Abstract
Large intergenic non-coding RNA ribonucleic acids-ROR (lincRNA-ROR) has been reported to exert impacts on the maintenance of induced pluripotent stem cells and embryonic stem cells, and play important roles in human hepatocellular cancer. It contributes to tumorigenesis and metastasis and functions as a competing endogenous RNA (ceRNA) by sponging miR-145 in breast cancer. However, its clinical significance and prognostic value in colon cancer remain unknown. The aim of the present study was to clarify the clinicopathological role and prognostic value of lincRNA-ROR and miR-145 in colon cancer. In the present study, qRT-PCR was performed to measure the expression levels of lincRNA-ROR in colon cancer tissues and cell lines. Then, the clinicopathological significance and prognostic value of lincRNA-ROR were analyzed. LincRNA-ROR expression correlated with pT stage, pN stage, AJCC stage and vascular invasion. Knockdown of lincRNA-ROR restored the expression of miR-145, and had a significant influence on colon cancer cell proliferation, migration and invasion. Patients of the high lincRNA-ROR/low miR-145 group had significantly poorer outcomes than those of the low lincRNA-ROR/high miR-145 group. Taken together, Overexpression of lincRNA-ROR combined with depletion of miR-145 may exert crucial impact on colon cancer prognosis evaluation and treatment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As one of the most prevalent human malignance, colon cancer is the third leading cause of mortality worldwide [1, 2]. Despite progress in diagnosis and treatment of colon cancer in recent years, it is hardly benefited through systemic therapeutic strategies due to the poor prognosis [3]. At present, although a large number of biomarkers have been associated with the diagnosis and prognosis of colon cancer, including cell adhesion molecules, adenomatous polyposis coli (APC), growth factors, and more microRNAs [4–7], there still remains a need for biomarkers of tumor progression that can be utilized in a prevention and early detection strategy and effective individualized therapy.
Emerging evidence suggests a strong link between long non-coding RNAs (lncRNAs) and plenty of cancers [8–11]. As a class of non-coding transcripts longer than 200 nucleotides [12], lncRNAs have been shown to regulate gene expression via transcription, post-transcription processing, genomic imprinting and chromatin modification [13, 14]. Large intergenic non-coding RNA ribonucleic acids-ROR (lincRNA-ROR) is a type of lncRNAs, which has been identified as a promoter of human-induced pluripotent stem cells (iPSCs) and considered to inhibit the core transcription factors from miRNA-mediated suppression in human embryonic stem cell self-renewal [15]. It has been reported that as a competing endogenous RNA (ceRNA), lincRNA-ROR contribute to tumorigenesis and invasion via targeting miR-145 in triple-negative breast cancer [16]. Moreover, lincRNA-ROR has been considered to function as a ceRNA to against mediation of the differentiation of endometrial cancer stem cells by sponging miR-145 [17]. Although it is clear that lincRNA-ROR and miR-145 are one pair of ceRNA that lincRNA-ROR acts as a “miRNA sponge” to show reciprocal repression with miR-145, their possible clinical value in colon cancer is still unknown up to now. In the current study, we evaluated the expression of lincRNA-ROR and miR-145 in colon cancer tissues, as well as correlations between lincRNA-ROR and miR-145 and their clinicopathological significance in colon cancer.
Material and Methods
Specimens
A total of 152 tissue samples, including 60 fresh colon cancer tissues and their matched adjacent mucosa and another 32 lymph node metastatic tissues were collected from 60 patients with colon cancer permitted operation by the General Surgery Department, the Second People’s Hospital of Wuhu, China, from October 2003 to July 2005. There were 29 males and 31 female patients, with ages ranging from 34 to 82 years. All tissues were stored at −80 °C immediately following removal until use. The diagnosis of all specimens was histopathologically confirmed by pathologist and staging was determined according to the 7th edition of cancer staging criteria of the American Joint Committee On Cancer (AJCC). None of the patients had received any treatment before surgery. Specimen collection and study were approved by the Ethics Committee of the Second People’s Hospital of Wuhu, China, and informed consent was obtained from each patient enrolled in the study.
Cell Culture and Transfection
A total of six cell lines were purchased from Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China) for use in this study: human colon mucosal epithelial cell line (NCM460) and five colon cancer cell lines (RKO, Caco2, HCT8, SW480 and SW620). Cells were cultured in Dulbecco’s modified Eagle’s medium (Gibco BRL, Grand Island, NY, USA), supplemented with 10 % fetal bovine serum (Invitrogen, Camarillo, CA, USA). Cells were maintained in a humidified atmosphere at 37 °C with 5 % CO2. RKO cells were transfected with 100 nM si-lincRNA-ROR (GGAGA GGAAG CCTGA GAGT) or non-targeting Control (si-NC 5′-UUCUC CGAAC GUGUC ACGUtt-3′) employing Lipofectamine RNAiMAX (Life Technologies, Carlsbad, CA, USA) following the manufacturer’s instruction.
Total RNA Isolation and Quantitative Real-Time PCR (qRT-PCR)
Total RNA from all tissues was extracted by Trizol reagent (Ambition, Carlsbad, CA, USA). For lincRNA-ROR, RNA was reversed transcribed into cDNA by Prime-Script RT-PCR kit (TaKaRa, Dalian, China) following the manufacturer’s instructions. For miR-145, RNA was reverse transcription by M-MLV reverse transcriptase (Invitrogen, Carlsbad, CA). qRT-PCR was used to detect lincRNA-ROR and miR-145 expression levels by the SYBR Premix Dimmer Eraser kit (Takara). The primer sequences were used for qRT-PCR as follows: for lincRNA-ROR, 5′-CAAAC ACATC GCCAC TCTGC-3′ (forward), 5′-AGGAG TCAGG AGAAG GTGCT-3′ (reverse); for miR-145, 5′-AGAACAGTATTTCCAGGAAT-3′ (forward), 5′-GTCAA GTTTT CCCAG GAATC C-3′ (reverse); for GAPDH, 5′-CAGGA GGCAT TGCTG ATGAT-3′ (forward), 5′-GAAGG CTGGG GCTCA TTT-3′ (reverse).The conditions of thermal cycling were as follows: 95 °C at 10 min, 40 cycles at 95 °C for 15 s, 60 °C for 30s, and 72 °C for 30s. The quantitative PCR reaction was repeated in triplicate. GAPDH was used as reference for lncRNA or miRNA, Relative expression levels of lncRNA and miRNA were calculated by using the ΔCt method in which higher ΔCt means lower expression.
Serological Tumor Marker Analysis
The cutoff values for serum carcinoembryonic antigen (CEA) and carbohydrate antigen 19–9 (CA19–9) were 5 ng/ml and 35 U/ml, respectively.
Cell Proliferation Assays
A cell proliferation assay was performed with Cell Counting Kit-8 (Byotime, Haimen, China) according to the manufacturer’s protocol, and detected at 24, 48, 72 and 96 h.
Cell Migration and Invasion Assays
At 48 h after transfection, cells in serum-free media were placed into the upper chamber of an insert (8.0 μm pore size, millepore) for migration assays (without Martrigel) and invasion assays (with Martrigel). Media containing 10 % FBS was added to the lower chamber. After 48 h of incubation, cells that had migrated of invaded through the membrane were fixed and stained with methanol and 0.1 % crystal violet. Finally, the stained cells were counted under the microscope, and meanwhile, the images were captured.
Statistical Analysis
All statistical analyses were set with a significance level of p < 0.05. Data were analyzed by the Statistical Program for Social Sciences (SPSS) 19.0 software (SPSS, Chicago, IL, USA). The paired t-test, χ2 test, one-way ANOVA, the Kaplan-Meier method with log-rank test and Cox regression were used as appropriate. All graphs were plotted using GraphPad Prism 5.0 (GraphPad Software, La Jolla, CA, USA).
Results
Expression of lincRNA-ROR and miR-145 in Colon Cancer Tissues
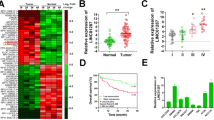
The result of qRT-PCR showed that lincRNA-ROR levels in colon cancer tissues were higher than those in the matched adjacent normal tissues (p < 0.001, Fig. 1a). Inversely, the miR-145 levels in colon cancer tissues were lower than those in the matched adjacent normal tissues (p < 0.001, Fig. 1b). In addition, a negative correlation between lincRNA-ROR and miR-145 was observed in colon cancer tissues (p < 0.001, R2 = 0.290, Fig. 1c). Additionally, lincRNA-ROR and miR-145 expression levels were evaluated in five colon cancer cell lines, RKO, Caco2, HCT8, SW480 and SW620, and all the levels were normalized to a normal colon mucosal epithelial cell line NCM460. Among all the cancer cell lines, lincRNA-ROR levels were higher with concurrent lower levels of miR-145 (Fig. 1d), consistent with the their expression in colon cancer tissues.
LincRNA-ROR and miR-145 expression levels in colon cancer. (a-b) Expression levels of LincRNA-ROR and miR-145 in colon cancer tissues were significantly higher or lower than those in the matched adjacent normal tissues, respectively (n = 60). c Negative correlation between the lincRNA-ROR levels and the miR-145 levels in 60 matched-tissues of colon cancer patients (p < 0.01, R2 = 0.290). d The higher levels of lincRNA-ROR with concurrent lower miR-145 levels in colon cancer cell lines compared with NCM460. The relative levels of LincRNA-ROR and miR-145 in all colon cancer cells were normalized to NCM460 cell by using the 2-ΔΔCt method. *p < 0.05, ***p < 0.001
All the colon cancer patients were divided into two groups in relation to the median value of relative lincRNA-ROR and miR-145 expression according to the references [18, 19]. As shown in Table 1, of the 60 colon cancer tissues, 32 (53.3 %) samples showed lincRNA-ROR high expression and 33 (55.0 %) samples showed miR-145 low expression. Of the 32 samples of lymph node metastatic tissue, 25 (78.1 %) samples showed lincRNA-ROR high expression and 26 (81.2 %) samples showed miR-145 low expression. It is noteworthy that lincRNA-ROR was elevated in lymph node metastatic tissues, with significant means between lymph node metastatic tissues and primary cancer tissues. Inversely, miR-145 was significantly reduced in lymph node metastatic tissues when compared with primary cancer tissues.
Association of lincRNA-ROR and miR-145 Expression with Clinicopathological Characteristics in Colon Cancer
Whether lincRNA-ROR and miR-145 expression levels were associated with the clinicopathological features of patients with colon cancer was further explored. As shown in Table 2, the lincRNA-ROR level in cancer tissues was associated with depth of tumor invasion (pT stage, p = 0.011), lymph node metastasis (pN stage, p < 0.001), vessel invasion (p = 0.005) and AJCC stage (p < 0.001). MiR-145 level correlated inversely with pT stage (p = 0.030), pN stage (p < 0.001), vessel invasion (p = 0.002) and AJCC stage (p < 0.001). But no significant correlation was found with other clinicopathological features such as age, gender, tumor location, differentiation, and serum CEA or CA19–9.
LincRNA-ROR Promotes Proliferation, Migration and Invasion of Colon Cancer Cells
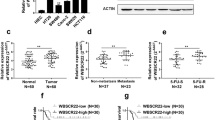
Given that lincRNA-ROR expression levels were significantly upregulated in RKO cells than those in the other colon cancer cell lines, RKO cells were treated with specific small interfering RNA and lincNRA-ROR expression levels were effectively knocked down by 65 %, meanwhile, the miR-145 levels were restored (Fig. 2a). CCK-8 assay revealed that cell proliferation was significantly impaired in RKO cells transfected with si-lincRNA-ROR (Fig. 2b). Moreover, in the Transwell migration assay, the transfection of lincRNA-ROR siRNA impeded the migratory ability of RKO cells by 53 % (Fig. 2c, 2d). Similarly, a corresponding effect on invasiveness was observed in a parallel invasion assay (Fig. 2e, 2f).
The influence of lincRNA-ROR on RKO cells proliferation, migration and invasion. a Expression levels of lincRNA-ROR and miR-145 after the transfection of si-lincRNA-ROR or si-NC. b Cell viability assay in si-lincRNA-ROR/si-NC RKO cells. (c-d) RKO cell migration following treatment with si-lincRNA-ROR or si-NC, the relative ratio of migrated cells per field is shown. (e-f) RKO cell invasion following treatment with si-lincRNA-ROR or si-NC, the relative ratio of invasive cells per field is shown. *p < 0.05, **p < 0.01
Overexpression of lincRNA-ROR Alone or Combined with Downregulation of miR-145 Predicts Poor Prognosis
As shown in Fig. 3, the Kaplan-Meier plot showed that high expression group (n = 32) of lincRNA-ROR had a poor OS (Fig. 3a) and DFS (Fig. 3 b) than the low expression group (n = 28). On the other hand, there was no difference between high expression group (n = 27) of miR-145 and low expression group (n = 33) in OS (Fig. 3c). However, miR-145 was significantly associated with DFS (Fig. 3d). Cases with both low lincRNA-ROR expression and high miR-145 expression (n = 18) showed a better OS (Fig. 3e) and DFS (Fig. 3f) than the cases with high lincRNA-ROR expression and low miR-145 expression (n = 23). Univariate and multivariate analysis suggested that increased lincRNA-ROR expression was a significant independent prognostic factor for decreased survival and increased disease recurrence. MiR-145 was found to be an independent prognostic factor for OS and DFS when combined with lincRNA-ROR expression in colon cancer (Table 3 and Table 4).
Kaplan-Meier analysis with a log rank test of survival. (a-b) Patients with a high lincRNA-ROR expression (n = 32) had a poor OS and DFS than the low lincRNA-ROR expression group (n = 28). (c-d) The DFS of miR-145 low group (n = 33) was shorter than that of the high group (n = 27), but showed no correlation with OS. (e-f) The patients with a lincRNA-ROR high/miR-145 low expression (n = 23) had shorter OS and DFS compared with lincRNA-ROR low/miR-145 high group (n = 18)
Discussion
Our study for the first time reports the association of lincRNA-ROR and miR-145 expression with colon cancer patients. We found that lincRNA-ROR expression was significantly associated with advanced colon cancer biology, and might serve as a suitable biomarker for prognosis of colon cancer when evaluated in conjunction with miR-145.
Based on previous reports, many lncNRAs have been reported highly or lowly expressed and play functional roles in human cancers. For instance, lncRNA H19 promotes carcinogenesis and considered as a potential prognostic indicator of renal cell caicinoma [11]. Homeobox transcript antisense intergenic RNA (HOTAIR) was increased in gastric cancer tissues and cell lines, it promotes gastric cancer metastasis through suppression of Poly r-Binding Protein 1 (PCBP 1) [20]. HNF1A antisense 1 (HNF1A-AS1) was involved in tumorigenesis and identified as a poor prognostic biomarker in lung adenocarcinoma [21]. Lee et al. reported that lncRNA snaR was dowanregulated in 5-fluorouracil resistant colon cancer cells, downregulation of snaR decreased colon cancer cell death after 5-fluorouracil treatment [22]. All these findings demonstrate that lncRNAs play a crucial regulatory impact on carcinogenesis and would provide a selective advantage in clinical application.
LincRNA-ROR has been first published in induced pluripotent stem cells (iPSCs) [23]. It has been reported to modulate chemosensitivity in human hepatocellular cancer and function as an important regulator of EMT in breast cancer [24, 25]. Recently, a model has been confirmed that there exist a widespread interaction network of ceRNAs, in which lncRNAs may fine-tune the concentration and biological functions of miRNAs [26, 27]. Similarly, lincRNA-ROR and miR-145 have been validated as one pair ceRNA, and they show reciprocal repression to each other by sharing miRNA response elements with other mRNAs [16, 17].
In the present study, we found that overexpression of lincRNA-ROR is a frequent event in colon cancer tissues. As a tumor suppressor, miR-145 has been reported to exert suppressive impacts on various tumors [28–31]. Consistent with the published reports, our findings revealed that miR-145 was decreased in colon cancer tissues. Moreover, its levels were found to be negatively correlated with lincRNA-ROR expression. Our further investigation confirmed that lincRNA-ROR expression was higher in Lymph node metastatic tissues than in primary tumors, which implies that upregulation of lincRNA-ROR may participate in the metastatic process of colon cancer. Despite some clinicopathological features, including tumor differentiation, clinical stage, vessel invasion and distant metastasis can be utilized to predict the progression of tumor [32], the valid prognostic biomarkers for colon cancer were still not completely known. In order to explore whether lincRNA-ROR could serve as a potential effective biomarker of colon cancer, the correlations between lincRNA-ROR, miR-145 expression and clinicopathological features were analyzed. As a result, lincRNA-ROR expression was not significantly associated with age, gender and differentiation of colon cancer patients, but was closely correlated with pT stage, pN stage, AJCC stage and vascular invasion. Our further investigation revealed that lincRNA-ROR expression was upregulated in invaded lymph nodes in comparison with primary colon cancers. Consistent with the above findings, our functional studies confirmed that lincRNA-ROR depletion resulted in the restoration of miR-145 expression, and inhibited cell viability, migration and invasion effectively. These findings suggested that lincRNA-ROR plays a direct role in the modulation of colon cancer progression. Furthermore, we found that the expression level of lincRNA-ROR influence the survival and recurrence of patients. When colon cancer patients were divided into two groups according to the relative expression of lincRNA-ROR and miR-145, lincRNA-ROR high/miR-145 low group had worse OS and DFS than lincRNA-ROR low/miR-145 high group. Univariate and multivariate analysis showed that lincRNA-ROR expression was strongly linked to OS and DFS, on the other hand, miR-145 expression affect OS and DFS in univariate analysis, but not multivariate analysis. All the results suggested that lincRNA-ROR alone or in combination with miR-145 expression could be considered as an independent prognostic factor for OS and DFS in colon cancer.
To summarize, we presented a new insight into the prognostic value of lincRNA-ROR. It was identified as a novel oncogene and exerts its impact by associating with miR-145 in colon cancer. Further investigation will advance our understanding linking the molecular mechanism of lincRNA-ROR mediated colon cancer development.
References
Brenner H, Kloor M, Pox CP (2014) Colorectal cancer. Lancet 383:1490–1502
Sieqel R, Desantis C, Jemal A (2014) Colorectal cancer statistics, 2014. CA Cancer J Clin 64:104–117
Edwards BK, Ward E, Kohler BA, Eheman C, Zauber AG, Anderson RN, et al. (2010) Annual report to the nation on the status of cancer, 1975-2006, featuring colorectal cancer trends and impact of interventions (risk factors, screening, and treatment) to reduce future rates. Cancer 116:544–573
Jankowski JA, Odze RD (2009) Biomarkers in gastroenterology: between hope and hype comes histopathology. Am J Gastroenterol 104:1093–1096
Modjtahedi H, Essapen S (2009) Epidermal growth factor receptor inhibitors in cancer treatment: advances, challenges and opporunities. Anti-Cancer Drugs 20:851–855
Aslam MI, Taylor K, Pringle JH, Jameson JS (2009) MicroRNAs are novel biomarkers of colorectal cancer. Br J Surg 96:702–710
Kiersem JB, Iksahl T, Lingjaerde OC, Guren T, Tveit KM, Kure EH (2014) Plasma microRNAs predicting clinical outcome in metastatic colorectal cancer patients receiving first-line oxaliplatin-based treatment. Mol Oncol 8:59–67
Yang F, Xue X, Bi J, Zheng L, Zhi K, Gu Y, et al. (2013) Long noncoding RNA CCAT1, which coild be activated by c-Myc, promotes the progression of gastric carcinoma. J Cancer Res Clin Oncol 139:437–445
Zhang EB, Yin DD, Sun M, Kong R, Liu XH, You LH, et al. (2014) P53-regulated long non- coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell Death Dis 5:e1243
Wu Y, Zhang L, Zhang L, Wang Y, Li H, Ren X, et al. (2015) Long non-coding RNA HOTAIR promotes tumor cell invasion and metastasis by recruiting EZH2 and repressing E-cadherin in oral squamous cell carcinoma. Int J Oncol 46:2586–2594
Wang L, Cai Y, Zhao X, Jia X, Zhang J, Liu J, et al. (2015) Down-regulated long non-coding RNA H19 inhibits carcinogenesis of renal cell carcinoma. Neoplasma 62:412–418
Wapinski O, Chang HY (2011) Long noncoding RNAs and human disease. Trends Cell Biol 21:354–361
Khalil AM, Guttman M, Huarte M, Garber M, Rai A, Rivea Morales D, et al. (2009) Many human large intergenic noncoding RNAs associate with chromatin modifying complexes and affect gene expression. Proc Natl Acad Sci U S A 106:11667–11672
Guttman M, Rinn JL (2012) Modular regulatory principles of large non-coding RNAs. Nature 482:339–346
Wang Y, Xu Z, Jiang J, Xu C, Kang J, Xiao L, et al. (2013) Endogenous miRNA sponge lincRNA-ROR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell 25:69–80
Eades G, Wolfson B, Zhang Y, Li Q, Yao Y, Zhou Q (2015) LincRNA-ROR and miR-145 regulate invasion in triple-negative breast cancer via targeting ARF6. Mol Cancer Res 13:330–338
Zhou X, Gao Q, Wang J, Zhang X, Liu K, Duan Z (2014) Linc-RNA-ROR acts as a “spong” against mediation of the differentiation of endometrial cancer stem cells by microRNA-145. Gynecol Oncol 133:333–339
Iguchi T, Uchi R, Nambara S, Saito T, Komatsu H, Hirata H, et al. (2015) A long noncoding RNA, lncRNA-ATB, is involved in the progression and prognosis of colorectal cancer. Anticancer Res 35:1385–1388
Xu TP, Huang MD, Xia R, Liu XX, Sun M, Yin L, et al. (2014) Decreased expression of the long non-coding RNA FENDRR is associated with poor prognosis in gastric cancer and FENDRR regulates gastric cancer cell metastasis by affecting fibronectin1 expression. J Hematol Oncol 7:63
Zhang ZZ, Shen ZY, Shen YY, Zhao EH, Wang M, Wang CJ, et al. (2015) HOTAIR long noncoding RNA promotes gastric cancer metastasis through suppression of poly r(c)-binding protein (PCBP) 1. Mo Cancer Ther 14:1162–1170
Wu Y, Liu H, Shi X, Yao Y, Yang W, Song Y (2015) The long non-coding RNA HNF1A-AS1 regulates proliferation and metastasis in lung adenocarcinoma. Oncotarget 6:9160–9172
Lee H, Kim C, Ku JL, Kim W, Yoon SK, Kuh HJ, et al. (2014) A long non-coding RNA snaR contributes to 5- fluorouracil resistance in human colon cancer cells. Mol Cell 37:540–546
Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, et al. (2010) Large intergenic non-coding RNA-ROR modulates reprogramming of human induced pluripotent stem cells. Nat Genet 42:1113–1117
Takahashi K, Yan IK, Koqure T, Haqa H, Patel T (2014) Exreacellular vesicle-mediated transfer of long non-coding RNA ROR modulates chemosensitivity in human hepatocellular cancer. FEBS Open Bio 4:458–467
Hou P, Zhao Y, Li Z, Yao R, Ma M, Gao Y, et al. (2014) LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis 5:e1287
Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP (2011) A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146:353–358
Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, et al. (2011) Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell 147:344–357
Yu CC, Tsai LL, Wang ML, Yu CH, Lo WL, Chang YC, et al. (2013) miR-145 targets the SOX9/ADAM17 axis to inhibit tumor-initiating cells and IL-6-mediated paracrine effects in head and neck cancer. Cancer Res 73:3425–3440
Noh JH, Chang YG, Kim MG, Jung KH, Kim JK, Bae HJ, et al. (2013) MiR-145 functions as a tumor suppressor by directly targeting histone deacetylase 2 in liver cancer. Cancer Lett 335:455–462
Zhang Y, Lin Q (2015) MicroRNA-145 inhibits migration and invasion by down-regulating FSCN1 in lung cancer. Int J Clin Exp Med 8:8794–8802
Qin J, Wang F, Jiang H, Xu J, Jiang Y, Wang Z (2015) MicroRNA-145 suppresses cell migration and invasion by targeting paxillin in human colorectal cancer cells. Int J Clin Exp Pathol 8:1328–1340
Foltran L, Maglio GD, Pella N, Ermacora P, Aprile G, Masiero E, et al. (2015) Prognostic role of KRAS, NRAS, BRAF and PIK3CA mutations in advanced colorectal cancer. Future Oncol 11:629–640
Acknowledgments
We thank Bin Zhang and Jiangming Chen for their technical assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of Interest
The authors declare that there is no conflict of interests regarding the publication of this paper.
Rights and permissions
About this article
Cite this article
Zhou, P., Sun, L., Liu, D. et al. Long Non-Coding RNA lincRNA-ROR Promotes the Progression of Colon Cancer and Holds Prognostic Value by Associating with miR-145. Pathol. Oncol. Res. 22, 733–740 (2016). https://doi.org/10.1007/s12253-016-0061-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12253-016-0061-x