitates the activation of signal transducers and transcriptional activators 3 (STAT3), which induces a cell process cycle to accelerate hepatoblastoma cell proliferation and metastasis [6]. On the contrary, lncRNA MT1JP expedites glioma cells’ apoptosis and hampers their proliferation, invasion, and migration through PTEN/Akt signaling pathway activation [7]. lncRNA LBX2-AS1, a novel cancer-concerned lncRNA, can serve as a crucial regulator for cancer development [8]. For instance, LBX2-AS1 boosts the proliferation of gastric cancer cells and attenuates their apoptosis through miR-219a-2-3p/FUS/LBX2 axis regulation [9]. Moreover, LBX2-AS1 bolsters the proliferation, migration, and EMT of esophageal squamous cell carcinoma (ESCC) by modulating ZEB1 and ZEB2 [10]. Notwithstanding, the mechanism of LBX2-AS1 in colon cancer remains poorly understood, and more investigation should be done.
miRNAs, endogenous non-coding small RNAs (18 to 25 nucleotides in length), play a prominent role in tumor occurrence and development [11]. For instance, miR-515-5p exerts a carcinostatic impact on breast cancer through mARK4/Hippo signaling pathway knockdown [12]. Similarly, miR-296-5p dampens cancer growth in the context of non-small cell lung cancer by targeting PLK1 [13]. As an important miRNA molecule, miR-627-5p, situated at 15q15.1 and 97 bp long, makes a great contribution to a variety of tumors. For instance, miR-627-5p exerts a carcinostatic effect on hepatocellular carcinoma (HCC) by weakening BCL3 [14]. Besides, the negative regulation of miR-627-5p by LINC00958 cramps oral squamous cell carcinoma (OSCC) development through experiments in vivo and in vitro, providing new insights into targeted molecular therapies for OSCC [15]. Nevertheless, the mechanism of miR-627-5p in regulating colon cancer development is surprisingly limited.
RAC1, a generally expressed Rho GTPase located on human chromosome 7p22, is a small molecular GTPase protein that has been confirmed to participate in the growth of multiple tumors [16]. Through luciferase activity experiments, Jowenland et al. have revealed that miR-224 targets the 3' untranslated region of RAC1. At the same time, they have uncovered that miR-224 defends dental pulp stem cells (DPSC) from apoptosis by down-regulating RAC1 through experiments both in vivo and in vitro [17]. What is more, miR-485-5p dampens Schwann cell proliferation and myelin formation by targeting CDC42 and RAC1 [18]. Numerous studies have substantiated that the PI3K/AKT signaling pathway is activated in most cancers, including colon cancer [19]. Take UNC5B-AS1 for example. It impedes the development and metastasis of colon cancer via miR-622 knockdown and AMPK/PI3K/AKT pathway inhibition [20]. Further, pharmacological inhibition of the mTOR/PI3K/AKT signaling pathway suppresses colorectal cancer growth [21]. Nevertheless, it remains to be seen whether LBX2-AS1 and RAC1 can influence colon cancer growth by modulating the PI3K/AKT signaling pathway.
Given the above findings, we hypothesized the irreplaceable status of LBX2-AS1 in colorectal cancer. Here, we discovered that LBX2-AS1 expression was uplifted in the tissues and cell lines of colorectal cancer patients. Moreover, functional experiments confirmed that it accelerated the proliferation and metastasis of colorectal cancer as a carcinogenic gene and repressed apoptosis. Further investigation of the mechanism revealed that LBX2-AS1, as a competitive endogenous RNA (ceRNA), targeted miR-627-5p, which acted on the 3 'UTR end of RAC1 and curbed its expression. The present study was designed to explore the mechanism of LBX2-AS1 and miR-627-5p/RAC1/PI3K/AKT in colorectal cancer development, further investigate its molecular mechanism, and provide a referential molecular marker for clinical treatment and prognosis.
Methods and materials
Clinical specimen collection and processing
The cancer tissues of 48 colorectal cancer patients who underwent colon cancer resection in Wuhan Fourth Hospital & PuAi Hospital of Tongji Medical College, Huazhong University of Science and Technology (December 2015 to December 2016) were obtained. Prior to operation, the patients did not receive chemotherapy, radiotherapy, or other adjuvant treatment. The control group samples were harvested from the same patients’ adjacent tissues (at least 3 cm away from the surgical resection edge), and no cancer cells were found in the postoperative pathological examination. The diagnosis of colon cancer was confirmed pathologically as per the world health organization (WHO) criteria. All specimens removed were immediately stored in -196 ℃ liquid nitrogen until they were utilized to extract RNA. Our experiments had received the imprimatur from the research ethics committee of Wuhan Fourth Hospital & PuAi Hospital with the informed consent of all patients involved (approval number: 2021–20-51). The clinical features of the colon cancer patients are exhibited in Table 1.
Cell culture and transfection
The human mucosal epithelial cell line NCM460 and colon cancer cell lines (SW480, HCT116, HCT-8, SW1116, HT29), ordered from the American Type Culture Collection (ATCC, Rockville, MD, USA), were used for the experiments. These cells were cultured with 10% fetal bovine serum (FBS) (Thermo Fisher Scientific, MA, USA) and 1% (Invitrogen, CA, USA) RPMI1640 (Thermo Fisher Scientific, MA, USA) in an incubator (37 ℃, 5% CO2). During the logarithmic growth phase, cells underwent trypsinization and passage with 0.25% trypsin (Thermo Fisher, HyClone, Utah, USA). Through detection, LBX2-AS1 expression was found to reach the lowest in SW480 cells and the highest in HT29. Hence, SW480 and HT29 cells were adopted as research objects in subsequent experiments. LBX2-AS1 overexpression plasmid, small inference RNA targeting LBX2-AS1 (si-LBX-AS1), miR-627-5p mimics, si-RAC1, and the corresponding negative control group were supplied by GenePharma (Shanghai, China) and FulenGen (Guangzhou, China). SW480 and HT29 cells were taken and inoculated onto 24-well cell culture plates (3 × 105 cells/well in density). After 24-h incubation at 37 ℃ with 5% CO2, the cells were transfected. Lipofectamine® 3000 (Invitrogen; ThermoFisherScientific, Inc.) was applied to transfect SW480 and HT29 cells in accordance with the supplier’s instructions. Then, we performed RT-PCR to evaluate the transfection efficiency. Finally, the cells were incubated for 24 h (37 °C, 5% CO2) in preparation for further analysis. The sequences of relevant siRNAs are as follows: miR-627-5p mimics: 5ʹ-GUGAGUCUCUAAGAAAAGAGGA-3ʹ; si-NC 5ʹ-UUCUCCGAACGUGUCACGUTT-3ʹ; si-LBX2-AS1#1 5ʹ-AGGAATGTTTGCTGAATTAATGG-3ʹ; si-LBX2-AS1#2 5ʹ-CCCAAGTTATAAAACTATAATGC-3ʹ; si-RAC1#1 5ʹ-GCAAAGUGGUAUCCUGAAG-3ʹ; si-RAC1#2 5ʹ-AATATATCCCTACTGTCTTTG-3ʹ.
RT-PCR
As per the manufacturer’s instructions (Invitrogen, Waltham, MA, USA), TRIzol reagent was taken to extract total RNA from tissues or cells. The RNA concentration and purity were determined with Nanodrop spectrophotometer. In line with the manufacturer’s protocol, complementary DNA (cDNA) was synthesized from 1 µg total RNA using the PrimeScript -RT Kit (Madison, WI, USA). Next, SYBR® Premix-Ex-Taq ™ (Takara, TX, USA) and the ABI7300 system were employed for quantitative reverse-transcription polymerase chain reaction (qRT-PCR). The PCR system had a total volume of 30 µL, and each sample contained 300 ng cDNA. The amplification process included 10 min’ initial denaturation at 95 °C, followed by 45 cycles: 10 s at 95 °C, 30 s at 60 °C, and 20 s at 85 °C. All fluorescence data were converted into relative quantification: GAPDH served as the endogenous control of lncRNA LBX2-AS1 and RAC1, while U6 acted as the endogenous control of miR-627-5p. All RT-PCT reactions were duplicated three times.
lncRNA LBX2-AS1 primers:
upstream 5ʹ-AAAACAAATGGAGTGGGGCC-3ʹ;
downstream 5ʹ-TGCAGCTTCCCTTTTGTTCC-3ʹ;
miR-627-5p primers: upstream 5ʹ-AAGCGCCTGTGAGAGTCTCTAAGAA-3ʹ, downstream 5ʹ-CAGTGCAGGGTCCGAGGT-3ʹ;
Primers for RAC1: upstream 5ʹ-TAAGCCCAGATTCACCGGTT-3ʹ,
downstream 5ʹ-CATCAAGTGTGTGTGTGGTGG-3ʹ;
GAPDH primers: upstream 5ʹ-CCACATCGCTCAGACACCAT-3ʹ,
downstream 5ʹ-TGACAAGCTTCCCGTTCTCA-3ʹ;
U6 primers: upstream 5ʹ-CTCGCTTCGGCAGCACA-3ʹ,
Downstream 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ.
CCK-8
After trypsinization, SW480 and HT29 cells in the logarithmic growth phase were adjusted to 2 × 103/mL and inoculated on 96-well plates with 100 μL cell suspension in each well and 3 replicates in each group. Then, the plates were placed in an incubator for further culture. Twenty-four hours later, each well was given 10 μL of CCK-8 solution (Hubei Baios biotechnology co., LTD.) for 1-h incubation. When the culture was terminated, the 96-well plates were placed in an enzyme marker to measure the absorbance (OD value) of each well at the wavelength of 450 nm. At last, the cell absorbance was measured on the 24th, 48th, and 72nd hours.
Transwell assay
According to the requirements of Chemicon company ECM550 series specifications, the preparation of matrix Transwell Chambers (Corting, NY, USA) was done. We respectively inoculated 200 μL SW480 and 200 μL HT29 cells into Transwell chambers, adjusted the density to 5 × 105/mL, administered 700 μL culture solution with FBS to the lower chamber with a 24-well plate, and cultivated them for 24 h. Afterwards, we aspirated the culture medium in the chamber, gently wiped off the matrigel with a cotton swab, washed it with PBS once, and air-dried it. After being soaked in the chamber with formaldehyde for 20 min, it was flushed once with PBS and air-dried. Violet staining was performed for 20 min. Following staining, the cells were rinsed with PBS 3 times, air-dried, and then imaged under an upright microscope (Olympus, Tokyo, Japan).
RIP experiment
We collected 2 × 107 cell samples, added 200 µL RIP lysis buffer to lyse the cells on ice for 5 min. The cells went through 15 min’ centrifugation at 1500 rpm to produce the supernatant. Next, 5 µg target protein-specific RIP and the corresponding IgG antibody-coated magnetic beads were mixed. The samples after cleavage were divided into two parts, respectively conflated with RIP antibody and the corresponding IgG antibody-coated magnetic beads, and incubated overnight at 4 ℃. Next, when the supernatant was discarded, the magnetic beads were flushed 5 times with a washing buffer. Then, we administered protease K lysates to the magnetic beads at 55 ℃ for 30 min. The supernatant was placed in a new centrifuge tube, and total RNA was extracted through phenol–chloroform-isoamyl alcohol extraction and purified via isopropanol centrifugation. The profiles of lncRNA LBX2-AS1 and RAC1 were estimated via RT-PCR.
Dual-luciferase reporter gene assay
The Dual-luciferase reporter assay system (Promega, Madison, WI, USA) was applied to measure luciferase reporter genes. The target fragments of wild-type LBX2-AS1 and mutant LBX2-AS1 were built and integrated into pGL3 vector (Promega, Madison, WI, USA) to construct pGL3-LBX2-AS1-wild type (LBX2-AS1-wt) and pGL3-LBX2-AS1-mutant (LBX2-AS1-mut) reporter vectors. Similarly, the target fragments of wild-type RAC1 and mutant RAC1 were constructed and integrated into pGL3 vector (Promega, Madison, WI, USA) to construct pGL3-RAC1-wild type (RAC1-WT) and pGL3-RAC1-mutant (RAC1-MUT) reporter vectors. LBX2-AS1/RAC1-WT or LBX2-AS1/RAC1-MUT was co-transfected with miR-627-5p or the negative control into SW480 cells. The luciferase activity was determined in conformity with the supplier’s instructions 48 h subsequent to transfection. All experiments were done in triplicate and duplicated three times.
Western blot
When the cell treatment came to an end, the culture medium was removed, protein lysates (Roche) were added, and the total protein was separated. Then, 50 g total protein was added to 12% polyacrylamide gel and subjected to 2 h’ 100 V electrophoresis. The protein was then electrically moved onto polyvinylidene fluoride (PVDF) membranes. After being flushed three times in TBST (10 min each) and sealed with 5% skimmed milk powder for an hour at room temperature (RT), the membranes were incubated along with primary antibodies Anti-RAC1 antibody (ab155938, 1: 1000, Abcam, MA, USA), Anti-PI3K antibody (ab191606, 1: 1000, Abcam, MA, USA), Anti-PI3k (phosphoY607) antibody (ab182651, 1: 1000, Abcam, MA, USA), Anti-pan-AKT antibody (ab18785, 1: 1000, Abcam, MA, USA), Anti-AKT (phospho T308) antibody (ab38449, 1: 1000, Abcam, MA, USA), Anti-Bax antibody (ab32503, 1: 1000, Abcam, MA, USA), Anti-Bcl-2 antibody (ab59348, 1: 1000, Abcam, MA, USA), Anti-Caspase-3 antibody (ab13847, 1: 1000, Abcam, MA, USA), Anti-E-cadherin antibody (ab16505, 1: 1000, Abcam, MA, USA), Anti-Vimentin antibody (ab92547, 1: 1000, Abcam, MA, USA), and Anti-N-cadherin antibody (ab18203, 1: 1000, Abcam, MA, USA) overnight at 4 °C. After TBST washing, the membranes underwent 1-h incubation with the horseradish peroxidase (HRP)-labeled goat anti-rabbit antibody (ab150077, 1:3000, 1:1000, Abcam, MA, USA) at RT. The membranes were rinsed with TBST 3 times, 10 min each. Finally, the Western blot reagent (Invitrogen) was adopted for color imaging, and Image J was introduced to analyze the gray value of each protein.
Tumor xenograft experiment
Twenty BALB/c nude mice (6 weeks of age, 16–18 g in weight) were supplied by the Animal Center of Tongji Medical College (Wuhan, China) and kept under specific pathogen-free conditions. SW480 cells transfected with NC or LBX2-AS1 overexpression plasmid were harvested and adjusted to 1 × 108 cells/mL in density. Next, 100 μL of tumor cell suspension was injected into BALB/c nude mice subcutaneously. From the fifth day of injection, the volume of the formed tumor tissues was gauged. The mice were euthanized 5 weeks later, and the tumor xenograft was surgically resected. The severed tumor tissues were kept in liquid nitrogen for use. A Vernier caliper was taken to measure the longest diameter (a) and the shortest diameter (b) of the tumors. The tumor volume was calculated as per this formula: V = ab2 × 0.5. A tumor growth curve was developed, and the tumor mass was weighed 5 weeks later. In order to construct a mouse lung-metastasis model, each mouse was injected with 100 μL tumor cell suspension (1 × 105 cells) through tail vein [22]. Then the lung metastasis of mice was observed by H&E staining. All animal procedures have been approved by the Ethics Review Board of Wuhan Fourth Hospital (approval number: 2021-20-51), and they were conducted according to the standards established by the Guidelines for the Care and Use of Laboratory Animals by Wuhan Fourth Hospital.
Hematoxylin and eosin (H&E) staining and Immunohistochemistry
The tumor and cancer tissues of mice were embedded in paraffin and sectioned (4 μm), dewaxed, hydrated, stained with hematoxylin and eosin, sealed with neutral resin, observed under microscope and photographed for analysis. For immunohistochemistry, after dewaxing and hydration, the sections were incubated with 3% H2O2 for antigen retrieval for 15 min, washed with PBS 3 times; After being incubated with primary antibodies, including Ki67 (1:200), E-cadherin (1:200), or Vimentin (1:200) overnight at 4 ℃, the sections were washed with PBS 3 TIMES, incubated with HRP-labeled secondary antibody at room temperature for 2 h. Diaminobenzidine (DAB) was used for color development. After hematoxylin counterstaining, it was dehydrated and transparent. The number of positive cells under 400 times visual field was counted and the average value was calculated.
TdT-mediated dUTP nick end labeling (TUNEL)
Paraffin embedded mouse tissue sections were dewaxed and hydrated. The operation was carried out according to the instructions of TUNEL apoptosis detection kit (Beyotime biotechnology, Shanghai, China). Protease K working solution was added and reacted at 37 ℃ for 15 min. Add 50 μL TUNEL test solution, incubate in the dark at 37 ℃ for 60 min, and then counterstain DAPI. Add quencher, seal the film and keep away from light. Detect and take photos with Leica (DM2500) fluorescence microscope. Ten high-power visual fields were randomly selected and the number of positive cells was calculated.
Statistical analysis
The SPSS17.0 statistical software (SPSS Inc., Chicago, IL, USA) was introduced for analysis. The measurement statistics were presented as mean ± standard deviation. The statistical data were presented in four tables (or percentages), and the differences between the two groups were analyzed using χ2. P < 0.05 was regarded as statistically meaningful.
Results
LBX2-AS1 was highly expressed in colon cancer tissues and cells
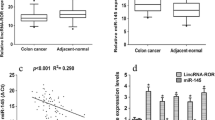
To probe the features of LBX2-AS1 expression in colon cancer tissues, we conducted RT-PCR and discovered that LBX2-AS1 expression was notably enhanced in colon cancer tissues than in adjacent normal tissues (P < 0.05, Fig. 1A). Other results illustrated that LBX2-AS1 expression was remarkably uplifted in colon cancer cell lines (SW480, HCT116, HCT-8, ADAM17, HT29) than in NCM460 human colon mucosal epithelial cells (P < 0.05, Fig. 1B). More importantly, colon cancer patients with a higher level of LBX2-AS1 displayed a more advanced tumor stage and a higher rate of distant metastasis (Table 1). Additionally, LBX2-AS1 up-regulation was associated with the poorer survival rate of colon cancer patients (Fig. 1c), which was consistent with the data of colon cancer patients shown in the Starbase database (http://starbase.sysu.edu.cn/) (Fig. 1D). The image of metastasis-free survival exhibited that highly-expressed LBX2-AS1 pertained to the rate of distant metastasis (Fig. 1E). The above results indicated that LBX2-AS1 was associated with the malignant phenotype of colon cancer cells and might have carcinogenic effects.
LBX2-AS1 was highly expressed in colon cancer cells and tissues. A qRT-PCR was taken to detect LBX2-AS1 expression in colon cancer and adjacent normal tissues, ***P < 0.001 (vs. the Normal group). N = 48. B qRT-PCR was performed to detect LBX2-AS1 expression in colon cancer cell lines (SW480, HCT116, HCT-8, ADAM17, HT29) and NCM460 human colon mucosal epithelial cells. ***P < 0.001 (vs. the NCM460 group). N = 3. C The survival rate of colon cancer patients with different levels of LBX2-AS1 was analyzed by Kaplan–Meier (KM) plotter assay. D The Starbase database validated the prognosis of LBX2-AS1 in colon adenocarcinoma (COAD) patients. E KM plotter assay was used for analyzing the correlation of LBX2-AS1 expression with metastasis-free survival (MFS) in human colon cancer. MFS was defined as the time between the date of surgery and the date of metastatic recurrence or last reported follow-up
LBX2-AS1 overexpression boosted the proliferation, metastasis, and EMT transformation of colon cancer cells and weakened their apoptosis
With a view to investigating the impact of LBX2-AS1 on colon cancer progression, we constructed the LBX2-AS1 overexpression and knockdown models in the colon cancer cell lines SW480 and HT29, respectively (P < 0.05, Fig. 2A). CCK-8 demonstrated that cell proliferation level was remarkably heightened after LBX2-AS1 overexpression, while LBX2-AS1 knockdown suppressed the proliferation of colon cancer (P < 0.05, Fig. 2B, C). Transwell revealed that LBX2-AS1 overexpression evidently enhanced cell invasion, whereas LBX2-AS1 knockdown considerably weakened the invasion ability (P < 0.05, Fig. 2D). Western blot was used to evaluate the effect of LBX2-AS1 regulation on apoptosis. The statistics disclosed that LBX2-AS1 overexpression dramatically drove up the expression level of the apoptotic protein Bcl-2 and lowered the expression levels of pro-apoptotic proteins Bax and Caspase-3, while LBX2-AS1 knockdown resulted in the opposite landscape (Fig. 2E). In order to explore the regulatory impact of LBX2-AS1 on EMT markers E-cadherin, Vimentin, and N-cadherin in colon cancer cells, we performed Western blot and uncovered that the protein profile of E-cadherin was notably brought down by LBX2-AS1 overexpression, while the protein profiles of Vimentin and N-cadherin were drastically elevated. After LBX2-AS1 knockdown, E-cadherin protein expression was dramatically uplifted, whereas Vimentin and N-cadherin protein profiles were remarkably reduced (P < 0.05, Fig. 2F). These statistics illustrated that LBX2-AS1 partook in the occurrence and growth of colon cancer, facilitated the proliferation, invasion, and EMT of tumor cells, and weakened their apoptosis.
LBX2-AS1 overexpression boosted the proliferation, metastasis, and EMT transformation of colon cancer cells and reduced their apoptosis. A The LBX2-AS1 overexpression and knockdown models were engineered in colon cancer cell lines SW480 and HT29, respectively. B, C CCK-8 tracked cell proliferation. D. Western blot examined apoptosis-concerned proteins. E. Transwell was conducted to test cell invasion. F. Western blot was performed to measure the effect of LBX2-AS1 on colon cancer cell EMT. ns P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001 (vs. the NC or si-NC group). N = 3
LBX2-AS1 promoted colon cancer cell growth in vivo
To further verify the role of LBX2-AS1 in tumor cell growth, we carried out a tumor xenograft experiment. The results indicated that LBX2-AS1 overexpression markedly enhanced the tumor volume, growth, and weight (compared with the NC group) (Fig. 3A–C). The level of LBX2-AS1 in the tumor tissues was also markedly uplifted in the LBX2-AS1 group (vs. the NC group) (Fig. 3D). Additionally, we performed RT-PCR, Western blot, and IHC to detect apoptosis and EMT-concerned genes or proteins. As a result, LBX2-AS1 up-regulation restrained Bax, C-caspase3, and E-cadherin levels but enhanced Bcl2, Vimentin, and N-cadherin expressions (Fig. 3E–I). Next, lung metastasis model was constructed in nude mice by injecting SW480 into the tail vein of the mice. Hematoxylin and eosin (H&E) staining showed that LBX2-AS1 overexpression enhanced the number of tumor nodules in the lung (Fig. 3J). H&E staining of the tumors indicated that there were apoptotic cells (noted by the circle) in the NC group, while enhanced neovascularization (noted by the arrows) was observed in the LBX2-AS1 group (Fig. 3K). TUNEL assay and IHC data displayed that LBX2-AS1 overexpression lowered the apoptosis rate, and enhanced cell proliferation (marked by KI67) of colon cancer cells (Fig. 3L, M). The above data revealed that LBX2-AS1 exerted an oncogenic effect in vivo.
LBX2-AS1 promoted colon cancer cell growth in vivo. A tumor xenograft experiment was conducted to check the growth of SW480 cells transfected with NC or LBX2-AS1 overexpression plasmid. A–C The tumor volume (A), images (B), and weight (C). D qRT-PCR determined the level of LBX2-AS1 in the tumor tissues. E qRT-PCR was used for detecting apoptosis-related genes (including Caspase3, 6, 7, 8, 9, Bak, Bad, Bax, BCL2, BCL2L1) in the tumor tissues. The data are shown by heat map. F Western blot examined apoptosis-associated proteins (including Bax, Bcl2 and Caspase3) in the tumor tissues. G qRT-PCR was used for detecting EMT phenotype genes (including N-cadherin, Vimentin, Snail1, Twist, MMP-2, MMP-3, MMP-9, Zeb1, E-cadherin, ZO-1) in the tumors, and the data are shown by heat map. H Western blot was carried out for evaluating EMT-associated proteins (including E-cadherin, Vimentin, and N-cadherin) in the tumor tissues. I IHC was carried out for detecting E-cadherin and Vimentin in the tumor tissues. Scale bar = 50 μm. J SW480 cells were injected into the tail vein of mice for constructing lung-metastasis model. H&E staining gauged the metastasis lesions in the lung. The average number of metastasis nodules (shown by the arrows) in five versions was counted. Scale bar = 100 μm. K. H&E staining was used for pathological examination of tumors. The circle indicates apoptotic cells, and the arrows indicate growing blood vessels. L TUNEL assay was used for detecting apoptotic cells (marked by red fluorescence) Scale bar = 50 μm. M IHC was carried out for detecting KI67 in the tumors. Scale bar = 25 μm. ***P < 0.001. N = 5
LBX2-AS1 enhanced the profile of RAC1/PI3K/AKT
The GEPIA database (http://gepia.cancer-pku.cn/) discovered a correlation between the profiles of LBX2-AS1 and RAC1/PI3K/AKT and demonstrated that they were positively associated with each other (Fig. 4A). Pearson correlation analysis showed that LBX2-AS1 was positively correlated with RAC1 in colon cancer tissues (R2 = 0.494, P < 0.0001, Fig. 4B). Furthermore, we determined the impact of LBX2-AS1 expression regulation on RAC1/PI3K/AKT via Western blot. The statistics reflected that RAC1 and p-PI3K/p-AKT expressions were notably heightened after overexpression of LBX2-AS1, but they were remarkably lowered after LBX2-AS1 knockdown (Fig. 4C, D). These results indicated that in colon cancer, LBX2-AS1 was positively associated with the profile of the RAC1/PI3K/AKT pathway, and overexpression of LBX2-AS1 could enhance the profile of RAC1/PI3K/AKT.
LBX2-AS1 bolstered the profile of the RAC1/PI3K/AKT axis. A: The GEPIA database (http://gepia.cancer-pku.cn/) illustrated the correlation between LBX2-AS1 and RAC1, PI3K, or AKT. B: Pearson correlation analysis revealed the correlation between LBX2-AS1 and RAC1. C and D: Western blot examined the altered RAC1 (C) and the PI3K/AKT pathway regulated by LBX2-AS1. ** P < 0.01, *** P < 0.001. N = 3
RAC1 inhibition attenuated colon cancer progression and PI3K/AKT activation
To confirm the function of RAC1 in colon cancer development, we engineered a RAC1 knockdown model in SW480 and HT29 cells (P < 0.05, Fig. 5A). CCK-8 and Transwell examined the influence of RAC1 knockdown on cell proliferation and invasion, respectively. As a result, the proliferation of RAC1 cells was considerably abated after RAC1 knockdown (P < 0.05, Fig.5B, C). RAC1 down-regulation lowered the level of Bcl2 and heightened the levels of Bax and C-caspase3 (FIG. 5 D-E). Furthermore, the results of Transwell and Western blot displayed that RAC1 inhibition mitigated the invasion and EMT of colon cancer cells (Fig. 5F–H). We further performed Western blot to evaluate the activation of the PI3K/AKT signaling pathway. It transpired that RAC1 knockdown notably hindered PI3K/AKT activation (Fig. 5I, J). The above findings unveiled that RAC1 inhibition attenuated colon cancer proliferation, invasion, and EMT and suppressed PI3K/AKT pathway activation.
Inhibition of RAC1 impeded colon cancer progression and PI3K/AKT activation. A. A knockdown model of RAC1 was established in SW480 and HT29 cell lines. B-C: CCK-8 checked cell proliferation. D-E. Western blot examined apoptosis-correlated proteins. F. Transwell was conducted to test cell invasion. G-J. Western blot was performed to examine EMT-correlated proteins and the PI3K/AKT pathway in colon cancer cells. ns P > 0.05, * P < 0.05, **P < 0.01, ***P < 0.001 (vs. the si-NC group)
The targeted correlation among LBX2-AS1, miR-627-5p, and RAC1
Given the lncRNA-miRNA-mRNA regulatory network diagram, we were curious about the regulatory mechanism of LBX2-AS1 and RAC1. By searching the targeted miRNA of LBX2-AS1 and RAC1 through the Starbase, miRmap, and microT databases, we discovered that miR-627-5p, miR-3612, and miR-650 were the three candidate miRNA targets of LBX2-AS1 and RAC1 (Fig. 6A). Interestingly, our results of RT-PCR exhibited that miR-627-5p was down-regulated when LBX2-AS1 was overexpressed (Fig. 6B). The binding sites of miR-627-5p with LBX2-AS1 and RAC1 are displayed in Fig. 6C. To clarify the targeted relationship among the three, we implemented dual luciferase activity assay and RIP. The results revealed that miR-627-5p mimics obviously cramped the luciferase activity of LBX2-AS1-WT and RAC1-WT but exerted no conspicuous effect on LBX2-AS1-MUT and RAC1-MUT (P < 0.05, Fig. 6D, E). In addition, following the transfection of miR-627-5p mimics, the levels of LBX2-AS1 and RAC1 precipitated in the Ago2 antibody group were notably higher than those in the IgG group, suggesting that LBX2-AS1 and RAC1 were bound to Ago2 protein through miR-627-5p (P < 0.05, Fig. 6F, G). Taken together, the above statistics indicated that miR-627-5p contained the binding sites with LBX2-AS1 and RAC1.
The targeted correlation among LBX2-AS1, miR-627-5p, and RAC1. A The Starbase, miRmap, and microT databases were introduced to predict the targeted miRNAs of LBX2-AS1 and RAC1. miR-627-5p, miR-3612, and miR-650 were the three candidate miRNA targets of LBX2-AS1 and RAC1. B qRT-PCR was used to detect miR-627-5p, miR-3612, and miR-650 in SW480 cells with LBX2-AS1 overexpression. C The binding sites of miR-627-5p with LBX2-AS1 and RAC1. D and E Dual luciferase activity assay was used to confirm the binding correlation between miR-627-5p and LBX2-AS1, miR-627-5p and RAC1, in SW480 cells. F and G SW480 cells were transfected with miR-627-5p mimics or NC. RIP was conducted, and the enrichment of LBX2-AS1 and RAC1 in the lysates was examined by qRT-PCR. NS P > 0.05, **P < 0.01, ***P < 0.001. N = 3
LBX2-AS1 repressed the anti-tumor impact of miR-627-5p on colon cancer
To assess the LBX2-AS1/miR-627-5p axis in colon cancer development, we conducted rescue experiments. LBX2-AS1 overexpression in the miR-627-5p overexpression group attenuated miR-627-5p’s level (Fig. 7A). Then, the proliferation, apoptosis, invasion, and EMT of colon cancer cells were examined. The results revealed that compared with the miR-627-5p group, the proliferation, invasion, and EMT of colon cancer cells were substantially attenuated, whereas their apoptosis was boosted (Fig. 7B–E). However, forced overexpression of LBX2-AS1 in the miR-627-5p group markedly mitigated the effects induced by miR-627-5p (P < 0.05, Fig.7B–E). Moreover, RAC1 and the PI3K/AKT pathway were down-regulated following miR-627-5p up-regulation, but such an effect was abolished by LBX2-AS1. Therefore, miR-627-5p dampened colon cancer development, while LBX2-AS1 repressed miR-627-5p-mediated effects.
LBX2-AS1 suppressed the anti-tumor effect of miR-627-5p on colon cancer. SW480 cells were transfected with miR-627-5p mimics and/or LBX2-AS1 overexpression plasmid. A qRT-PCR was used to detect miR-627-5p in SW480 cells. B CCK-8 examined cell proliferation. C Transwell was conducted to test cell invasion. D–F Western blot was implemented to check apoptosis- (D) and EMT-related proteins (E) as well as the RAC1/PI3K/AKT pathway in colon cancer cells. ns P > 0.05, * P < 0.05, **P < 0.01, ***P < 0.001 (vs. the NC group). ns P > 0.05, &&P < 0.01, &&&P < 0.001 (vs. the miR-627-5p group). N = 3
Discussion
As a regulatory RNA discovered in recent years, lncRNA has gradually become a research hotspot in the tumor biology field [23]. Prior studies have confirmed that lncRNA FOXD2-AS1, drastically increased in colorectal cancer tissues, expedites the proliferation, migration, and invasion of colorectal cancer cells by negatively targeting miR-185-5p [24]. What is more, lncRNA MALAT1 steps up the proliferation and metastasis of colon cancer cells by targeting the negative regulation of miR-129-5p expression [25]. Given the above statistics, we ventured to speculate that LBX2-AS1 might also play an oncogenic function in colon cancer. Surprisingly, we discovered that LBX2-AS1 expression was uplifted in colon cancer tissues. Further, functional experiments demonstrated that LBX2-AS1 boosted colon cancer proliferation, invasion, and EMT transformation but suppressed cell apoptosis. Therefore, LBX2-AS1 was substantiated to be a sound diagnostic marker and a potential target for colon cancer.
miRNAs make a great contribution to cell proliferation regulation, apoptosis, and tumorigenesis [26]. For instance, miR-378 represses proliferation and boosts apoptosis in colon cancer cells by targeting SDAD1 expression [27]. Additionally, miRNA-185 is identified to be a prognostic biomarker and mitigates the migration and invasion of colon cancer cells through targeting Wnt1 [28]. The above findings represented that a variety of miRNAs contribute to tumor inhibition in colon cancer growth. Moreover, lncRNAs have been reported to influence tumor progression by modulating miR-627-5p expression. For instance, researches have revealed that lncRNA UCA1 overexpression heightens NR2C2 expression by dampening miR-627-5p, thereby promoting the malignant behaviors of glioma cells [29]. Besides, lncRNA SNHG15 activates CDK6 and SOX-2 through repressing miR-627-5p, thus enhancing the progression of glioma [30]. These discoveries mean that miR-627-5p may be involved in tumor development as a suppressor and is regulated by lncRNAs. Consistent with the above findings, here, we uncovered that miR-627-5p impeded the proliferation, invasion, and EMT transformation of colon cancer cells and facilitated their apoptosis. Bioinformatics analysis uncovered the targeted binding correlation between miR-627-5p and LBX2-AS1, and LBX2-AS1 prominently dampened miR-627-5p-mediated anti-tumor functions. These facts denoted that miR-627-5p was a tumor suppressor of colon cancer and was negatively modulated by LBX2-AS1.
RAC1 is one of the important members in the GTPases Rho family. Reportedly, its positive expression is notably elevated in colon cancer cells, and the mechanism of colon cancer infiltration and metastasis may be related to the activation of CDC42, Smad4, ERK/JNK, and other signals, thus promoting EMT [31, 32]. In colon cancer, RAC1 has also been recognized to exert a vital function. For instance, RAC1, an upstream mediator of PAK1 in colon cancer cells, steps up β-catenin phosphorylation and accumulation, thus boosting the proliferation of colon cancer cells [33]. Meanwhile, several studies have also unveiled that some drugs can frustrate colon cancer development by down-regulating RAC1 expression. For instance, overexpression of RAC1 culminated in accelerated cell migration, invasion, and EMT in colon cancer. Diallyl disulfide (DADS), a component of garlic oil, weakened colon cancer metastasis by attenuating the RAC1-mediated PI3K/AKT pathway [34]. What is more, preceding studies have revealed that the RAC1/PI3K/AKT signaling pathway participates in the development of multiple tumors. For instance, Neo1 positively regulates ZEB1 expression by initiating the RAC1/PI3K/AKT pathway, thus facilitating the proliferation, movement, and adhesion of gastric cancer [35]. What is more, epidermal growth factors induce the activation of the RAC1-PI3K/Akt pathway and promote the migration of breast cancer cells [36]. Consistent with the above research conclusions, in the present study, we discovered that in contrast with the normal tissues adjacent to carcinoma, RAC1 expression rose significantly in colon cancer tissues, and RAC1 inhibition remarkably attenuated colon cancer growth and PI3K/AKT activation. All the outcomes indicated that RAC1 expression might participate in the malignant evolution of biological processes in colon cancer cells. Besides, bioinformatics analysis discovered a targeted relationship between miR-627-5p and RAC1, and Western blot demonstrated that interference with miR-627-5p negatively modulated the profile of RAC1/PI3K/AKT. Meanwhile, LBX2-AS1 was positively correlated with RAC1 expression, and LBX2-AS1 overexpression bolstered RAC1/PI3K/AKT expression. All these findings displayed that in colon cancer, LBX2-AS1 regulated RAC1/PI3K/AKT via miR-627-5p expression regulation, thereby affecting cell proliferation, metastasis, and EMT transformation.
In summary, our work has confirmed that LBX2-AS1 is highly expressed in colon cancer tissues and cells and conspicuously correlated with tumor staging and early distant metastasis in patients. It means LBX2-AS1 can serve as an adverse prognostic factor for colon cancer and modulate colon cancer growth by regulating the miR-627-5p /RAC1/PI3K/AKT pathway. These findings provide a new intervention target for colon cancer treatment and prognosis and lay a necessary experimental foundation for better immunobiological adoptive therapies. However, more molecular mechanisms need to be further investigated.
Availability of data and material
The data sets used and analyzed during the current study are available from the corresponding author on reasonable request.
References
Seetha A, Devaraj H, Sudhandiran G. Indomethacin and juglone inhibit inflammatory molecules to induce apoptosis in colon cancer cells. J Biochem Mol Toxicol. 2020;34(2): e22433.
Song M, Chan AT, Sun J. Influence of the gut microbiome, diet, and environment on risk of colorectal cancer. Gastroenterology. 2020;158(2):322–40. https://doi.org/10.1053/j.gastro.2019.06.048.
Tsuji Y, Sugihara K. Adjuvant chemotherapy for colon cancer: the difference between Japanese and western strategies. Expert Opin Pharmacother. 2016;17(6):783–90.
Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36(41):5661–7.
Renganathan A, Felley-Bosco E. Long noncoding RNAs in cancer and therapeutic potential. Adv Exp Med Biol. 2017;1008:199–222.
Zhenqin L, Peiguo C. Long noncoding RNA PVT1 promotes hepatoblastoma cell proliferation through activating STAT3. Cancer Manag Res. 2019;11:8517–27.
Ye Z, Rui S, Yi C, et al. Long noncoding RNA MT1JP inhibits proliferation, invasion, and migration while promoting apoptosis of glioma cells through the activation of PTEN/Akt signaling pathway. J Cell Physiol. 2019;234:19553–64.
Fan Wu, Zheng Z, Ruichao C, et al. Expression profile analysis of antisense long non-coding RNA identifies WDFY3-AS2 as a prognostic biomarker in diffuse glioma. Cancer Cell Int. 2018;18:107.
Yang Z, Dong X, Pu M, et al. LBX2-AS1/miR-219a-2-3p/FUS/LBX2 positive feedback loop contributes to the proliferation of gastric cancer. Gastric Cancer. 2020;23(3):449–63.
Yanshan Z, Weizuo C, Tingting P, et al. LBX2-AS1 is activated by ZEB1 and promotes the development of esophageal squamous cell carcinoma by interacting with HNRNPC to enhance the stability of ZEB1 and ZEB2 mRNAs. Biochem Biophys Res Commun. 2019;511:566–72.
Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–22.
Kun Q, Shipeng N, Lin W, et al. LINC00673 is activated by YY1 and promotes the proliferation of breast cancer cells via the miR-515-5p/MARK4/Hippo signaling pathway. J Exp Clin Cancer Res. 2019;38:418.
Xu C, Li S, Chen T, Hu H, Ding C, Xu Z, Chen J, Liu Z, Lei Z, Zhang HT, Li C, Zhao J. miR-296-5p suppresses cell viability by directly targeting PLK1 in non-small cell lung cancer. Oncol Rep. 2016;35(1):497–503.
Wang J, Chen T, Wang L, et al. MicroRNA-627-5p inhibits the proliferation of hepatocellular carcinoma cells by targeting BCL3 transcription coactivator. Clin Exp Pharmacol Physiol. 2020;47(3):485–94.
Fuyang C, Liu M, Yixiu Y, et al. LINC00958 regulated miR-627-5p/YBX2 axis to facilitate cell proliferation and migration in oral squamous cell carcinoma. Cancer Biol. 2019;20:1270–80.
Ze Ji, Xing P, Yan S, et al. KIF18B as a regulator in microtubule movement accelerates tumor progression and triggers poor outcome in lung adenocarcinoma. Tissue Cell. 2019;61:44–50.
Wenlan Q, Dong Li, Qing S, et al. miR-224-5p protects dental pulp stem cells from apoptosis by targeting RAC1. Exp Ther Med. 2020;19:9–18.
Zhang Z, Li X, Li A, Wu G. miR-485-5p suppresses Schwann cell proliferation and myelination by targeting cdc42 and RAC1. Exp Cell Res. 2020;388(1): 111803.
Yan Y, Huang H. Interplay among PI3K/AKT, PTEN/FOXO and AR signaling in prostate cancer. Adv Exp Med Biol. 2019;1210:319–31.
Yan Z, Zhen Li, Zhi L. Silencing lncRNAUNC5B represses growth and metastasis of human colon cancer cells via raising miR-622. Artif Cells Nanomed Biotechnol. 2020;48:60–7.
Li G, Zhang C, Liang W, Zhang Y, Shen Y, Tian X. Berberine regulates the Notch1/PTEN/PI3K/AKT/mTOR pathway and acts synergistically with 17-AAG and SAHA in SW480 colon cancer cells. Pharm Biol. 2021;59(1):21–30.
Shimaoka H, Takeno S, Maki K, Sasaki T, Hasegawa S, Yamashita Y. A cytokine signal inhibitor for rheumatoid arthritis enhances cancer metastasis via depletion of NK cells in an experimental lung metastasis mouse model of colon cancer. Oncol Lett. 2017;14(3):3019–27.
Wang L, Cho KB, Li Y, Tao G, Xie Z, Guo B. Long noncoding RNA (lncRNA)-mediated competing endogenous rna networks provide novel potential biomarkers and therapeutic targets for colorectal cancer. Int J Mol Sci. 2019;20(22):5758.
Zhu Y, Qiao L, Zhou Y, et al. Long non-codingRNA FOXD2-AS1 contributes to colorectal cancer proliferation through its interaction with miR-185-5p. Cancer Sci. 2018;109(7):2235–42.
Wu Q, Meng WY, Jie Y, Zhao H. LncRNA MALAT1 induces colon cancer development by regulating miR-129-5p/HMGB1 axis. J Cell Physiol. 2018;233(9):6750–7.
Ahmed FE. miRNA as markers for the diagnostic screening of colon cancer. Expert Rev Anticancer Ther. 2014;14(4):463–85.
Zeng M, Zhu L, Li L, Kang C. miR-378 suppresses the proliferation, migration and invasion of colon cancer cells by inhibiting SDAD1. Cell Mol Biol Lett. 2017;22:12.
Zhang W, Sun Z, Su L, et al. miRNA-185 serves as a prognostic factor and suppresses migration and invasion through Wnt1 in colon cancer. Eur J Pharmacol. 2018;825:75–84.
Zirong F, Jian Z, Yixue X, et al. NR2C2-uORF targeting UCA1-miR-627-5p-NR2C2 feedback loop to regulate the malignant behaviors of glioma cells. Cell Death Dis. 2018;9:1165.
Zhenzhe Li, Jixing Z, Hongshan Z, et al. Modulating lncRNA SNHG15/CDK6/miR-627 circuit by palbociclib, overcomes temozolomide resistance and reduces M2-polarization of glioma associated microglia in glioblastoma multiforme. J Exp Clin Cancer Res. 2019;38:380.
André S, Singh T, Lacal JC, Smetana K Jr, Gabius HJ. Rho GTPase RAC1: molecular switch within the galectin network and for N-glycan α2,6-sialylation/O-glycan core 1 sialylation in colon cancer in vitro. Folia Biol (Praha). 2014;60(3):95–107.
Witte D, Otterbein H, Förster M, et al. Negative regulation of TGF-β1-induced MKK6-p38 and MEK-ERK signalling and epithelial-mesenchymal transition by Rac1b. Sci Rep. 2017;7(1):17313.
Zhu G, Wang Y, Huang B, et al. A RAC1/PAK1 cascade controls β-catenin activation in colon cancer cells. Oncogene. 2012;31(8):1001–12.
Xia L, Lin J, Su J, et al. Diallyl disulfide inhibits colon cancer metastasis by suppressing RAC1-mediated epithelial-mesenchymal transition. Onco Targets Ther. 2019;12:5713–28.
Qu H, Sun H, Wang X. Neogenin-1 promotes cell proliferation, motility, and adhesion by up-regulation of zinc finger e-box binding homeobox 1 via activating the RAC1/PI3K/AKT pathway in gastric cancer cells. Cell Physiol Biochem. 2018;48(4):1457–67.
Yang Y, Du J, Hu Z, et al. Activation of RAC1-PI3K/Akt is required for epidermal growth factor-induced PAK1 activation and cell migration in MDA-MB-231 breast cancer cells. J Biomed Res. 2011;25(4):237–45.
Funding
This work was supported by Huazhong University of Science and Technology “Double Top” Construction Project of International Cooperation (grant 540–5001540013 to Feng JP) and Wuhan Municipal Health Commission (WX19D65 to Jing Fang).
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: JF, JF. Performed the experiments: JF, Junyuan Yang. Statistical analysis: HC, WS. Formal analysis: LX. Funding acquisition: Jing Fang, JF. Wrote the paper: JF. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical statement
Our study was approved by the Ethics Review Board of Wuhan Fourth Hospital.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Fang, J., Yang, J., Chen, H. et al. Long non-coding RNA LBX2-AS1 predicts poor survival of colon cancer patients and promotes its progression via regulating miR-627-5p/RAC1/PI3K/AKT pathway. Human Cell 35, 1521–1534 (2022). https://doi.org/10.1007/s13577-022-00745-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13577-022-00745-x